Abstract
Hyaluronan (HA), the only non-sulfated glycosaminoglycan, is involved in morphogenesis, wound healing, inflammation, angiogenesis, and cancer. In mammals, HA is synthesized by three homologous HA synthases, HAS1, HAS2, and HAS3, that polymerize the HA chain using UDP-glucuronic acid and UDP-N-acetylglucosamine as precursors. Since the amount of HA is critical in several pathophysiological conditions, we developed a non-radioactive assay for measuring the activity of HA synthases (HASs) in eukaryotic cells and addressed the question of HAS activity during intracellular protein trafficking. We prepared three cellular fractions: plasma membrane, cytosol (containing membrane proteins mainly from the endoplasmic reticulum and Golgi), and nuclei. After incubation with UDP-sugar precursors, newly synthesized HA was quantified by polyacrylamide gel electrophoresis of fluorophore-labeled saccharides and high performance liquid chromatography. This new method measured HAS activity not only in the plasma membrane fraction but also in the cytosolic membranes. This new technique was used to evaluate the effects of 4-methylumbeliferone, phorbol 12-myristate 13-acetate, interleukin 1β, platelet-derived growth factor BB, and tunicamycin on HAS activities. We found that HAS activity can be modulated by post-translational modification, such as phosphorylation and N-glycosylation. Interestingly, we detected a significant increase in HAS activity in the cytosolic membrane fraction after tunicamycin treatment. Since this compound is known to induce HA cable structures, this result links HAS activity alteration with the capability of the cell to promote HA cable formation.
Hyaluronan (HA)3 is the only non-sulfated linear polymer belonging to the family of glycosaminoglycans (GAGs). HA is an unbranched polymer of alternating GlcNAc and GlcUA residues linked by alternate β(1→4) and β(1→3) bonds. Native HA is typically larger than other GAGs, reaching molecular mass values between 106 and 107 Da.
HA is a major component of extracellular matrices and in pericellular spaces, particularly in tissues with rapid cell proliferation and cell migration (1). Through interactions with cell surface receptors, notably CD44 and RHAMM (receptor for HA-mediated motility), HA has important roles in regulating cell behavior, including signal transduction, cell adhesion, proliferation, migration, and differentiation (2). Recently, novel interactions involving HA and Toll-like receptors 4 and 2 have been described that have important roles in inflammation (3, 4). Moreover, HA has been implicated in morphogenesis (5–8), wound healing (9), angiogenesis (10), malignancies, cancer growth, and tumor invasion (11).
In mammals, HA is normally synthesized at the plasma membrane and extruded directly into the extracellular space by three isoforms of HA synthases (HASs), HAS1, -2, and -3. The three HAS isoforms differ in tissue distribution, regulation, and enzymatic properties (12); nevertheless, they are similar in amino acid sequences and molecular structures.
HA biosynthesis is under the control of a wide variety of cytokines and growth factors (13). The changes in HA synthesis can be related to HAS mRNA expression (14), to availability of the UDP-sugar precursors (15, 16), or to modulation by phosphorylation of HAS (17–19) in response to cytokines and growth factors. Moreover, HA chain synthesis can be controlled by additional mechanisms, such as cell type, intracellular environment, or HAS accessory proteins (20). Cultures of smooth muscle cells isolated from human colon increase synthesis of HA after treatment with a viral mimetic molecule (poly(I-C)) (21). The HA is organized into novel cable-like structures, and their synthesis may be initiated in the perinuclear and/or the endoplasmic reticulum (ER) membranes (22). Furthermore, HA interstitial deposition is correlated with inflammatory processes (23, 24) in which HA-CD44 interactions stimulate leukocyte adhesion in order to generate an inflammatory response (25).
Cytokines and growth factors, such as IL-1β and platelet-derived growth factor BB (PDGF-BB), as well as 4-methylumbeliferone (4-MU) and the tumor promoter phorbol 12-myristate 13-acetate (PMA), also modulate HA synthesis (26–29). In order to elucidate how these different effectors affect HAS activity, it is important to purify and solubilize the HAS enzymes as previously underlined in studies on eukaryotic cell lines (30). In this context, Itano and Kimata (31) used a mammalian transient expression system to characterize the three different HAS isoforms in either cells or cellular membrane extracts. On the other hand, Spicer (32) described three relatively simple procedures for the detection of HA synthase activity in cultured mammalian cell lines. In all of these studies, the enzyme activity was measured by incubating cellular membrane extracts with radiolabeled UDP-sugar precursors, and the final analysis of the products was done by liquid scintillation counting. Various other strategies and methods can be used to determine the HA biosynthetic capacity of cells, although they are always based on the use of radiolabeled UDP-sugar precursors (33–35).
In our previous studies, we described methods of polyacrylamide gel electrophoresis of fluorophore-labeled saccharides (PAGEFS) and high performance liquid chromatography (HPLC) for the analysis of disaccharides derived from HA and chondroitin sulfate (33–38). In order to improve the sensitivity of this method, a derivatization with 2-aminoacridone (AMAC) was done, followed by fluorescence detection (39). In this study, we modified this method to address the question of localization of HAS activity during intracellular trafficking, since HA has been detected inside cells in previous studies (40–42). This new non-radioactive method was used to quantify HAS activity on cell membranes fractionated by sucrose gradient methods. To test the robustness of our approach, we analyzed the effect of 4-MU, PMA, IL-1β, PDGF-BB, and tunicamycin on cell cultures. In particular, we found that tunicamycin induced an increase of HA synthesis in both plasma and internal cell membranes in EVC cells, whereas it increased HA synthesis only in the internal cell membranes in the OVCAR-3 cells. The results suggest that post-translational modulation of HAS activity is responsible for the increased HA synthesis inside the cells. Moreover, since tunicamycin induced HA cable structures in the OVCAR-3 cells, we correlated the altered intracellular HAS activity with the capability to promote HA cable formation.
EXPERIMENTAL PROCEDURES
Cell Culture
The ECV 304 cell line (43) was obtained from “Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna” (Brescia, Italy), the OVCAR-3 cell line from human ovarian adenocarcinoma was kindly donated by Prof. Roberto Taramelli (University of Insubria), and the U937 cell line derived from a human histiocytic lymphoma was a gift of Carol A. de la Motte (Cleveland Clinic). ECV cells were grown in EGM2 complete culture medium (Cambrex, Baltimore, MD) supplemented with 10% fetal bovine serum (FBS), OVCAR-3 cells were cultured in Dulbecco's modified Eagle's medium (Euroclone) supplemented with 10% FBS, and U937 cells were grown in RPMI 1640 medium (Cambrex) supplemented with 5% FBS. Cells were seeded at 2.5 × 105 cells in 25-cm2 culture flasks and were grown for 96 h in their culture medium in an atmosphere of humidified 95% air, 5% CO2 at 37 °C.
In some experiments, ECV and OVCAR-3 cells were treated with 2 mm 4-MU (Sigma) or with 2 nm PMA (Sigma) for 18 h of incubation; with IL-1β (Peprotech) or PDGF-BB (Calbiochem), both at 10 ng/μl, for 18 h; with 200 nm PMA (a non-toxic concentration) for 5 or 10 min of incubation; or with IL-1β or PDGF-BB, both at 10 ng/μl, for 5 or 10 min. Further, ECV and OVCAR-3 cells were incubated for 18 and 24 h with 10 μg/ml tunicamycin from Streptomyces sp. (Sigma), with 10 μg/ml cycloheximide (Sigma), or with 20 μg/ml suramin (Sigma). The above mentioned concentrations were chosen from preliminary experiments in which several doses were tested.
Membrane Fractionation
Membrane fractionations were done by ultracentrifugation on a discontinuous sucrose gradient by modifying previous protocols (44–46). Briefly, cells were washed with lysis buffer (10 mm Tris, 1.5 mm MgCl2, 10 mm KCl, pH 7.4) (32) and, subsequently, were incubated with 1 ml of ice-cold lysis buffer with protease inhibitor mixture (Complete Mini EDTA-free; Roche Applied Science) for 10 min. After incubation, cells were scraped into the lysis buffer and disrupted by three strokes of 6 s each at 80% microns amplitude using a high intensity ultrasonic liquid processor (Sonics & Materials). The suspensions were left on ice during treatment. Nuclei were pelleted by spinning at 3,000 rpm for 5 min at 4 °C. The obtained supernatants were mixed with an equal volume of 85% sucrose (w/v) in 10 mm Tris-HCl, pH 7.5, placed at the bottom of a discontinuous sucrose concentration gradient (40–5%) in the same buffer, and centrifuged (150,000 × g) for 24 h at 4 °C. After ultracentrifugation, 20 fractions were collected, starting from the top of the tube. Each fraction was tested for protein content by the Bradford method and characterized by Western blot analysis before use.
Western Blotting
Western blot analyses were done to check the purity of each fraction after membrane purification steps. Twenty μg of proteins were separated by SDS-PAGE, blotted onto polyvinylidene difluoride membrane (Amersham Biosciences), and probed with polyclonal anti-CD44 monoclonal antibodies (Sigma), polyclonal anti-glyceraldeyde-3-phosphate dehydrogenase antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), polyclonal anti-GM130 (Sigma), and polyclonal anti-calnexin (Sigma), as described by Vigetti et al. (47).
Non-radioactive Assay of HA Synthesis
The HAS activity assay was essentially developed following the method described by Spicer (32) without adding any radiolabeled substrate. Twenty μg of proteins from selected membrane fractions were mixed with 10 μl of 10× HA synthase buffer (50 mm DTT, 150 mm MgCl2, 250 mm HEPES, protease inhibitor mixture, pH 7.1). One μl of 100 mm UDP-GlcNAc and 1 μl of 5 mm UDP-GlcUA were added and brought to 100 μl with lysis buffer with protease inhibitor mixture. The mixtures composed of membranes and UDP-sugar precursors were incubated for 1 h at 37 °C in a thermomixer.
After incubation, the samples were digested by hyaluronidase SD (Seikagaku Kogyo, Tokyo, Japan) at 37 °C for 1 h at a concentration of 100 milliunits/ml. A 100-milliunit/ml solution of chondroitinase ABC (Seikagaku Kogyo, Tokyo, Japan) was then added, and the mixture was incubated at 37 °C for 3 h according to our previous procedure of analysis of HA and CS (36). Subsequently, SDS (Bio-Rad) was added to each sample at a final concentration of 1%, and they were heated at 100 °C for 5 min. The samples were then frozen at −80 °C and lyophilized. In some experiments, 2 mm 4-MU or 200 nm PMA, 10 ng/μl IL-1β, or 10 ng/μl PDGF-BB at final concentrations were added into the mixture as controls.
Derivatization and Disaccharide Quantification
The disaccharides obtained with hyaluronidase SD and chondroitinase ABC digestions were derivatized with AMAC (Molecular Probes) and quantified by using PAGEFS, as previously described by Karousou et al. (36). After the run, gels were scanned in UV light using a CCD camera and Gel Doc 2000 software (Bio-Rad). Analysis of the Δdi-nonSHA band was done by comparison of its migration and intensity with standard Δ-disaccharides in the same gel. Subsequently, Δdi-nonSHA quantification in each sample was standardized to total protein mass. AMAC-derivatized disaccharides were also quantified by using HPLC (34).
Cell Adhesion Assay
The quantification of leukocyte adhesion was done as described (48) with some modifications. Briefly, ECV and OVCAR-3 cells were grown to 80% confluence in 6-well plates and then treated with the test compounds for 18 h. U937 cells were labeled for 1 h at 37 °C with 5 μm CellTrackerTM Green 5-chloromethylflourescein diacetate (Invitrogen). The labeled cells were washed extensively with PBS and resuspended at a concentration of 5 × 106 cells/ml in RPMI medium without FBS. The monocytes were added to the ECV or OVCAR-3 cells (106/well) and incubated at 4 °C for 1 h. Nonadherent monocytes were removed by washing the wells gently at 4 °C with PBS. To verify that cell adhesion was dependent on HA, we treated some wells with hyaluronidase SD (2 units/μl) or with exogenous added high molecular weight HA (4 × 106 Da; Healon). The quantification of adhered monocytes was done by fluorescence microscopy (Olympus) counting six independent fields.
Immunofluorescence
ECV304 and OVCAR-3 cells grown on coverslips were rinsed with PBS and fixed in 2% paraformaldehyde. The coverslips were preincubated with PBS containing 2% FBS and then incubated in the same solution containing biotinylated HA-binding protein (Seikagaku) (5 μg/ml) and a CD44 monoclonal antibody (clone A3D8; 10 μg/ml; Sigma) overnight at 4 °C. The coverslips were washed with PBS and then incubated with a solution containing fluorescein-tagged streptavidin (1:500) and CY2.2-conjugated anti-mouse Ig (H+L) (1:1000) in PBS containing 2% FBS. After washing in PBS, the coverslips were mounted using mounting medium (Vector Laboratories). The images were obtained using a confocal microscope (SP5; Leica). In some experiments, cells were pretreated with hyaluronidase for 30 min at 37 °C to remove cell-associated HA, fixed with cold methanol (in order to permeabilize the cells), and probed as described above. To inhibit endocytosis during the hyaluronidase treatment, lower incubation temperatures were used (i.e. 15 and 25 °C), always obtaining similar results.
Quantitative RT-PCR
Total RNA from both ECV304 and OVCAR-3 cells was extracted using Tri-reagent solution (Ambion). To remove DNA contamination, DNase (Ambion) treatment was done in all samples. One μg of total RNA was retrotranscribed using the High Capacity cDNA synthesis kit (Applied Biosystems, Foster City, CA) for 2 h at 37 °C. Quantitative RT-PCR was done on an Abi Prism 7000 instrument (Applied Biosystems) using Taqman Universal PCR Master Mix (Applied Biosystems). TaqMan gene expression assays were done for HAS1 (Hs00155410), HAS2 (Hs00193435), HAS3 (Hs00193436), and β-actin (Hs99999903) by amplification following the manufacturer's protocol (Applied Biosystems). Comparison of the amount of each gene transcript among different samples was made using β-actin as the reference and the ΔΔCt method, as previously described (15, 47, 49). After treatment with tunicamycin, the presence of ER stress markers (i.e. CHOP and ATF4) was confirmed by RT-PCR, as described previously (50).
Statistical Analysis
Statistical analysis of the data was done using analysis of variance (ANOVA), followed by post hoc tests (Bonferroni) using Origin 7.5 software (Microcal Software, Studio City, CA).
RESULTS
Measurement of HAS Activity by a Non-radioactive Assay
Contrary to other GAGs, HA synthesis normally takes place on the plasma membrane, where active HAS enzymes are located. HAS proteins are synthesized in the ER, and, through the Golgi apparatus, they reach the plasma membrane. Previous studies have indicated that HAS enzymes associated with ER/Golgi membranes are not active (51), whereas other studies highlight the presence of HA inside the cells (40, 52) and show that HA cable structures can arise from internal cell compartments (22).
To investigate the possibility that HAS enzymes associated with ER/Golgi membranes can synthesize HA, we fractionated ECV 304 cells (43), which are known to synthesize and secrete HA into the culture medium, in order to isolate fractions enriched in plasma membranes, ER/Golgi membranes, and nuclear membranes. ECV 304 cells are described as a spontaneously transformed line derived from human umbilical vein endothelial cells, although a genetic characterization revealed that this cell line is not appropriate to study endothelial cell biology (53).
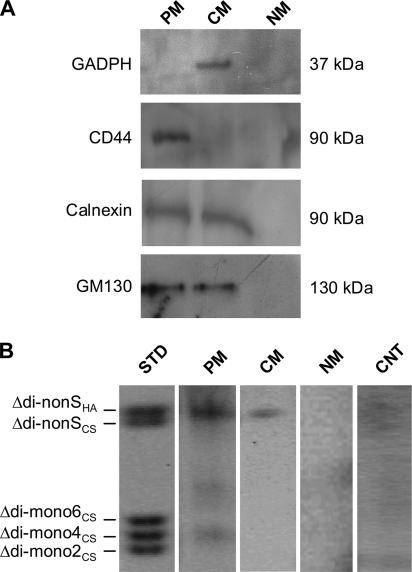
Among 20 subcellular fractions obtained from discontinuous sucrose gradient separation (supplemental Fig. S1), we isolated three fractions representative of plasma membrane (PM), cytosolic membranes (CM), and nuclear membranes (NM). To check the purity of these representative fractions, we did Western blot analyses using antibodies against CD44 (a plasma membrane-specific marker), glyceraldeyde-3-phosphate dehydrogenase (a cytosolic membrane marker), GM130 (a Golgi membrane marker), and calnexin (an ER membrane marker). Fig. 1A shows that a 90 kDa band corresponding to CD44 was present only in the PM fraction, indicating that no plasma membrane protein contaminations were present in the other two fractions. The 37 kDa band corresponding to the glyceraldeyde-3-phosphate dehydrogenase protein was only detected in the CM fraction. On the other hand, GM130 and calnexin were present in both the PM and CM fractions but were absent in the NM fraction. This suggests that the PM fraction contains some Golgi and ER membrane components.
FIGURE 1.
A, Western blot analyses on PM, CM, and NM fractions isolated after sucrose gradient cell fractionation by using anti-CD44, anti-glyceraldeyde-3-phosphate dehydrogenase (GAPDH), anti-GM130, and anti-calnexin antibodies that are plasma membrane, cytosol, Golgi, and ER markers, respectively. B, PAGEFS analyses of GAGs using the new in vitro non-radioactive method to quantify HAS activity. The STD lane represents 100 pmol of a standard mix of AMAC-disaccharides. The PM, CM, and NM lanes represent the disaccharide analyses of GAGs synthesized by 20 μg of ECV membrane proteins from PM, CM, and NM fractions, respectively. Each sample was incubated with 0.05 mm UDP-GlcUA and 1 mm UDP-GlcNAc at 37 °C for 1 h, followed by hyaluronidase SD and chondroitinase ABC digestions and AMAC derivatization. The CNT lane contains 20 μg of ECV membrane proteins from PM without incubation with the UDP-sugars. The results shown are representative of three independent experiments.
Previous studies used radioactive precursors to measure HAS activity in subcellular fractions (30–34). We recently showed that PAGEFS and HPLC techniques are simple, sensitive, and low cost procedures to quantify Δ-disaccharides derived from digested GAGs (36–38). Therefore, we incubated 20 μg of proteins contained in PM, CM, and NM fractions and incubated them at 37 °C for 1 h with 0.05 mm UDP-GlcUA and 1 mm UDP-GlcNAc (final concentrations), as previously reported for the radioactive HAS activity assay (32). After enzyme heat inactivation, the samples were digested with hyaluronidase SD and chondroitinase ABC in order to generate the Δ-HA and Δ-CS disaccharides, which were then derivatized with AMAC. The AMAC-disaccharides were separated and quantified by PAGEFS (Fig. 1B). A specific AMAC-labeled Δ-HA disaccharide band was identified in the PM and, surprisingly, also in the CM fraction lanes by comparison with a standard mix of disaccharides. Nuclear membranes did not synthesize any HA detectable by PAGEFS after incubation with the UDP-precursors. A control (CNT) shows an analysis of a hyaluronidase/chondroitinase digest of an equivalent sample of PM that was incubated without the UDP-sugar substrates and indicates that endogenous HA contributed minimally to the amounts generated by the PM and CM samples incubated with the substrates.
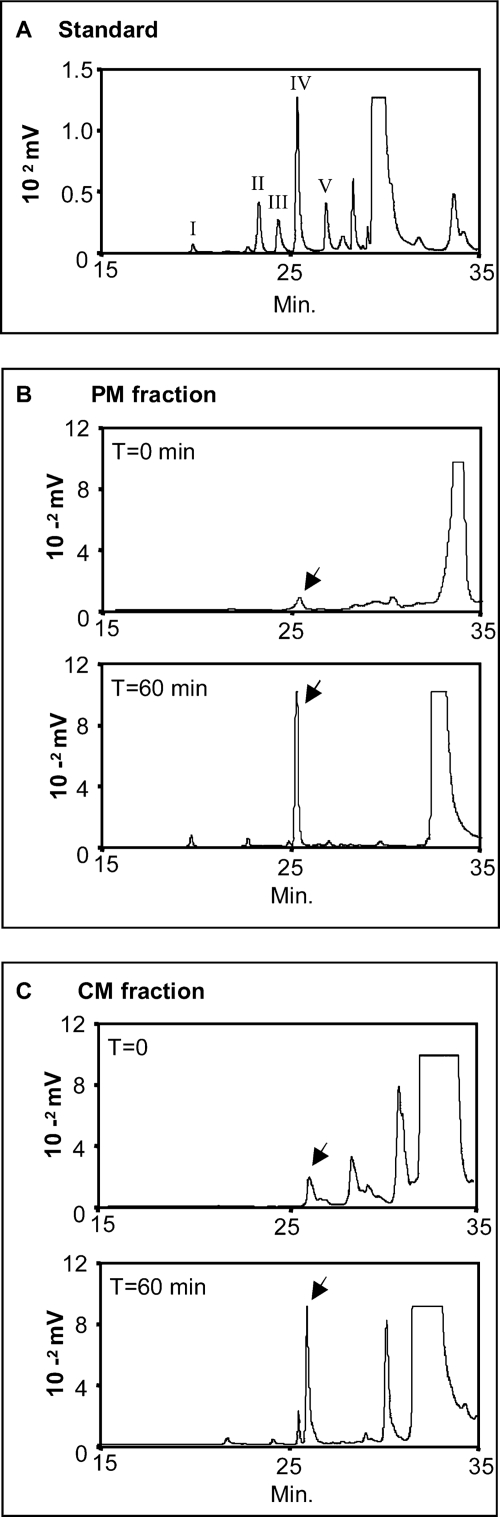
We also quantified AMAC-labeled Δ-disaccharides by HPLC analyses, as shown in Fig. 2. Fig. 2A shows an HPLC analysis to determine the retention times of a standard mix of disaccharides. Subsequently, we analyzed PM sample disaccharides before (T = 0; Fig. 2B) and after (T = 60 min; Fig. 2B) incubation with the UDP-sugar precursors. The analyses showed a clear increase of ∼8-fold of the HA disaccharide peak area after 1 h of incubation. Interestingly, the sensitivity of HPLC revealed that some HA was present in the PM before incubation with the UDP-sugars, most likely reflecting the presence of HA localized on the extracellular plasma membrane attached to HAS enzymes. Similar determinations were done on the CM fraction (Fig. 2C). The HA disaccharide peak area after 1 h of incubation with the UDP-sugar precursors increased ∼3-fold, indicating that some HA synthetic activity was apparently present inside the cell.
FIGURE 2.
HPLC analyses of glycosaminoglycans synthesized using the in vitro non-radioactive method to quantify HAS activity. A, 100 pmol of a standard mix of AMAC-derivatives Δdi-di(2,4)SCS (Δ-disaccharides of chondroitin 2,4-sulfated) (I), Δdi-mono4SCS (Δ-disaccharides of chondroitin 4-sulfated) (II), Δdi-mono6SCS (Δ-disaccharides of chondroitin 6-sulfated) (III), Δdi-nonSHA (IV), and Δdi-nonSCS (Δ-disaccharides of CS (non-sulfated)) (V). B, analysis of HA synthetic activity present in 20 μg of PM fraction proteins before incubation (T = 0) and after 1 h of incubation (T = 60 min) with UDP-GlcUA and UDP-GlcNAc after hyaluronidase SD and chondroitinase ABC digestions and AMAC derivatization. C, analysis of HA synthetic activity present in 20 μg of CM fraction proteins before incubation (T = 0) and after 1 h of incubation (T = 60 min) with UDP-sugar precursor as in B. The arrows indicate Δdi-nonSHA peaks in the PM and CM chromatograms. The results shown are representative of three independent experiments.
The specific HA synthase activities (determined from the increases between 0 and 60 min) were 3.7 × 10−3 ± 3.3 × 10−4 and 8.4 × 10−4 ± 5 × 10−4 pmol of Δ-HA/μg of membrane protein/min in the PM fraction and CM fractions, respectively.
Effects of 4-MU, PMA, IL-1β, and PDGF-BB on HA Synthesis Activity
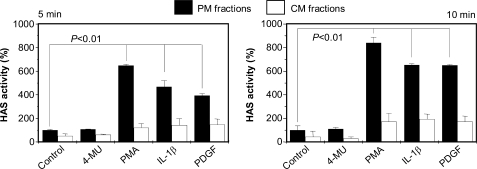
HA synthesis can be modulated by different molecules that can regulate HAS activity. Cells treated with PMA, IL-1β, or PDGF-BB increase the amount of HA released into the culture medium (29, 54, 55), whereas treating cells with 4-MU decreases synthesis of HA (56). We tested the effects of these compounds on HAS activity in the PM and CM fractions. ECV cells were treated with 2 mm 4-MU, 200 nm PMA, 10 ng/μl IL-1β, or 10 ng/μl PDGF-BB for 5 and 10 min. The three subcellular fractions were then isolated, and 20 μg of proteins in PM and CM fractions were incubated with the UDP-sugar precursors to quantify the HAS enzymatic activity (Fig. 3). 4-MU did not inhibit HA synthetic activity in either the PM or CM fraction, indicating that this compound does not directly interact with HAS proteins, which confirms the finding of Nakamura et al. (27). Interestingly, the treatments with PMA, IL-1β, and PDGF-BB increased HAS activity compared with untreated samples mainly in PM fractions, since the slight increase in CM fractions was not statistically significant. As a control, aliquots of the PM fraction from untreated cultures were exposed to the same concentrations of the reagents in the presence of the UDP-sugar substrates. In all cases, we detected no increase in the amount of synthesized HA, indicating that the reagents did not directly alter HAS activity (data not shown).
FIGURE 3.
Relative HAS activity in PM (black bars) or CM (white bars) fractions prepared from untreated ECV cells (controls) or treated for 5 or 10 min with 4-MU (2 mm final concentration), PMA (200 nm final concentration), IL-1β (10 ng/μl final concentration), and PDGF-BB (10 ng/μl final concentration). Eighty percent confluent cells were treated with the compounds cited above for the indicated times, membrane fractions were prepared, and HAS activity was assessed by using the non-radioactive method with PAGEFS and HPLC analyses. HAS activity is expressed as a percentage relative to control PM. Data represent mean ± S.E. from two independent experiments of triplicate determinations.
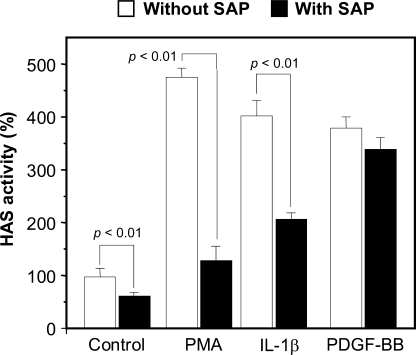
Since PMA, IL-1β, and PDGF-BB modified HAS activity only when added to the viable cells, these findings suggest that some rapid post-translational modifications are responsible for the enzymatic activity change. The 5–10-min time frame makes translational production of new HAS protein unlikely. To investigate whether HAS phosphorylation could have happened after cell treatments, we isolated the PM fraction from cells treated with PMA, IL-1β, or PDGF-BB for 5 min and incubated them with or without shrimp alkaline phosphatase (SAP) before measuring HAS activity. Fig. 4 shows that SAP treatment significantly reduced HAS activity in each sample except for PDGF-BB-treated cells. The results indicate that phosphorylation of HAS or of some auxiliary protein that interacts with HAS after PMA or IL-1β treatment affects HAS enzymatic activity. Interestingly, PM fractions purified from control cells and treated with SAP also showed a decreased HAS activity (Fig. 4), suggesting that the presence of a certain degree of protein phosphorylation supports HA synthesis without adding any modulators.
FIGURE 4.
Relative HAS activity in PM membrane fraction after SAP treatment. Two hundred nm PMA, 10 ng/μl IL-1β, and 10 ng/μl PDGF-BB were added to 80% confluent ECV cells. After 10 min of incubation, the ECV membranes were purified, and 20 μg of PM fraction proteins were preincubated or not with 0.04 units/μl SAP for 30 min and subsequently incubated with the UDP-precursors and analyzed for HAS activity, as described in the legend to Fig. 3. *, p < 0.01 (ANOVA) without SAP versus with SAP. Values are given as percentages of the control value without SAP. Data represent mean ± S.E. from two independent experiments of triplicate determinations.
Effect of Tunicamycin on HA Synthesis and Cable Formation
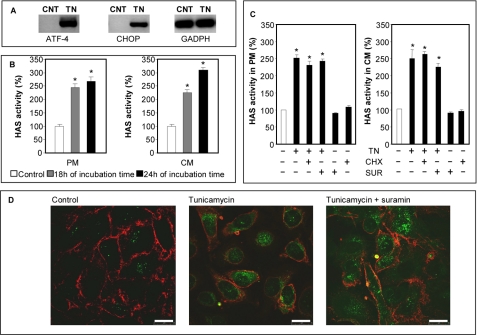
Tunicamycin inhibits N-glycosylation of proteins and initiates ER stress (also known as unfolded protein response) (57), and it is known to modify HA metabolism (23). ECV cell cultures were treated with 10 μg/ml tunicamycin for 18 and 24 h, and ER stress was confirmed by RT-PCR detecting specific markers expressed only in tunicamycin-treated cells (ATF-4 and CHOP) (Fig. 5A). PM and CM fractions were prepared and tested for HAS activity. Fig. 5B shows that tunicamycin induced an increase of HAS activity of about 3-fold in the PM fractions after 18 h of incubation (from about 3.6 × 10−3 to about 1.1 × 10−2 pmol of HA disaccharide/μg of membrane protein/min). Interestingly, HAS activity in the CM fraction also significantly increased after 24 h of incubation, reaching the level of that in PM membranes. The induction of HAS activity in the fractions could not be ascribed to an increased HAS translation, because ER stress drastically reduces protein synthesis (53) and because we quantified mRNAs coding for HASs after tunicamycin treatments by quantitative RT-PCR and found a clear messenger reduction (result not shown). The HAS activity increment in the CM fraction was also detected in cells treated with other ER stress-inducing compounds (i.e. DTT and ethanol), whereas serum starvation, a nonspecific stress, did not induce ER stress (supplemental Fig. S2). These data suggest that increased HAS activity could be a specific response to ER stress.
FIGURE 5.
A, RT-PCR amplification of ER stress markers (CHOP and ATF-4) and housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase; GAPDH) starting from cDNA obtained from control (CNT)- and 10 μg/ml tunicamycin (TN)-treated ECV for 18 h. The results are representative of six independent experiments. B, relative HAS activity in PM or CM fractions after different treatments. Eighty percent confluent ECV cells were left untreated (control; white bars) or treated with a 10 μg/ml final concentration of tunicamycin (gray and black bars). After 18 and 24 h of incubation, the ECV membranes were purified, and 20 μg of PM or CM fraction proteins were incubated with the UDP-precursors and analyzed for HAS activity, as described previously. *, p < 0.01 (ANOVA), treated PM or CM versus control. Data represent mean ± S.E. from two independent experiments of triplicate determinations. C, 80% confluent ECV cells were left untreated or treated with 10 μg/ml tunicamycin (TN), 10 μg/ml cycloheximide (CHX), or 20 μg/ml suramin (SUR) for 18 h. Membrane fractions were obtained, and the relative HAS activity was calculated as described above. *, p < 0.01 (ANOVA), treated PM or CM versus control. Data represent mean ± S.E. from two independent experiments of triplicate determinations. D, confocal immunolocalization of HA (green) and CD44 (red) on untreated, tunicamycin-treated (10 μg/ml final concentration), or tunicamycin + suramin-treated (20 μg/ml) ECV pretreated with hyaluronidase and fixed with methanol. The microphotographs show representative results of three different independent experiments.
To confirm that the increased HAS activity was not due to increased translation of HAS messenger RNAs, ECV cells were treated with a combination of 10 μg/ml tunicamycin and 10 μg/ml cycloheximide for 18 h, and the HAS activity was quantified in PM and CM fractions. The combination of cycloheximide with tunicamycin gave the same results as tunicamycin treatment alone (Fig. 5C). Moreover, to rule out the possibility that the increment of HAS activity in the CM fraction could be due to internalization of plasma membrane-resident HAS enzymes, ECV cells were treated with a combination of tunicamycin and 20 μg/ml suramin for 18 h, and the HAS activity was quantified in the PM and CM fractions. Suramin is known to interfere with internalization, recycling, and degradation of cell surface proteoglycans (i.e. glypican-1) in the same cell line used in our experiments (58). We used suramin to attempt to block HAS protein recycling. Since the commercial HAS antibodies had poor sensibility in our conditions, to confirm the inhibitory effect on membrane recycling, we investigated the localization of CD44 by means of immunofluorescence and confocal microscopy and found an intensification of plasma membrane-associated CD44 signal in cells treated with suramin, supporting the inhibition of recycling (supplementary Fig. S3). As shown in Fig. 5C, suramin in combination with tunicamycin did not alter HAS activity significantly compared with tunicamycin alone in either PM or CM fractions. Neither cycloheximide nor suramin treatment alone increased HA synthesis in PM or CM fractions compared with untreated cells. An experiment was done to demonstrate the effects of tunicamycin and suramin in intact cells. ECV cells were untreated or treated with tunicamycin or tunicamycin plus suramin for 24 h, and the pericellular HA was removed with hyaluronidase for 30 min at 37 °C. After fixation with methanol in order to solubilize cell membranes, the cells were stained for HA and visualized by fluorescence and confocal microscopy. As shown in Fig. 5D, the green intracellular spots corresponding to HA were clearly increased after tunicamycin or tunicamycin plus suramin treatments, supporting the increased HA synthesis in internal membranes. Since HA fragments generated by hyaluronidase could increase endocytosis, we performed control experiments inhibiting endocytosis by reducing the incubation temperature (59) and demonstrating that the intracellular HA cannot be ascribed to endocytosis of external HA (result not shown). There was no apparent change in CD44 distribution in ECV cells (Fig. 5D) and OVCAR-3 cells (data not shown) after the tunicamycin treatments.
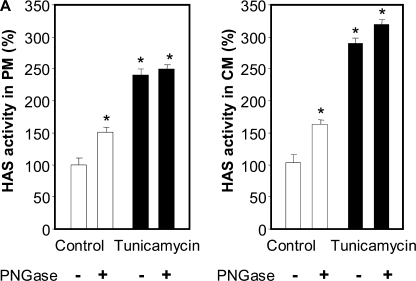
Since tunicamycin inhibits N-glycosylation, we incubated PM and CM fractions obtained from control and tunicamycin-treated cells with N-glycosidase F in order to remove accessible N-oligosaccharides. Interestingly, the removal of N-oligosaccharides in PM and CM fractions prepared from untreated cells significantly increased HAS activity, whereas such activity remained unvaried in PM and CM fractions obtained from tunicamycin-treated cells (Fig. 6).
FIGURE 6.
Relative HAS activity in PM or CM fractions after different treatments. A, 20 μg of PM or CM fraction proteins from control (white bars) or 24-h tunicamycin-treated cells (black bars) were preincubated or not with 3 units/μl N-glycosidase F (PNGase; New England Biolabs) for 1 h and assessed for HAS activity. *, p < 0.01 (ANOVA) control without N-glycosidase F versus other samples. Data represent mean ± S.E. from two independent experiments of triplicate determinations.
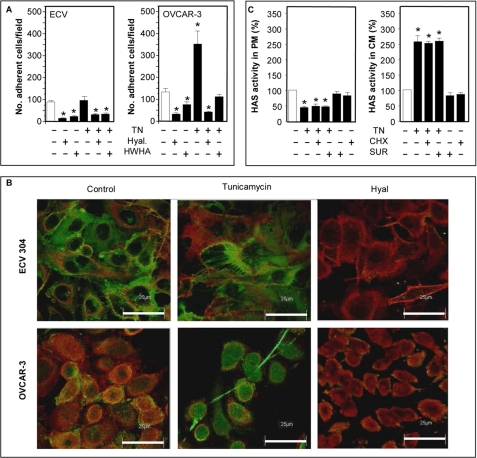
Previous studies have shown that ER stress induces the formation of HA cable-like structures that are adhesive for leukocytes (23) but without describing the molecular mechanism. Hascall et al. (22) hypothesized that HA cable synthesis could originate from intracellular compartments. Therefore, we determined whether the increment of HAS activity in the CM fraction could be related to HA cable formation. We tested the adhesion of U937 monocytes to ECV and OVCAR-3 (a cell line derived from an ovarian cancer (60)) after treatment with 10 μg/ml tunicamycin for 18 h. We used these two cell types because, by quantitative RT-PCR, we found that ECV and OVCAR-3 expressed different HAS isoform transcripts, HAS3 and HAS2, respectively (results not shown). U937 cells were marked with a green fluorescent vital dye and added to tunicamycin-treated cells. After an incubation of 1 h and the removal of unbound U937 cells by washing, the monocytes bound to ECV and OVCAR-3 cells were directly counted under a fluorescent microscope. As shown in Fig. 7A, tunicamycin induced a strong increase in monocyte adhesion compared with control only in the OCVAR-3 cell line. The slight increase in U937 adhesion in tunicamycin treated ECV was not significant. Interestingly, the interaction of U937 cells with ECV and OVCAR-3 cells was HA-dependent, because hyaluronidase treatment before adding monocytes inhibited their binding. Moreover, the addition of exogenous high molecular weight HA (Healon; average molecular mass 4 × 106 Da) also strongly inhibited U937 binding, indicating a competition between cell-derived HA and exogenously added HA. A similar adhesion experiment was done with ECV and OVCAR-3 cells by treating them with DTT and ethanol. Only OVCAR-3 cells treated with DTT and ethanol showed an increment of monocyte adhesion, whereas cells incubated without serum showed results comparable with the controls (supplemental Fig. S4). Such results suggest that the U937 cell HA-adhesive properties can be modulated in a specific manner by the ER stress response, possibly depending in part on the HAS isoform in the cells.
FIGURE 7.
A, U937 adhesion on tunicamycin-treated ECV or OVCAR-3 cells. Eighty percent confluent cells were left untreated or treated with a 10 μg/ml final concentration of tunicamycin. After 18 h of incubation, 1 × 106/well fluorescent U937 cells were added for 60 min. After washing with PBS, adherent U937 cells were counted under a fluorescent microscope, and the number of cells/field is reported. In some experiments, before adding U937, ECV and OVCAR-3 cells were treated with 2 units/μl hyaluronidase for 20 min (Hyal), or U937 cells were added with 50 μg/ml high molecular weight HA (HWHA). *, p < 0.05 (ANOVA), untreated versus treated samples. Data represent mean ± S.E. from two independent experiments of triplicate determinations. B, confocal immunolocalization of HA (green) and CD44 (red) on untreated or tunicamycin (10 μg/ml final concentration) treated ECV or OVCAR-3 cells. As a control, 2 units/μl hyaluronidase was added before the immunolocalization (Hyal). The microphotographs show representative results of three different independent experiments. The white bars correspond to 25 μm. Original magnification was ×630. C, 80% confluent OVCAR-3 cells were left untreated or treated with 10 μg/ml tunicamycin (TN), 10 μg/ml cycloheximide (CHX), or 20 μg/ml suramin (SUR) for 18 h. Membrane fractions were obtained, and the relative HAS activities were calculated as described above. *, p < 0.01 (ANOVA), treated PM or CM versus control. Data represent mean ± S.E. from two independent experiments of duplicate determinations.
By using immunofluorescence and confocal microscopy, we studied the distribution of HA in untreated and tunicamycin-treated ECV cells cultured for 18 h without finding any cable-like structures (Fig. 7B). We tried several combinations of tunicamycin concentrations and incubation times with the same result (data not shown). The fluorescence relative to HA was distributed in a uniform manner around the cells both in control and treated cells. In contrast, when we treated OVCAR-3 with 10 μg/ml of tunicamycin (final concentration) for 18 h, we found HA filaments typical of the cable-like structures (Fig. 7B).
In order to relate HAS activity and cable formation, we prepared the subcellular membrane fractions from untreated and tunicamycin-treated OVCAR-3 cells and assayed them for HAS activity, as described previously. Tunicamycin treatment induced a significant increase of HAS-specific activity in the CM fraction and a significant reduction of HAS specific activity in the PM fraction (Fig. 7C), whereas tunicamycin-treated ECV cells showed increased HAS activity in both PM and CM fractions (Fig. 5C). Experiments with tunicamycin and either cycloheximide or suramin did not alter the results with tunicamycin alone (Fig. 7C), similar to the results obtained with ECV cells (Fig. 5C). An experiment was done to demonstrate the effects of tunicamycin and suramin in intact OVCAR-3 cells. Cultures were untreated or treated with tunicamycin or tunicamycin plus suramin for 24 h, and pericellular HA was removed with hyaluronidase. After fixation with methanol, HA was stained and visualized by fluorescence and confocal microscopy. As shown in supplemental Fig. S5, the green intracellular spots corresponding to HA were clearly observable after tunicamycin or tunicamycin plus suramin treatments, supporting the increased synthesis of HA in internal membranes, as was observed in the ECV cells (Fig. 5D).
DISCUSSION
Active HASs normally reside on the plasma membrane, and their presence in active form in the intracellular compartments may represent either the newly synthesized HASs during the sorting process through ER and Golgi to the plasma membrane or recycled HASs from the cell surface due to an endocytic pathway. In this context, the critical issue is whether the HASs could be active in different cell compartments.
This study shows the presence of HA synthetic activity both in plasma membrane and in membranes from ER and/or Golgi. HAS enzymatic activity was quantified in purified membranes with a new non-radioactive method. In this procedure, we added non-radioactive UDP-sugar precursors to the membrane fractions in contrast to previously reported methods that used 14C- or 3H-labeled UDP-precursors (12, 31, 32, 61, 62) and determined HAS activity by counting radioactivity in newly synthesized HA. These classical radioactive assays were very sensitive, but, on the other hand, they involved radioactive manipulations with all of the problems that this involves. In order to quantify the newly synthesized HA in our non-radioactive method, we applied PAGEFS, an electrophoretic method to measure Δ-disaccharides obtained from digestion of newly synthesized HA (36). This method requires reducing end labeling with a fluorophore (AMAC), and the procedure has comparable sensitivity to radioactive methods. Moreover, since AMAC labels the reducing end of the disaccharides, it does not label the excess of UDP-sugar precursors, which is an issue with the classical radioactive method. While we were working on this project, another method to assay HAS activity without using radioisotopes was developed and published (63).
By centrifugation, we prepared three main fractions containing the PM, CM, and NM. Although the PM fraction contained some ER and Golgi proteins, CM preparations were negative for plasma membrane marker, and the absence of CD44 contaminations in the CM membrane preparation allowed us to ascribe the cytosolic HA production only to HASs present and active in the internal membranes of the cells.
The evidence of HAS activity in cytosol membrane fractions raises the question of HA synthesis inside the cells in vivo. In fact, previous studies (40, 41) described a significant amount of HA inside cells, which was proposed to represent internalization from pericellular HA due to hyaluronidase activity (40). On the other hand, other authors (51, 64) proposed that HASs move throughout the intracellular membrane organelles before reaching the plasma membrane, and intracellular HA can represent the product of HASs located in ER/Golgi vesicles. Recently it has been shown that in dividing cells in hyperglycemic growth medium, HA synthesis initiates inside the cell, which appears to induce autophagy and extrusion of a monocyte-adhesive matrix (52). It is to be noted that the observation of intracellular HA by Evanko and Wight in 1999 was made in hyperglycemic conditions (40).
Previous studies demonstrated that in various biological systems, HAS activity could be stimulated with a variety of compounds, such as cyclic AMP, calf serum (65), epidermal growth factor (66), vanadate (67), PMA (68), insulin-like growth factor-I (69, 70), PDGF-BB, fibroblast growth factor, and TGF-β1 (54). In all reported studies, HAS activity was tested by using a radioactive approach, and we confirmed such data with our method. In the case of PMA, IL-1β, and PDGF-BB, the mechanisms involved in HAS activation are most likely mediated by phosphorylation induced by protein kinase C, c-AMP-dependent protein kinases, or calcium ion-dependent protein kinases (61). The effect of phosphorylation on HAS activity was also confirmed by our experiments with phosphatases, which demonstrated reduced activity. Only the increased HAS activity induced by PDGF-BB treatment was found to be resistant to phosphatase. This suggests that that the modifications induced by PDGF-BB could be inaccessible to SAP or reflect an alternate pathway to activate HAS. Further, our experiments cannot exclude the possibility that phosphorylation involves other protein or proteins located in proximity to the HASs that could have a role in regulating their function after the treatments.
4-MU inhibits HA synthesis by lowering both the cytosolic concentration of UDP-GlcUA and the expression of HASs (71). We confirmed that 4-MU reduced HA synthesis in intact cells (result not shown) without affecting the HAS activity when added to membrane preparations, as already shown by Nakamura et al. (27).
Interestingly, all of the above substances used to treat the cells for short times drastically modified HA synthetic activity of HASs on plasma membrane but not that of HASs in the interior of the cells. In contrast, tunicamycin affected the activity of both HASs located in the plasma membrane and in ER/Golgi membranes. The effect of tunicamycin is to induce ER stress by inhibiting N-glycosylation (57). Protein N-glycosylation is known to affect protein folding, oligomerization, quality control, sorting, and transport, moving through ER via the Golgi complex to their final destination inside and/or outside of the cells (72). Our data obtained after tunicamycin treatment suggest that HAS proteins are glycosylated. In fact, the enzymatic removal of N-oligosaccharides in proteins isolated from untreated cells increased HAS activity to levels similar to those in proteins prepared from tunicamycin-treated cells in which N-glycosylation was inhibited by the treatment. It is to be noted that, as for phosphorylation, N-glycosylation could involve other not yet identified HAS-regulating proteins.
Tunicamycin treatment requires 18–24 h of incubation to reach a maximum effect. Although ER stress inhibits protein synthesis, we used cycloheximide, which inhibits protein synthesis rapidly, and showed that its combination with tunicamycin gave no difference from tunicamycin alone, confirming that new HAS synthesis does not contribute to the increased HA synthesis. Moreover, a similar increase of HAS activity in CM fractions was observed when suramin, an inhibitor of plasma membrane recycling, was used in combination with tunicamycin, indicating that internalization of HAS enzymes from plasma membrane is unlikely to contribute to the increased HA synthesis activity in the ER/Golgi membranes. Although suramin has been reported to prevent endocytosis of proteoglycans, the mechanisms involving HA endocytosis may be different. The fact that CD44 was stopped on the plasma membrane would suggest that specific internalization of HA is inhibited, but we cannot exclude the possibility of HA pinocytosis or an alteration of the HA lysosomal degradation pathway due to suramin treatment.
Cable-like HA structures were described after inducing ER stress by treating with proinflammatory cytokines both in vitro and in vivo (73). Such HA structures are adhesive for immune cells and therefore have a central role in inflammation and recruitment for monocytes (21). More detailed morphological analyses suggested that these HA cables seem to have contacts not only with the surface of the cells where active HAS enzymes are located but also with intracellular vesicular structures that could correspond to Golgi, ER, or the nuclear envelope (22). Since tunicamycin has been shown to induce HA cable formation, we tested whether the changes in intracellular HA synthesis could be involved in HA cable formation. As indirect proof of HA cable formation, we counted the monocytes adherent to tunicamycin-treated ECV cells but found that the number was not increased. This suggests that ECV cells did not produce cables. In contrast, OVCAR-3, a cell line derived from a human carcinoma strongly increased monocyte adhesion after tunicamycin treatment, which suggests that this cell line was able to synthesize HA cable in our conditions. The direct evidence of HA cable was obtained by confocal microscopy, which confirmed that tunicamycin-stimulated ECV cells did not, whereas OVCAR-3 cells did, produce HA filaments.
The mechanisms that distinguish the different abilities of ECV and OVCAR-3 cells to form HA cable structures although both increase HA synthesis in response to tunicamycin are not known but are crucial for understanding how the cables are formed. Our hypothesis is that alteration of HAS activity could have a role in cable formation, and we found that both cell lines increased HA synthesis activity in membranes enriched in ER and Golgi membranes. Interestingly, HAS activity in the plasma membrane decreased in response to tunicamycin in OVCAR-3 cells, which are able to produce HA cables, but not in ECV cells, which suggests a more complex synthetic mechanism. On the other hand, the different expression of HAS isoforms in the two cell lines, HAS2 in ECV cells and HAS3 in OVCAR-3 cells, could have a role in this issue. Interestingly, HAS2 and HAS3 sequence analyses through bioinformatic tools (available on the World Wide Web) show different N-glycosylation sites that could have different roles. Such analysis revealed that only HAS2 has two hypothetical asparagine residues (82 and 178 in the human HAS2 sequence deposited in the public data base with accession number NP_005319) that could be N-glycosylated. However, these two residues are in the cytosolic region of the protein, making them unsuitable for this modification. This issue could support the hypothesis that such modification does not happen directly on the HAS enzyme but in other not yet identified protein(s) that could interact and regulate HAS activity.
The different properties of HAS enzymes were previously studied on renal tubular cells showing that overexpression of HAS3 increased HA cable formation, whereas overexpression of HAS2 reduced it (73, 74).
In conclusion, by using the new non-radioactive method to assess HAS activity in vitro, we were able to address the question about the HAS activity in Golgi and ER membrane. Our findings highlighted the role of N-glycosylation and phosphorylation to modulate HAS activity and the possible involvement of intracellular HASs in the HA cable formation.
Supplementary Material
Acknowledgments
We are grateful to Prof. Floriana Rosati (University of Siena, Italy) for antibodies, Prof. Roberto Taramelli for the OVCAR-3 cell line, Luisa Guidali for confocal microscope imaging, and Andrea Moriondo for imaging analysis. We gratefully acknowledge the “Centro Grandi Attrezzature per la Ricerca Biomedica” Università degli Studi dell'Insubria, for instrument availability.
This work was supported by Fondazione Comunitaria del Varesotto-ONLUS (to D. V.) and PRIN 2007 (to G. D. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HA
- hyaluronan
- 4-MU
- 4-methylumbeliferon
- AMAC
- 2-aminoacridone
- CS
- chondroitin sulfate
- ER
- endoplasmic reticulum
- DTT
- dithiothreitol
- GAG
- glycosaminoglycan
- HAS
- hyaluronan synthase
- IL-1β
- interleukin 1β
- PAGEFS
- polyacrylamide gel electrophoresis of fluorophore-labeled saccharides
- PDGF-BB
- platelet-derived growth factor-BB
- PMA
- phorbol 12-myristate 13-acetate
- SAP
- shrimp alkaline phosphatase
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- ANOVA
- analysis of variance
- PM
- plasma membrane
- CM
- cytosolic membrane(s)
- NM
- nuclear membrane(s)
- Δdi-nonSHA and Δ-HA
- Δ-disaccharides of HA (non-sulfated)
- Δdi-nonScs
- disaccharides of CS (non-sulfated)
- Δdi-mono6cs
- chondroitin 6, sulfate disaccharide
- Δdi-mono4cs
- chondroitin 4, sulfate disaccharide
- Δdi-mono2cs
- chondroitin 2, sulfate disaccharide.
REFERENCES
- 1.Toole B. P. (1990) Curr. Opin. Cell Biol. 2, 839–844 [DOI] [PubMed] [Google Scholar]
- 2.Lee J. Y., Spicer A. P. (2000) Curr. Opin. Cell Biol. 12, 581–586 [DOI] [PubMed] [Google Scholar]
- 3.Jiang D., Liang J., Fan J., Yu S., Chen S., Luo Y., Prestwich G. D., Mascarenhas M. M., Garg H. G., Quinn D. A., Homer R. J., Goldstein D. R., Bucala R., Lee P. J., Medzhitov R., Noble P. W. (2005) Nat. Med. 11, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 4.Scheibner K. A., Lutz M. A., Boodoo S., Fenton M. J., Powell J. D., Horton M. R. (2006) J. Immunol. 177, 1272–1281 [DOI] [PubMed] [Google Scholar]
- 5.Camenisch T. D., Spicer A. P., Brehm-Gibson T., Biesterfeldt J., Augustine M. L., Calabro A., Jr., Kubalak S., Klewer S. E., McDonald J. A. (2000) J. Clin. Invest. 106, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ori M., Nardini M., Casini P., Perris R., Nardi I. (2006) Development 133, 631–640 [DOI] [PubMed] [Google Scholar]
- 7.Raio L., Cromi A., Ghezzi F., Passi A., Karousou E., Viola M., Vigetti D., De Luca G., Bolis P. (2005) Matrix Biol. 24, 166–174 [DOI] [PubMed] [Google Scholar]
- 8.Spicer A. P., Tien J. Y. (2004) Birth Defects Res. C Embryo Today 72, 89–108 [DOI] [PubMed] [Google Scholar]
- 9.Mack J. A., Abramson S. R., Ben Y., Coffin J. C., Rothrock J. K., Maytin E. V., Hascall V. C., Largman C., Stelnicki E. J. (2003) FASEB J. 17, 1352–1354 [DOI] [PubMed] [Google Scholar]
- 10.Gao F., Cao M., Yang C., He Y., Liu Y. (2006) J. Biomed. Mater. Res. B Appl. Biomater. 78, 385–392 [DOI] [PubMed] [Google Scholar]
- 11.Heldin P. (2003) Braz. J. Med. Biol. Res. 36, 967–973 [DOI] [PubMed] [Google Scholar]
- 12.Itano N., Sawai T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., Miyauchi S., Spicer A. P., McDonald J. A., Kimata K. (1999) J. Biol. Chem. 274, 25085–25092 [DOI] [PubMed] [Google Scholar]
- 13.Campo G. M., Avenoso A., Campo S., Angela D., Ferlazzo A. M., Calatroni A. (2006) Mol. Cell Biochem. 292, 169–178 [DOI] [PubMed] [Google Scholar]
- 14.Recklies A. D., White C., Melching L., Roughley P. J. (2001) Biochem. J. 354, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigetti D., Ori M., Viola M., Genasetti A., Karousou E., Rizzi M., Pallotti F., Nardi I., Hascall V. C., De Luca G., Passi A. (2006) J. Biol. Chem. 281, 8254–8263 [DOI] [PubMed] [Google Scholar]
- 16.Jokela T. A., Jauhiainen M., Auriola S., Kauhanen M., Tiihonen R., Tammi M. I., Tammi R. H. (2008) J. Biol. Chem. 283, 7666–7673 [DOI] [PubMed] [Google Scholar]
- 17.Goentzel B. J., Weigel P. H., Steinberg R. A. (2006) Biochem. J. 396, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldin P., Asplund T., Ytterberg D., Thelin S., Laurent T. C. (1992) Biochem. J. 283, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prehm P. (1983) Biochem. J. 211, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de La Motte C. A., Hascall V. C., Calabro A., Yen-Lieberman B., Strong S. A. (1999) J. Biol. Chem. 274, 30747–30755 [DOI] [PubMed] [Google Scholar]
- 21.de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., Strong S. A. (2003) Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hascall V. C., Majors A. K., De La Motte C. A., Evanko S. P., Wang A., Drazba J. A., Strong S. A., Wight T. N. (2004) Biochim. Biophys. Acta 1673, 3–12 [DOI] [PubMed] [Google Scholar]
- 23.Majors A. K., Austin R. C., de la Motte C. A., Pyeritz R. E., Hascall V. C., Kessler S. P., Sen G., Strong S. A. (2003) J. Biol. Chem. 278, 47223–47231 [DOI] [PubMed] [Google Scholar]
- 24.Teder P., Vandivier R. W., Jiang D., Liang J., Cohn L., Puré E., Henson P. M., Noble P. W. (2002) Science 296, 155–158 [DOI] [PubMed] [Google Scholar]
- 25.Milinkovic M., Antin J. H., Hergrueter C. A., Underhill C. B., Sackstein R. (2004) Blood 103, 740–742 [DOI] [PubMed] [Google Scholar]
- 26.Jacobson A., Brinck J., Briskin M. J., Spicer A. P., Heldin P. (2000) Biochem. J. 348, 29–35 [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T., Funahashi M., Takagaki K., Munakata H., Tanaka K., Saito Y., Endo M. (1997) Biochem. Mol. Biol. Int. 43, 263–268 [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M., Asplund T., Yamashita H., Heldin C. H., Heldin P. (1995) Biochem. J. 307, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Boom M., Sarbia M., von Wnuck Lipinski K., Mann P., Meyer-Kirchrath J., Rauch B. H., Grabitz K., Levkau B., Schrör K., Fischer J. W. (2006) Circ. Res. 98, 36–44 [DOI] [PubMed] [Google Scholar]
- 30.Ng K. F., Schwartz N. B. (1989) J. Biol. Chem. 264, 11776–11783 [PubMed] [Google Scholar]
- 31.Itano N., Kimata K. (1996) J. Biol. Chem. 271, 9875–9878 [DOI] [PubMed] [Google Scholar]
- 32.Spicer A. P. (2001) Methods Mol. Biol. 171, 373–382 [DOI] [PubMed] [Google Scholar]
- 33.Brinck J., Heldin P. (1999) Exp. Cell Res. 252, 342–351 [DOI] [PubMed] [Google Scholar]
- 34.Chao H., Spicer A. P. (2005) J. Biol. Chem. 280, 27513–27522 [DOI] [PubMed] [Google Scholar]
- 35.Tlapak-Simmons V. L., Baron C. A., Gotschall R., Haque D., Canfield W. M., Weigel P. H. (2005) J. Biol. Chem. 280, 13012–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karousou E. G., Militsopoulou M., Porta G., De Luca G., Hascall V. C., Passi A. (2004) Electrophoresis 25, 2919–2925 [DOI] [PubMed] [Google Scholar]
- 37.Karousou E. G., Porta G., De Luca G., Passi A. (2004) J. Pharm. Biomed. Anal. 34, 791–795 [DOI] [PubMed] [Google Scholar]
- 38.Karousou E. G., Viola M., Genasetti A., Vigetti D., Luca G. D., Karamanos N. K., Passi A. (2005) Biomed. Chromatogr. 19, 761–765 [DOI] [PubMed] [Google Scholar]
- 39.Mitropoulou T. N., Lamari F., Syrokou A., Hjerpe A., Karamanos N. K. (2001) Electrophoresis 22, 2458–2463 [DOI] [PubMed] [Google Scholar]
- 40.Evanko S. P., Wight T. N. (1999) J. Histochem. Cytochem. 47, 1331–1342 [DOI] [PubMed] [Google Scholar]
- 41.Furukawa K., Terayama H. (1977) Biochim. Biophys. Acta 499, 278–289 [DOI] [PubMed] [Google Scholar]
- 42.Furukawa K., Terayama H. (1979) Biochim. Biophys. Acta 585, 575–588 [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K., Sawasaki Y., Hata J., Mukai K., Goto T. (1990) In Vitro Cell Dev. Biol. 26, 265–274 [DOI] [PubMed] [Google Scholar]
- 44.Bonifacino J. S., Dasso M., Harford J. B., Lippincott-Schwartz J., Yamada K. M. (2004) in Short Protocols in Cell Biology: A Compendium of Methods from Current Protocols in Cell Biology, pp. 3.1–3.88, John Wiley & Sons, Inc., New York [Google Scholar]
- 45.Poole R. J., Briskin D. P., Krátký Z., Johnstone R. M. (1984) Plant Physiol. 74, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prinetti A., Chigorno V., Tettamanti G., Sonnino S. (2000) J. Biol. Chem. 275, 11658–11665 [DOI] [PubMed] [Google Scholar]
- 47.Vigetti D., Moretto P., Viola M., Genasetti A., Rizzi M., Karousou E., Pallotti F., De Luca G., Passi A. (2006) FASEB J. 20, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 48.Vigetti D., Rizzi M., Viola M., Karousou E., Genasetti A., Clerici M., Bartolini B., Hascall V. C., De Luca G., Passi A. (2009) Glycobiology 19, 537–546 [DOI] [PubMed] [Google Scholar]
- 49.Vigetti D., Viola M., Karousou E., Rizzi M., Moretto P., Genasetti A., Clerici M., Hascall V. C., De Luca G., Passi A. (2008) J. Biol. Chem. 283, 4448–4458 [DOI] [PubMed] [Google Scholar]
- 50.Carracedo A., Lorente M., Egia A., Blázquez C., García S., Giroux V., Malicet C., Villuendas R., Gironella M., González-Feria L., Piris M. A., Iovanna J. L., Guzmán M., Velasco G. (2006) Cancer Cell 9, 301–312 [DOI] [PubMed] [Google Scholar]
- 51.Müllegger J., Rustom A., Kreil G., Gerdes H. H., Lepperdinger G. (2003) Biol. Chem. 384, 175–182 [DOI] [PubMed] [Google Scholar]
- 52.Ren J., Hascall V. C., Wang A. (2009) J. Biol. Chem. 284, 16621–16632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown J., Reading S. J., Jones S., Fitchett C. J., Howl J., Martin A., Longland C. L., Michelangeli F., Dubrova Y. E., Brown C. A. (2000) Lab. Invest. 80, 37–45 [DOI] [PubMed] [Google Scholar]
- 54.Heldin P., Laurent T. C., Heldin C. H. (1989) Biochem. J. 258, 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rilla K., Pasonen-Seppänen S., Rieppo J., Tammi M., Tammi R. (2004) J. Invest. Dermatol. 123, 708–714 [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T., Takagaki K., Shibata S., Tanaka K., Higuchi T., Endo M. (1995) Biochem. Biophys. Res. Commun. 208, 470–475 [DOI] [PubMed] [Google Scholar]
- 57.Lee A. S. (2001) Trends Biochem. Sci. 26, 504–510 [DOI] [PubMed] [Google Scholar]
- 58.Ding K., Jonsson M., Mani K., Sandgren S., Belting M., Fransson L. A. (2001) J. Biol. Chem. 276, 3885–3894 [DOI] [PubMed] [Google Scholar]
- 59.Sudhakaran P. R., Prinz R., Von Figura K. (2007) J. Biosci. 4, 413–418 [Google Scholar]
- 60.Hamilton T. C., Young R. C., McKoy W. M., Grotzinger K. R., Green J. A., Chu E. W., Whang-Peng J., Rogan A. M., Green W. R., Ozols R. F. (1983) Cancer Res. 43, 5379–5389 [PubMed] [Google Scholar]
- 61.Klewes L., Prehm P. (1994) J. Cell. Physiol. 160, 539–544 [DOI] [PubMed] [Google Scholar]
- 62.Itano N., Sawai T., Atsumi F., Miyaishi O., Taniguchi S., Kannagi R., Hamaguchi M., Kimata K. (2004) J. Biol. Chem. 279, 18679–18687 [DOI] [PubMed] [Google Scholar]
- 63.Kyossev Z., Weigel P. H. (2007) Anal. Biochem. 371, 62–70 [DOI] [PubMed] [Google Scholar]
- 64.Rilla K., Siiskonen H., Spicer A. P., Hyttinen J. M., Tammi M. I., Tammi R. H. (2005) J. Biol. Chem. 280, 31890–31897 [DOI] [PubMed] [Google Scholar]
- 65.Tomida M., Koyama H., Ono T. (1977) J. Cell. Physiol. 91, 323–328 [DOI] [PubMed] [Google Scholar]
- 66.Lembach K. J. (1976) J. Cell. Physiol. 89, 277–288 [DOI] [PubMed] [Google Scholar]
- 67.Ohashi Y., Honda A., Iwai T., Mori Y. (1988) Biochem. Int. 16, 293–302 [PubMed] [Google Scholar]
- 68.Ullrich S. J., Hawkes S. P. (1983) Exp. Cell Res. 148, 377–386 [DOI] [PubMed] [Google Scholar]
- 69.Honda A., Iwai T., Mori Y. (1989) Biochim. Biophys. Acta 1014, 305–312 [DOI] [PubMed] [Google Scholar]
- 70.Honda A., Noguchi N., Takehara H., Ohashi Y., Asuwa N., Mori Y. (1991) J. Cell Sci. 98, 91–98 [DOI] [PubMed] [Google Scholar]
- 71.Kakizaki I., Kojima K., Takagaki K., Endo M., Kannagi R., Ito M., Maruo Y., Sato H., Yasuda T., Mita S., Kimata K., Itano N. (2004) J. Biol. Chem. 279, 33281–33289 [DOI] [PubMed] [Google Scholar]
- 72.Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 73.Selbi W., de la Motte C. A., Hascall V. C., Day A. J., Bowen T., Phillips A. O. (2006) Kidney Int. 70, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 74.Selbi W., Day A. J., Rugg M. S., Fülöp C., de la Motte C. A., Bowen T., Hascall V. C., Phillips A. O. (2006) J. Am. Soc. Nephrol. 17, 1553–1567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.