Abstract
Combinatorial libraries of designed ankyrin repeat proteins (DARPins) have been proven to be a valuable source of specific binding proteins, as they can be expressed at very high levels and are very stable. We report here the selection of DARPins directed against a macromolecular multiprotein complex, the baseplate BppU·BppL complex of the lactococcal phage TP901-1. Using ribosome display, we selected several DARPins that bound specifically to the tip of the receptor-binding protein (RBP, the BppL trimer). The three selected DARPins display high specificity and affinity in the low nanomolar range and bind with a stoichiometry of one DARPin per BppL trimer. The crystal structure of a DARPin complexed with the RBP was solved at 2.1 Å resolution. The DARPin·RBP interface is of the concave (DARPin)-convex (RBP) type, typical of other DARPin protein complexes and different from what is observed with a camelid VHH domain, which penetrates the phage p2 RBP inter-monomer interface. Finally, phage infection assays demonstrated that TP901-1 infection of Lactococcus lactis cells was inhibited by each of the three selected DARPins. This study provides proof of concept for the possible use of DARPins to circumvent viral infection. It also provides support for the use of DARPins in co-crystallization, due to their rigidity and their ability to provide multiple crystal contacts.
Lactococcus lactis is a Gram-positive bacterium widely used by the dairy industry for the production of an array of fermented milk products. Several industrial strains are sensitive to various distinct bacteriophages, mostly belonging to the Siphoviridae family. The lactococcal phage population is divided in at least 10 genetically distinct groups, of which the 936, c2, and P335 groups are prominent (1, 2). These L. lactis-infecting phages are considerably problematic in causing milk fermentation failures and resulting in decreased yields as well as low quality products (3). Preventing these infections has proven to be difficult because of lactococcal phage ubiquity, biodiversity, and genomic plasticity (4).
Phage infection is initiated by binding of the phage receptor-binding protein (RBP),5 located within the baseplate at the distal part of the tail, to its receptor on the host cell surface (5). We have previously solved the crystal structures of the three RBPs of the lactococcal phages p2 (936) (6), bIL170 (936) (7), TP901-1 (P335) (8), and their chimera (9) as well as characterized their saccharide binding sites (10). The RBPs of these phages have a similar homotrimeric architecture related by a 3-fold axis. They comprise three domains: the N terminus shoulder domain, the interlaced β-prism neck domain, and the jellyroll head domain at the C terminus. The head domain has a saccharide binding site likely involved in host recognition. The lactococcal phage TP901-1 contains a double-disk-shaped baseplate at the tip of its tail which is made of a lower baseplate protein (BppL) and an upper baseplate protein (BppU) (11).
One strategy to minimize bacteriophage infections is to competitively block phage adsorption by adding a protein that specifically binds to the phage RBP. A neutralizing llama VHH domain recognizing the head domain of the phage p2 RBP has been used to block L. lactis phage infection in milk fermentation (12). Lactococcal phages could readily escape neutralization by generating mutations interfering with VHH binding over the large interaction surface while keeping the central polysaccharide receptor binding pocket intact (10). Designed ankyrin repeat proteins (DARPins) may be another tool to neutralize viral infection, as they display distinct characteristics from VHHs and contain the required properties in terms of stability and facility of expression (13).
Ankyrin repeat proteins are found in virtually all phyla and mediate specific protein-protein interactions in all cell compartments (14). The ankyrin elementary module is composed of 33 amino acids structured as a β-turn followed by two antiparallel α-helices and a loop connected to the β-turn of the next repeat. The repeats are stacked in a rigid manner. In creating a DARPin library, residues in each repeat were subdivided in two groups; (i) randomized residues constituting potential target interaction points and (ii) framework residues, important for maintaining the ankyrin fold (13). Libraries with varying repeat numbers were assembled and named according to the constituent repeat number; N2C and N3C libraries were used in this study, with two and three internal repeats inserted between the N and C capping repeats, respectively. DARPins are a powerful alternative to the use of antibodies, notably because of their very high expression rates in Escherichia coli, their high stability paired with high affinity, and successful reports of their use in co-crystallization (15–19). Their architecture results in a very rigid structure that facilitates multiple crystal contacts and may promote crystal formation of the protein of interest by providing additional surfaces for such crystal contacts.
We report here the selection and analysis of DARPin binders directed against a macromolecular multiprotein ensemble, the TP901-1 baseplate BppU·BppL protein complex. Ribosome display selection, ELISA screening, and surface plasmon resonance (SPR) measurements allowed us to isolate and characterize three N2C DARPins that recognized the RBP (BppL of the BppU·BppL complex) with high specificity and affinity. Further studies showed that the three DARPins bound to a unique area of the RBP at the tip of the head domain. QELS, MALS, UV, and refractometry coupled online with a size exclusion chromatography (SEC) column allowed us to monitor complex formation in solution as well as to estimate DARPin binding stoichiometry. Crystals of one of these selected DARPins in complex with the RBP were obtained, and the x-ray structure was solved at 2.1 Å resolution. This constitutes the first structure of a DARPin complex originating from the N2C library and the highest resolution for a DARPin complex structure reported to date. Finally, phage adsorption inhibition experiments demonstrated that the three N2C DARPins strongly inhibited L. lactis infection by TP901-1. We describe the DARPin·RBP interface and compare it to other DARPin interfaces. We also compare it to the p2 RBP·VHH5 complex, a previously selected llama VHH domain inhibiting p2 phage adsorption (12), to highlight the different binding mode of these two types of binders.
EXPERIMENTAL PROCEDURES
Target Protein Preparation
BppU and BppL coding sequences from phage TP901-1 were amplified, cloned into a bicistronic operon, and expressed and purified as described elswhere,6 leading to two distinct fractions: the BppU·BppL complex and an excess of free BppL. The BppU coding sequence alone was also amplified, cloned, and expressed as described elswhere.6 The BppU·BppL complex was biotinylated using Sulfo-NHS-SS-Biotin (Pierce). First, the protein buffer was exchanged to 50 mm K2HPO4/KH2PO4, pH 7.8 (4 °C), 100 mm NaCl with the aid of a NAP5 column (GE Healthcare). The biotin reagent was mixed with the protein in a 40:1 molar ratio and incubated for 4 h on ice. The protein buffer was then exchanged to a TBS buffer (50 mm Tris, pH 7.6 (4 °C), 150 mm NaCl) using a NAP5 column and extensive dialysis. The resulting complex is termed (BppU·BppL)biot.
Ribosome Display
Both N2C and N3C DARPin libraries (13) were used to select for proteins binding to the BppU·BppL complex using three rounds of ribosome display on plates coated alternately with neutravidin or streptavidin (20, 21). Decreasing coating concentrations of (BppU·BppL)biot were used in successive rounds: 68, 20, and 10 nm, respectively. The DARPin libraries were incubated with the target for 1 h at 4 °C. Six washes were performed at each round with a buffer containing 50 mm Tris acetate pH 7.6 (4 °C), 100 mm NaCl, 50 mm magnesium acetate, and 0.05% Tween 20. Total wash times were increased from 6 to 45 to 60 min in successive rounds. In the third round, the translation mix containing the ternary mRNA-ribosome-DARPin complexes was first pre-panned for 30 min at 4 °C in one identical well without the biotinylated target. After washing, mRNAs were eluted twice with 100 μl of elution buffer (50 mm Tris acetate, pH 7.6 (4 °C), 100 mm NaCl, 25 mm EDTA). The number of PCR cycles after reverse transcription was reduced in successive rounds from 40 to 30 to 25, adjusting to the yield due to progressive enrichment of binders in each round.
Binder Identification
MaxiSorp plates (Nunc) were coated at 4 °C overnight with 100 μl of 66 nm neutravidin in TBS, blocked with TBSTB (TBST supplemented with 0.2% bovine serum albumin) for 1 h at room temperature, and 100 μl of (BppU·BppL)biot was added at a final concentration of 10 nm. Single clones from each of the two DARPin libraries (N2C and N3C) were screened by crude extract ELISA as previously described (22). A 5-μl volume of cell lysate was mixed with 95 μl of TBSTB and added directly to the target-containing wells or to a control well without immobilized target and incubated for 1 h at 4 °C with orbital shaking. After washing with TBST, the primary anti-RGS(H)4 antibody (Qiagen catalog no. 34650, 1:2000 dilution in TBSTB) was added and incubated for 1 h at 4 °C. The wells were washed with TBST (TBS supplemented with 0.05% Tween 20), and the secondary goat-anti-mouse-IgG-AP-conjugate antibody (Pierce catalog no. A3562, 1:10,000 dilution in TBSTB) was added. Binding was detected using 3 mm disodium 4-nitrophenyl phosphate (Fluka) in a buffer containing 50 mm NaHCO3 and 50 mm MgCl2. Absorbance at 430 nm was measured using a Genios plus plate reader (Tecan) after 12 h of incubation at room temperature. We used a competition ELISA setup to confirm that the binding of the three selected DARPins to immobilized (BppU·BppL)biot could be inhibited by the free BppU·BppL complex in solution. The protocol was the same as for the crude extract ELISA, except that crude extracts were replaced by 100 μl of 150 nm purified DARPins. In addition, before adding individual DARPins into each (BppU·BppL)biot-coated well, the appropriate DARPin was incubated for 1 h at 4 °C with several concentrations of free non-biotinylated target.
DARPins Purification and Complex Preparation
Enriched DARPins from the N2C and N3C libraries were cloned into pDST067, a modified version of the pQE30 vector (Qiagen) (22). This vector introduced a MRGSH6 tag at the N terminus. DARPins were expressed in E. coli XL1-Blue grown in 2×YT medium. Cells were grown to an A600 of 0.4, and DARPin expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. Cells were resuspended in lysis buffer (50 mm Tris, pH 8.0, 300 mm NaCl, 10 mm imidazole, 1 mm EDTA, 0.5 mg/ml lysozyme, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Complete EDTA-free anti-proteases, Roche Applied Science)), and stored at −80 °C. After thawing, DNase I and MgSO4 were added to final concentrations of 10 μg/ml and 20 mm, respectively. Cells were lysed by sonication and centrifuged at 20000 × g for 30 min. Purification was performed on an Äkta system using a Ni2+ column (HisTrap Ni2+ 5 ml, GE Healthcare). Pure proteins were mixed in a 5:1 DARPin·BppU·BppL or DARPin/BppL molar ratio and incubated for 1 h at room temperature. The complexes were purified by a SEC step using a Superose 6 16/60 or a Sephacryl S100 26/60 column (GE Healthcare), respectively, and run in a buffer containing 10 mm Hepes, pH 7.5, 150 mm NaCl. Purified complexes were then concentrated up to 5–7 mg/ml with an Amicon Ultra centrifugal filter device with a molecular mass cutoff of 30 kDa (Millipore). For BIAcore and phage adsorption inhibition experiments, the three selected DARPins were each further purified by a SEC polishing step using a Sephacryl S100 26/60 column and run in the same buffer as for complex purification.
SPR Measurements
Measurements were performed in 10 mm Hepes, pH 7.5, 150 mm NaCl, 3 mm EDTA, 0.005% detergent P-20 at 20 °C using a BIAcore X100 (BIAcore). We first coated a CM5 chip (BIAcore) with 100 response units of one of the three selected DARPins. We also used the inverse set-up, coating a CM5 chip with either 200 response units of BppL or 1000 response units of BppU·BppL. The signal from an uncoated reference cell as well as the buffer response was subtracted from all measurements. Initial binding assays were performed with the BppU·BppL complex, BppU alone, and BppL alone to estimate specificity, kinetic parameters, and/or affinities as well as to map DARPin binding epitopes. Final measurements were performed using single cycle kinetic assays to precisely characterize the binding of each of the three DARPins at 10, 5, 2.5, 1.25, and 0.625 nm concentrations (analytes) to either the BppU·BppL complex or BppL alone (ligand). The KD, kon and koff values were obtained using the fitting tool of the BIAevaluation software (BIAcore). A 1:1 binding model was assumed in all cases.
Stoichiometry Measurements
We characterized the mass of each DARPin-target complex using a combination of UV spectrophotometry, MALS, and refractometry coupled online with an analytical SEC column. For DARPin·BppU·BppL complexes, we also determined hydrodynamic radii using an online QELS. UV, MALS, QELS, and refractometry measurements were achieved with a Photo Diode Array 2996 (Waters), a MiniDawn Treos (Wyatt technology), a DynaPro (Wyatt technology), and an Optilab rEX (Wyatt technology), respectively (23). We used either a 24-ml Superose 6 10/30 column (GE Healthcare) run at 0.35 ml·min−1 or a 15-ml KW-804 column (Shodex) run at 0.5 ml·min−1 on an Alliance HPLC 2695 system (Waters). The buffer was 10 mm Hepes, pH 7.5, 150 mm NaCl, 0.02% NaN3. We injected either 100 μl (Superose 6) or 30 μl (KW-804) of each complex sample at concentrations of 5–7 mg/ml.
Crystallization
We used 5 mg/ml DARPin 20·BppL complex for crystallization experiments (the UV extinction coefficient of the DARPin was not included in the determination of concentration because of its very low value). Protein buffer was exchanged by dialysis to 10 mm Tris, pH 7.5, 100 mm NaCl. An initial hit was obtained in condition 10 of the Wizard screen (Emerald Biosystem) performed in 96-well Greiner plates (24, 25). This condition was optimized by varying pH and precipitant concentration as well as by adding a second buffer to the crystallization solution. Finally, the crystal exploited for data collection was grown within 1 week from a solution containing 40 mm Tris, 20 mm Bistris propane, pH 6.0, 25% polyethylene glycol 2000 monomethyl ether. The crystal was cryoprotected with the mother liquor supplemented with 10% glycerol and immediately flash-frozen under a stream of nitrogen.
Data Collection, Structure Determination, and Refinement
The data set used for structure determination was collected at beamline ID14–4 (European Synchrotron Radiation Facility, Grenoble, France). The transmission was set to 20%, and each frame was collected with an exposure time of 1 s and 1° oscillation using an ADSC Quantum Q315r detector. Ninety images were integrated and scaled using XDS and XSCALE (26). The structure was solved by molecular replacement using AmoRe software (27) with TP901-1 RBP and H10-2-G3 N2C DARPin structures as search models (Protein Data Bank entry 2F0C and 2JAB, respectively). Refinement was carried out using phenix.refine (28) alternating with manual building in Coot (29). TLS groups were generated with the TLS Motion Determination server (30). Data processing and refinement statistics as well as final model geometry evaluation are reported in Table 1. Figs. 3 and 4 were generated with Pymol. The coordinates have been deposited at the Protein Data Bank with the code 3HG0. The DARPin 20·RBP interface was analyzed using the Protein Interfaces, Surfaces, and Assemblies (PISA) server (31) as well as with the protein-protein interaction (ProtorP) server (32).
TABLE 1.
Summary of data collection and refinement statistics
| Data collection | |
| Space group | P212121 |
| Cell dimensions (Å) | 46.650, 80.440, 182.870 |
| Resolution (Å) | 19.89-2.1 (2.3–2.1) |
| Completeness (%) | 97.1 (91.8) |
| Redundancy | 3.58 (3.67) |
| I/σ(I) | 12.43 (2.48) |
| Rmerge (%) | 6.5 (51.8) |
| Refinement | |
| Resolution (Å) | 19.89-2.1 (2.3–2.1) |
| Protein/solvent atoms | 3933/446 |
| R/Rfree (%) | 20.79/24.27 |
| Root mean square deviations on bonds (Å)/angles (degrees) | 0.014/1.676 |
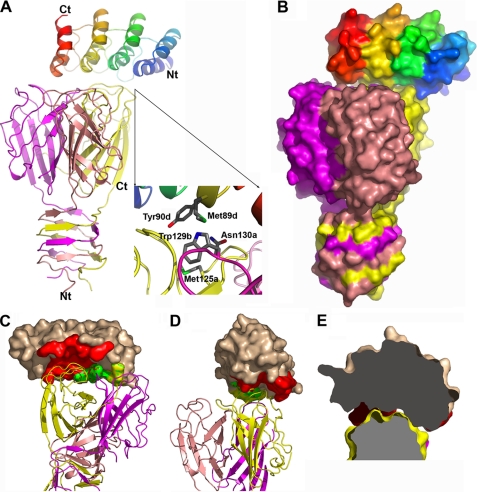
FIGURE 3.
Structure of the RBP·DARPin 20 complex. A, shown is the overall structure with both DARPin (rainbow-colored) and the RBP (the three chains colored in salmon, purple, and yellow) in schematic representation. The inset shows the detail of the interactions involving a central role of Tyr-90 from the DARPin; the small letters refer to DARPin (d) and the a, b, c chains of the RBP. B, shown is the overall structure of the two partners with molecular surface representation according to the same color scheme as in A. C, shown is the interface region with the buried surface area of the DARPin colored red, green, and yellow corresponding to interaction with chain A, B, and C of the RBP, respectively. D, shown is the same view as in C rotated approximately by 90°. E, shown is an illustration of the concave (DARPin)-convex (RBP) type of the interaction interface. Nt, N terminus; Ct, C terminus.
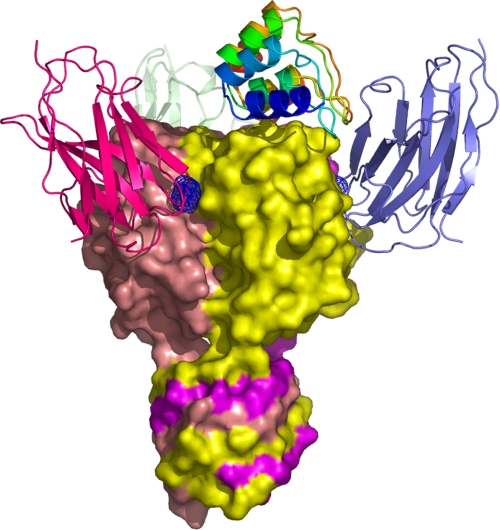
FIGURE 4.
Comparison of the phage TP901-1 RBP·DARPin 20 complex with the phage p2 RBP·VHH5 complex (6). The three VHH5 have been superimposed on the TP901-1 RBP structure for comparison, taking advantage of the three-dimensional similarity of the two RBPs. The blue grid locates the bound glycerol in the receptor binding site (10). Each VHH5 binds in a crevice between two subunits. The structure of the three VHHs was taken from the Protein Data Bank entry 2BSE.
Phage Inhibition Assay
First, L. lactis strains were grown at 30 °C in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17). For phage induction, L. lactis TP901-1 was grown to an A600 of 0.1, and mitomycin C was added to a final concentration of 1 μg/ml. After 20 h, the clear lysate was filtered (0.45 μm), and the phage TP901-1 titer was determined as follows. Ten μl of lysate were added to 3 ml of GM17 Top Agar (0.75% agar) containing 300 μl of an overnight culture of L. lactis 3107 (indicator host strain). The mixture was then poured onto a GM17 plate (1% agar) supplemented with 0.5% glycine and incubated overnight at 30 °C, and the plaques were counted. The protocol for the phage inhibition assay was adapted from the phage inactivation experiments described elsewhere (33, 34). Approximately 700 plaque-forming units of lactococcal phage TP901-1 (10 μl) were mixed with 10 μl of DARPin (1, 0.5, 0.1, 0.05, 0.01, 0.005 mg/ml) or buffer (10 mm Hepes, pH 7.5, 150 mm NaCl). After an incubation of 1 h at 30 °C, 20 μl of M17 was added to the mixture. Then the phage titer was determined in triplicate using 10 μl of the mixture. The percentage of inhibition was calculated by dividing the phage titer with DARPin by the phage titer in the buffer (no DARPin). The quotient was subtracted from 1 and multiplied by 100. The experiment was repeated three times.
RESULTS
Ribosome Display
Ribosome display selections were performed on (BppU·BppL)biot immobilized alternately via neutravidin or streptavidin coated on microtiter plates. Two DARPin libraries were used for the selection, N2C and N3C, with theoretical diversities of 5.2 × 1015 and 3.8 × 1023, respectively (13). We estimated that the initial DNA library diversity used for ribosome display was about 1012 individual members for each of the two libraries. We observed a strong enrichment for the N2C DARPin library after the third round of ribosome display. The enrichment for the N3C DARPin library was much less pronounced, probably because binder enrichment was slower on this target for this library format. Selected DARPins from each library were then cloned into the pDST067 vector (22) to screen single clones.
ELISA Screening
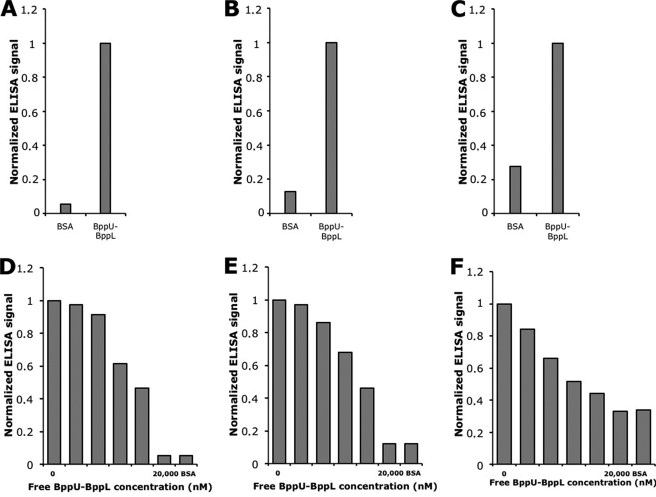
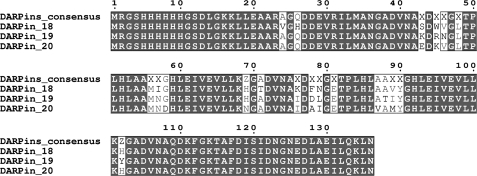
Crude extract ELISAs allowed us to identify three N2C DARPins binding to the immobilized BppU·BppL complex (Fig. 1, A–C) but no N3C DARPin. The three N2C DARPins were termed DARPin 18, DARPin 19, and DARPin 20. After sequence analysis (Fig. 2), the clones were ranked as independent, because many randomized positions were different for the three sequences and because they exhibited different framework mutations (average number of 2, consistent with previous observations (18)). We then performed a competition ELISA in which purified DARPins were preincubated with various concentrations of the free non-biotinylated target before adding the mixture to target-coated wells. We clearly observed that the binding of the three selected DARPins to the immobilized BppU·BppL complex was inhibited by the addition of the free target and that this inhibition was concentration-dependent (Fig. 1, D–F). This demonstrated that DARPins 18, 19, and 20 bound specifically to the BppU·BppL complex, as a well coated with only neutravidin and bovine serum albumin produced a low signal compared with target-coated wells, and that they bind BppU·BppL complex in solution.
FIGURE 1.
Top, single clone crude extract ELISAs. A, DARPin 18. B, DARPin 19. C, DARPin 20. DARPin binding was tested on a well with neutravidin-immobilized (BppU·BppL)biot and compared with the signal intensity obtained from a control well with only neutravidin and bovine serum albumin (BSA). Bottom, competition ELISAs. DARPins 18 (D), 19 (E), and 20 (F) were preincubated with increasing concentrations of free non-biotinylated BppU·BppL complex for 1 h at 4 °C before the addition to their respective ELISA wells.
FIGURE 2.
Sequence alignment of DARPins 18, 19, and 20. The DARPin consensus is displayed with X, indicating randomized positions (any amino acid residues except glycine, proline, and cysteine) and Z for the partially randomized positions (allowing histidine, tyrosine, or asparagine); all remaining positions are framework residues (13). This figure was created with Multalin (44).
SPR Kinetic/Affinity Parameters Determination and Epitope Mapping Studies of DARPin Binding
We measured the kinetic parameters and calculated the affinity of DARPins 18, 19, and 20 for the BppU·BppL protein complex as well as for the isolated RBP (BppL trimer) by SPR. Single cycle kinetic experiments yielded very similar values for kon and koff, and, thus, KD, for a given DARPin when comparing the binding to BppU·BppL and BppL. This observation was true for all three DARPins tested (Table 2). Affinities of the three DARPins for BppU·BppL and BppL were in the low nanomolar range, 3.3–22 nm, for the former and 3.5–16 nm for the latter. These results indicated that the three selected DARPins recognized only the RBP in the BppU·BppL complex despite their sequence divergence. Moreover, as expected from these results, no binding was detected for the BppU protein alone (data not shown). Competition experiments between the three different DARPins for binding to BppU·BppL revealed that each selected DARPin inhibited the binding of the other two to the target (data not shown). This led us to conclude that the epitopes of the three selected DARPins were overlapping or identical. We, thus, obtained high affinity (low nanomolar range) DARPins that recognized the RBP component of the BppU·BppL complex. This is the first report of the selection and characterization of DARPins directed against a macromolecular multiprotein complex.
TABLE 2.
DARPins 18, 19, and 20 affinities and kinetic parameters for BppU·BppL and BppL, determined by SPR (BIAcore)
| Proteins | BppU·BppL/BppL kon | BppU·BppL/BppL koff | BppU·BppL/BppL KD | χ2 |
|---|---|---|---|---|
| 1/Ms | 1/s | m | ||
| DARPin 18 | 2.63 × 106/2.40 × 106 | 1.60 × 10−2/1.80 × 10−2 | 6.00 × 10−9/7.47 × 10−9 | 0.295/0.124 |
| DARPin 19 | 1.40 × 107/1.70 × 107 | 5.80 × 10−2/5.90 × 10−2 | 3.30 × 10−9/3.50 × 10−9 | 2.41/0.004 |
| DARPin 20 | 4.10 × 106/N.D.a | 9.00 × 10−2/N.D. | 2.20 × 10−8/1.60 × 10−8 | 0.199/0.002 |
N.D., not determined.
Stoichiometry Studies of DARPin Binding
We investigated complex formation and stoichiometry on mixing of each of the DARPins with either BppU·BppL or BppL in solution using MALS/QELS/UV/refractometry coupled online with a SEC column (23). The addition of any DARPin to either target resulted in a shift of the elution peak to a lower retention volume, indicating that complex formation had occurred. Our results also indicated that the addition of DARPins to the BppU·BppL complex resulted in a 5% increase of the hydrodynamic radii of the eluted species (Tables 3 and 4). Finally, we found that all three DARPins could bind to the BppU·BppL complex in solution, and that one DARPin could bind per RBP independent of whether the RBP was alone or complexed to BppU (Tables 3 and 4). Thus, complex formation of BppL with BppU does not influence DARPin binding stoichiometry. This stoichiometry suggests that the DARPin binds close to the 3-fold axis to inhibit the simultaneous binding of other DARPins to the same epitope in the other subunits.
TABLE 3.
Stoichiometry of BppU·BppL·DARPin complexes
| Proteins | Theoreticalmolecular weight | Molecular weight measuredby MALS/refractometry/SEC | Stoichiometry(BppU3-BppL9):DARPins | (BppU3-BppL9)/DARPin R1h QELS |
|---|---|---|---|---|
| Da | Da | nm | ||
| (BppU3-BppL9) | 263,640 (9 × 17,970 + 3 × 33,970) | 255,000 | 6.0 | |
| (BppU3-BppL9) + DARPin 18 | 308,025 (263,640 + 3 × 14,795) | 291,000 | 1:3 | 6.6 |
| (BppU3-BppL9) + DARPin 19 | 307,971 (263,640 + 3 × 14,777) | 289,000 | 1:3 | 6.4 |
| (BppU3-BppL9) + DARPin 20 | 307,788 (263,640 + 3 × 14,716) | 285,000 | 1:3 | 6.4 |
TABLE 4.
Stoichiometry of BppL·DARPin complexes
| Proteins | Theoreticalmolecular weight | Molecular weightmeasured by MALS/refractometry/SEC | StoichiometryBppL3:DARPins |
|---|---|---|---|
| Da | Da | ||
| BppL3 | 53,910 (3 × 17 970) | 54,000 | |
| BppL3 + DARPin 18 | 53,910 + 14,795 | 63,627 | 1:1 |
| BppL3 + DARPin 19 | 53,910 + 14,777 | 60,370 | 1:1 |
| BppL3 + DARPin 20 | 53,910 + 14,716 | 61,618 | 1:1 |
Structure of the RBP in Complex with DARPin 20
An x-ray structure of DARPin 20 bound to the RBP was solved at a resolution of 2.1 Å (Fig. 3, A and B). This is the first reported structure of a N2C DARPin complexed with its target. The overall fold of the RBP is unchanged relative to the previously reported isolated RBP structure (8), with root mean square deviation values for the Cα atoms ranging between 0.381 and 0.429 Å for the three chains relative to the corresponding chain in the original structure. Nevertheless, a major difference is visible in the crystal structure described here; no electron density was observed before residue 32 (in the best case, chain C), suggesting that the RBP was proteolyzed and, thus, lacks the three-helix-bundle domain. Such a phenomenon was previously observed in the phage TP901-1 RBP structure lacking the first 16 residues (8) as well as in the phage p2 RBP structure in complex with VHH5, in which only the three-β-barrel domain of the RBP was found in complex with its antibody partner (6).
DARPin 20 binds to the top of the head domain at a ratio of 1:1 DARPin·RBP; that is, in agreement with the stoichiometry determined in solution. Crystal contacts are mediated both by the DARPin and the RBP. In the crystal lattice, each DARPin interacts with two RBPs in addition to the target RBP, whereas each RBP contacts four other RBPs and three DARPins in addition to its binding partner. Thus, DARPin 20 contributes extensively to the crystal packing, bearing out the promise from the extended rigid scaffold and underlining its utility in co-crystallization.
Analysis of the Interaction between the RBP and DARPin 20
DARPin 20 interacts with a convex surface formed by the three chains of the RBP via its own concave randomized surface (Fig. 3, C–E). However, most of the RBP surface area buried in the interface is contributed by chain A, reaching 512 Å2, whereas the value for the whole RBP is 737.3 Å2. All four DARPin 20 repeats interact with the RBP, i.e. the two randomized repeats and the two capping repeats, resulting in a buried surface area of 665.6 Å2. The first α-helix of each of the four repeats as well as the three β-turns separating the four repeats carry all the target-interacting residues. The interaction surface of the RBP is formed by residues located in the loops connecting the strands of the three β-barrels of the head domain. The interface between the two partners involves 20 residues from the DARPin and 20 residues from the RBP, resulting in a total interface buried surface area of 1402.9 Å2. Residues involved in the interaction interface are listed in Table 5. Among the 14 randomized positions of DARPin 20, 9 are involved in the interface, illustrating the robustness of the library design. The 11 remaining residues participating in the interaction with the RBP are conserved framework residues. None of the two introduced framework mutations directly contribute to the binding interface or influence the overall ankyrin fold.
TABLE 5.
Residues involved in the interaction interface between DARPin 20 and the RBP
| Chain | Residue | Chain | Residue |
|---|---|---|---|
| D | Glu-20 | A | Glu-93 |
| D | Arg-23 | A | Ile-94 |
| D | Asp-44 | A | Ser-95 |
| D | Val-46 | A | Ser-96 |
| D | Leu-48 | A | Ser-97 |
| D | Leu-53 | A | Leu-99 |
| D | Met-56 | A | Ala-100 |
| D | Asn-57 | A | Asn-101 |
| D | Asp-77 | A | Asn-130 |
| D | Ala-78 | A | Pro-147 |
| D | Ile-79 | A | Thr-148 |
| D | Glu-81 | A | Ala-149 |
| D | Leu-86 | A | Ser-150 |
| D | Met-89 | A | Ser-151 |
| D | Tyr-90 | B | Gly-127 |
| D | Phe-112 | B | Gly-128 |
| D | Lys-114 | B | Trp-129 |
| D | Asp-122 | B | Asn-130 |
| D | Asn-123 | B | Ser-151 |
| D | Gly-124 | C | Trp-129 |
The involvement of the three RBP protomers in forming the DARPin epitope explains the observed stoichiometry, illustrating that it would be impossible to bind a second DARPin to the same RBP. Hydrogen bonds and van der Waals contacts mediate the binding of DARPin 20 to the RBP, and no salt bridge is observed. Of particular interest is the unusual hydrogen bond formed between residues Tyr-90 of DARPin 20, a selected residue in a randomized position, and Trp-129 of RBP monomer B. The N-H group of the indole ring from Trp-129 interacts with the π electronic cloud of the phenol ring from Tyr-90 of DARPin 20 (Fig. 3A, inset). On one side, the Trp-129 side chain is maintained in a defined orientation by a stacking to residue Asn-130 and van der Waals contacts to Met-125, both provided by chain A of the RBP. On the other side, the Trp-129 side chain interacts with residues Trp-144 and Pro-147 from chain A as well as Met-89 from DARPin 20, still via van der Waals contacts. It should be noted that the latter residue is also located at a randomized position and was selected.
The convex-shaped epitope on the RBP fits perfectly with the concave-shaped paratope on the DARPin (Fig. 3, C–E), which is a general feature of DARPin interaction and is reminiscent of all other DARPin-target complex structures. The main part of the interaction interface (61% of the DARPin 20 interface buried surface area) is mediated through residues in randomized positions, explaining the high specificity of the selected molecules. In contrast to most DARPin-target complexes (with the exception of the AcrBDARPin 110818 complex), the C-terminal capping repeat is fully ordered and involved in the interaction surface in contacting RBP chain C. Because the first interacting residue is at position 20 and the last at position 124, the full width of the 136-amino acid residues of DARPin 20 is involved in this complex.
Neutralization of Phage TP901-1 by DARPins 18, 19, and 20
Finally, the binding of the three selected DARPins was tested against the whole phage TP901-1. All three DARPins inhibited in a concentration-dependent manner the infection of L. lactis cells by lactococcal phage TP901-1 (Table 6). Although all three DARPins were similarly effective, DARPin 19 was the most efficient inhibitor, whereas DARPin 20 was the least, reflecting measured affinities. Structural analysis of the DARPin 20 bound to the RBP and the location of the putative interaction site of polysaccharide with the RBP suggested that the inhibition was probably because of steric hindrance. Indeed, DARPin 20 did not directly protrude into the polysaccharide binding site but was slightly more distant. Nevertheless, the observed inhibition probably resulted from the fact that DARPin 20 binding to TP901-1 phage precluded its interaction with polysaccharides located on the surface of the Gram-positive bacterium, thus preventing the phage from anchoring to its host. Furthermore, the similar behavior of DARPins 18, 19, and 20 in phage infection inhibition experiments reinforces our hypothesis that they share a common epitope on the RBP.
TABLE 6.
Inhibition of lactococcal phage TP901-1 by DARPins
| DARPin concentration | DARPin 18 | DARPin 19 | DARPin 20 |
|---|---|---|---|
| mg/ml | % | % | % |
| 1 | 100.0 ± 0.0 | 100.0 ± 0.0 | 87.6 ± 10.8 |
| 0.5 | 99.6 ± 0.40 | 100.0 ± 0.0 | 75.9 ± 22.4 |
| 0.1 | 91.2 ± 7.8 | 95.8 ± 1.9 | 60.7 ± 17.2 |
| 0.05 | 69.1 ± 9.0 | 79.7 ± 12.1 | 61.2 ± 10.6 |
| 0.01 | 13.2 ± 8.5 | 31.2 ± 8.8 | 23.6 ± 20.8 |
| 0.005 | 4.5 ± 7.7 | 39.0 ± 10.2 | 5.8 ± 18.5 |
DISCUSSION
When we analyzed the crystal structure of the DARPin 20·RBP complex, we observed that most of the DARPin 20-interacting residues were conserved in DARPins 18 and 19. This information along with the results of the SPR competition experiments and the phage infection inhibition experiments allowed us to reasonably suggest that the three DARPins probably share identical epitopes on the RBP. Furthermore, the three selected DARPins exhibited a conserved Tyr residue at the randomized position 90. Thus, the N-H—π interaction between the Tyr-90 residue and Trp-129 of the RBP chain B seems to be conserved.
Comparison with previously obtained DARPins, for which co-crystallization in complex with a target was successful and led to structure determination, revealed two interesting features; (i) the measured binding affinities of the three selected N2C DARPins for the BppU·BppL complex or for the isolated RBP fell in the range of those previously reported for N3C DARPins (35); (ii) the overall buried surface area of the DARPin 20·RBP interface is also comparable with previously reported surface areas, but it is among the smallest (35). This value is also roughly identical to those reported for antibody-antigen complexes.
In contrast to DARPin 20, the structure of the lactococcal phage p2 RBP in complex with a llama antibody fragment (VHH5) revealed that VHH5 binds the RBP at the interface between two β-barrels (Fig. 4). This implied that three VHH5 proteins are bound to each RBP, because of its internal molecular symmetry, in contrast to the unique DARPin 20 interacting with one RBP at the top of the head domain. Each VHH5, thus, interacts with two RBP chains, whereas DARPin 20 contacts three RBP chains. However, the surface area buried in the interface is roughly comparable with DARPin 20; VHH and p2 RBP averaged buried surface areas are of 699.2 and 667.2 Å2, respectively. This yielded a total value of 1366.4 Å2, very similar to the DARPin 20/TP901-1 RBP interface. Furthermore, the reported affinity of VHH5 for lactococcal phage p2 RBP (1.4 nm) (36) is similar to the measured affinities of DARPins 18, 19, and 20 for the lactococcal phage TP901-1 RBP. The protruding paratope of VHH5 penetrates into a crevice-shaped epitope located between two RBP protomers. Both VHH5 and p2 RBP contribute to the interface with 20 residues each, mediating hydrogen bonds and Van der Waals contacts, as for the complex described in this study.
These two RBP binding molecules (VHH and DARPins), whose specificities are illustrated by the two complex structures, rely on very different scaffolds. VHHs have an immunoglobulin domain scaffold displaying three complementary determining region loops to achieve target binding (37–39). In contrast, DARPins combine both an extended flat surface (formed by the first α-helix of each repeat) and turns to bind their targets. VHH5 and DARPin 20 employ opposite interaction modes with their targets, the convex VHH5 interacting with a concave target structure, whereas DARPin 20 displays a concave surface that binds a convex epitope. DARPin binding mode is more rare and constitutes an elegant alternative approach to the antibody binding mode. Because almost all proteins contain domains that will present a convex surface, DARPin binding appears to be of a general nature.
The problems caused by phage infections in industrial dairy processes were identified and characterized many years ago (3), and ever since research has been directed at finding ways to circumvent these problems. One solution proposed was to add a neutralizing antibody fragment to inhibit the first event of phage infection; that is, RBP interaction with the host cell wall. A successful illustration of this principle was provided by the selection of the VHH5 antibody, which blocked adsorption of the p2 phage to its host (12, 36).
Given the non-optimized DARPin 20 expression yield in E. coli (185 mg/liter of shake flask culture), it is reasonable to believe that efforts to optimize this yield for industrial purposes would easily provide very large quantities of purified DARPin. The convenience of DARPin purification, providing pure protein with a single IMAC step, is of great benefit in applying such proteins as scalable and robust industrial tools.
Before looking at industrial applications and the government regulatory process, a number of additional experiments is needed to support this proof of concept study, for example, with regard to the broadness of the protection provided by the DARPin binders isolated in this study against other phages of the P335 group, which is a notoriously diverse group of phages (40). Moreover, other phage-neutralizing DARPin binders would have to be developed against other predominant lactococcal phage groups (936 and c2), which are genetically distinct from the P335 group (41). As lactococcal phages are known to mutate either by point mutation or recombination when facing selective pressure (42), it has to be demonstrated if these phages can easily mutate to circumvent the inhibition provided by DARPin binders. It is worth mentioning, as suggested previously (12), that a mutation in the gene encoding the phage structural protein recognized by the DARPin binders may lead to a change in the host range and may also prevent the adsorption of the mutated phage to its natural host.
CONCLUSIONS
We report here, for the first time, the selection of DARPins directed against a hetero-oligomeric macromolecular multiprotein complex; that is, the BppU·BppL complex, which comprises part of the baseplate of the L. lactis-infecting phage TP901-1. The crystal structure of one of the selected high affinity DARPins in complex with the TP901-1 RBP, determined at 2.1 Å resolution, showed that the concave DARPin binding site interacts with a convex surface of the RBP, such that one DARPin binds one RBP. This stoichiometry is consistent with measurements in solution. The observed DARPin binding mode differs significantly from that of a camelid VHH (VHH5) interacting with a concave RBP surface (6). DARPins and VHHs provide two classes of binders with opposite characteristics, thus allowing a large coverage of antigens surface. We also demonstrated that the three selected DARPins inhibited TP901-1 infection. It should be noted that we obtained low nanomolar DARPins with only three rounds of ribosome display. Two additional rounds with error prone PCR and selection for affinity (43) would probably result in the selection of picomolar DARPins, further reducing the required quantity of DARPin necessary to phage neutralization. Considering the high expression yield of DARPin 20 in E. coli as well as its low nanomolar affinity for the RBP, we believe that we have provided a proof of concept for the use of DARPins to circumvent phage infection. The use of DARPins as a tool to fight viral infections in general, including those affecting humans, might also be considered.
Acknowledgment
We thank Barbara-An Conway for editorial assistance.
This work was supported, in part, by grants from the Marseille-Nice Génopole (to J. L.), the CNRS, the Natural Sciences and Engineering Research Council of Canada through its strategic grogram (to S. M.), and by Agence Nationale de la Recherche (ANR-07-BLAN-0095).
The atomic coordinates and structure factors (code 3HG0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
V. Campanacci, J. Lichière, S. Blangy, G. Sciara, D. Veesler, S. Moineau, D. Van Sinderen, P. Bron, and C. Cambillau, manuscript in preparation.
- RBP
- receptor-binding protein
- ELISA
- enzyme-linked immunosorbent assay
- MALS
- multi-angle static light scattering
- QELS
- quasi-elastic light scattering
- SEC
- size-exclusion chromatography
- TBS
- Tris-buffered saline
- SPR
- surface plasmon resonance
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- DARPin
- designed ankyrin repeat protein.
REFERENCES
- 1.Moineau S., Pandian S., Klaenhammer T. R. (1993) Appl. Environ. Microbiol. 59, 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissonnette F., Labrie S., Deveau H., Lamoureux M., Moineau S. (2000) J. Dairy Sci. 83, 620–627 [DOI] [PubMed] [Google Scholar]
- 3.Moineau S. (1999) Antonie van Leeuwenhoek 76, 377–382 [PubMed] [Google Scholar]
- 4.Sturino J. M., Klaenhammer T. R. (2006) Nat. Rev. Microbiol. 4, 395–404 [DOI] [PubMed] [Google Scholar]
- 5.Dupont K., Vogensen F. K., Neve H., Bresciani J., Josephsen J. (2004) Appl. Environ. Microbiol. 70, 5818–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spinelli S., Desmyter A., Verrips C. T., de Haard H. J., Moineau S., Cambillau C. (2006) Nat. Struct. Mol. Biol. 13, 85–89 [DOI] [PubMed] [Google Scholar]
- 7.Ricagno S., Campanacci V., Blangy S., Spinelli S., Tremblay D., Moineau S., Tegoni M., Cambillau C. (2006) J. Virol. 80, 9331–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spinelli S., Campanacci V., Blangy S., Moineau S., Tegoni M., Cambillau C. (2006) J. Biol. Chem. 281, 14256–14262 [DOI] [PubMed] [Google Scholar]
- 9.Siponen M., Spinelli S., Blangy S., Moineau S., Cambillau C., Campanacci V. (2009) J. Bacteriol. 191, 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremblay D. M., Tegoni M., Spinelli S., Campanacci V., Blangy S., Huyghe C., Desmyter A., Labrie S., Moineau S., Cambillau C. (2006) J. Bacteriol. 188, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vegge C. S., Vogensen F. K., Mc Grath S., Neve H., van Sinderen D., Brøndsted L. (2006) J. Bacteriol. 188, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ledeboer A. M., Bezemer S., de Hiaard J. J., Schaffers I. M., Verrips C. T., van Vliet C., Düsterhöft E. M., Zoon P., Moineau S., Frenken L. G. (2002) J. Dairy Sci. 85, 1376–1382 [DOI] [PubMed] [Google Scholar]
- 13.Binz H. K., Stumpp M. T., Forrer P., Amstutz P., Plückthun A. (2003) J. Mol. Biol. 332, 489–503 [DOI] [PubMed] [Google Scholar]
- 14.Bork P. (1993) Proteins 17, 363–374 [DOI] [PubMed] [Google Scholar]
- 15.Kohl A., Amstutz P., Parizek P., Binz H. K., Briand C., Capitani G., Forrer P., Plückthun A., Grütter M. G. (2005) Structure 13, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 16.Schweizer A., Roschitzki-Voser H., Amstutz P., Briand C., Gulotti-Georgieva M., Prenosil E., Binz H. K., Capitani G., Baici A., Plückthun A., Grütter M. G. (2007) Structure 15, 625–636 [DOI] [PubMed] [Google Scholar]
- 17.Sennhauser G., Amstutz P., Briand C., Storchenegger O., Grütter M. G. (2007) PLoS Biol. 5, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binz H. K., Amstutz P., Kohl A., Stumpp M. T., Briand C., Forrer P., Grütter M. G., Plückthun A. (2004) Nat. Biotechnol. 22, 575–582 [DOI] [PubMed] [Google Scholar]
- 19.Bandeiras T. M., Hillig R. C., Matias P. M., Eberspaecher U., Fanghänel J., Thomaz M., Miranda S., Crusius K., Pütter V., Amstutz P., Gulotti-Georgieva M., Binz H. K., Holz C., Schmitz A. A., Lang C., Donner P., Egner U., Carrondo M. A., Müller-Tiemann B. (2008) Acta Crystallogr. D. Biol. Crystallogr. 64, 339–353 [DOI] [PubMed] [Google Scholar]
- 20.Hanes J., Plückthun A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4937–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zahnd C., Amstutz P., Plückthun A. (2007) Nat. Methods 4, 269–279 [DOI] [PubMed] [Google Scholar]
- 22.Steiner D., Forrer P., Plückthun A. (2008) J. Mol. Biol. 382, 1211–1227 [DOI] [PubMed] [Google Scholar]
- 23.Veesler D., Blangy S., Siponen M., Vincentelli R., Cambillau C., Sciara G. (2009) Anal. Biochem. 388, 115–121 [DOI] [PubMed] [Google Scholar]
- 24.Sulzenbacher G., Gruez A., Roig-Zamboni V., Spinelli S., Valencia C., Pagot F., Vincentelli R., Bignon C., Salomoni A., Grisel S., Maurin D., Huyghe C., Johansson K., Grassick A., Roussel A., Bourne Y., Perrier S., Miallau L., Cantau P., Blanc E., Genevois M., Grossi A., Zenatti A., Campanacci V., Cambillau C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 2109–2115 [DOI] [PubMed] [Google Scholar]
- 25.Vincentelli R., Bignon C., Gruez A., Canaan S., Sulzenbacher G., Tegoni M., Campanacci V., Cambillau C. (2003) Acc. Chem. Res. 36, 165–172 [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 27.Navaza J. (1993) Acta Crystallogr. D. Biol. Crystallogr. 49, 588–591 [DOI] [PubMed] [Google Scholar]
- 28.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Painter J., Merritt E. A. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 31.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32.Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 33.Geller B. L., Ngo H. T., Mooney D. T., Su P., Dunn N. (2005) J. Dairy Sci. 88, 900–907 [DOI] [PubMed] [Google Scholar]
- 34.Hultberg A., Tremblay D. M., de Haard H., Verrips T., Moineau S., Hammarström L., Marcotte H. (2007) BMC Biotechnol. 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sennhauser G., Grütter M. G. (2008) Structure 16, 1443–1453 [DOI] [PubMed] [Google Scholar]
- 36.De Haard H. J., Bezemer S., Ledeboer A. M., Müller W. H., Boender P. J., Moineau S., Coppelmans M. C., Verkleij A. J., Frenken L. G., Verrips C. T. (2005) J. Bacteriol. 187, 4531–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewert S., Cambillau C., Conrath K., Plückthun A. (2002) Biochemistry 41, 3628–3636 [DOI] [PubMed] [Google Scholar]
- 38.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R. (1993) Nature 363, 446–448 [DOI] [PubMed] [Google Scholar]
- 39.Muyldermans S., Cambillau C., Wyns L. (2001) Trends Biochem. Sci. 26, 230–235 [DOI] [PubMed] [Google Scholar]
- 40.Labrie S. J., Josephsen J., Neve H., Vogensen F. K., Moineau S. (2008) Appl. Environ. Microbiol. 74, 4636–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deveau H., Labrie S. J., Chopin M. C., Moineau S. (2006) Appl. Environ. Microbiol. 72, 4338–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labrie S. J., Moineau S. (2007) J. Bacteriol. 189, 1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahnd C., Spinelli S., Luginbühl B., Amstutz P., Cambillau C., Plückthun A. (2004) J. Biol. Chem. 279, 18870–18877 [DOI] [PubMed] [Google Scholar]
- 44.Corpet F. (1988) Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]