Abstract
Propionylation has been identified recently as a new type of protein post-translational modification. Bacterial propionyl-CoA synthetase and human histone H4 are propionylated at specific lysine residues that have been known previously to be acetylated. However, other proteins subject to this modification remain to be identified, and the modifying enzymes involved need to be characterized. In this work, we report the discovery of histone H3 propionylation in mammalian cells. Propionylation at H3 lysine Lys23 was detected in the leukemia cell line U937 by mass spectrometry and Western analysis using a specific antibody. In this cell line, the propionylated form of Lys23 accounted for 7%, a level at least 6-fold higher than in other leukemia cell lines (HL-60 and THP-1) or non-leukemia cell lines (HeLa and IMR-90). The propionylation level in U937 cells decreased remarkably during monocytic differentiation, indicating that this modification is dynamically regulated. Moreover, in vitro assays demonstrated that histone acetyltransferase p300 can catalyze H3 Lys23 propionylation, whereas histone deacetylase Sir2 can remove this modification in the presence of NAD+. These results suggest that histone propionylation might be generated by the same set of enzymes as for histone acetylation and that selection of donor molecules (propionyl-CoA versus acetyl-CoA) may determine the difference of modifications. Because like acetyl-CoA, propionyl-CoA is an important intermediate in biosynthesis and energy production, histone H3 Lys23 propionylation may provide a novel epigenetic regulatory mark for cell metabolism.
Introduction
Eukaryotic histones are rich in multiple post-translational modifications, the combinatory array of which is the basis for epigenetic regulation of gene expression (1). Appropriate histone modifications are required for normal cell growth and differentiation, whereas aberrant histone modifications contribute to tumor formation such as malignant hematopoiesis (2). For example, genome-wide alterations of histone modification patterns are found in prostate cancer and are predictive of clinical outcomes (3); histone H3 Lys79 (H3K79) hypermethylation and the activation of oncogenic HOX genes are prominent causes for leukemia with chromosome translocation involving the AF10 gene (4, 5), whereas global hypomethylation at H3K79 sites has been proposed to cause genome instability in AF10-related leukemia cancers (6). Alteration in histone modification patterns serves as a hallmark of cancer that provides clues for cancer diagnosis and treatment (7).
Lysine propionylation has recently been identified as a new type of post-translational modification. It was first discovered in human histone H4 enriched for acetylation using anti-acetyl H4 antibodies (9). The propionyl-CoA synthetase of Salmonella enterica was also found to be propionylated at lysine 592, which inactivates the enzyme activity in vivo (8). In vitro experiments showed that histone acetyltransferase p300 and CREB-binding protein (CBP) can propionylate histone H4, suggesting that propionylation and acetylation may share the common enzymes that recognize both propionyl-CoA and acetyl-CoA (9). The in vivo propionylation of propionyl-CoA synthetase is catalyzed by a protein related to GCN5 histone acetyltransferase (HAT), and the propionyl group can be removed by NAD+-dependent human Sir2 histone deacetylase (sirtuins) in vitro (8). Moreover, the GCN5-related mammalian HAT PCAF has the ability to propionylate histone H3 with similar efficiency and lysine specificity as it acetylates H3 in vitro (10). However, it is unclear so far whether propionylation is present in histone H3 in vivo.
In this work, we report the identification and characterization of histone H3 propionylation in mammalian cells. This modification was specifically detected at H3 Lys23 (H3K23) in the myeloid precursor leukemia cell line U937. The H3K23 propionylation level in U937 cells decreased significantly during monocytic differentiation, suggesting that this modification might be associated with lineage determination. The histone acetyltransferase p300 can catalyze this modification, whereas the histone deacetylase Sir2 catalyzes the removal of the propionyl group in vitro. Because propionyl-CoA and acetyl-CoA, both metabolic products of fatty acid, are differential cofactors for dual functional histone acetyltransferases, we suggest that histone propionylation might be associated with cellular metabolic status.
EXPERIMENTAL PROCEDURES
Cell Cultures
U937T (provided by Dr. Gerard Grosveld) is a derivative of U937 harboring the tet-VP16 fusion gene under the control of a tetracycline-inducible promoter (11). U937T stable cell lines expressing FLAG-AF10 or FLAG-AF10ΔOM-LZ (where “OM” is octapeptide motif and “LZ” is leucine zipper) were established using the pUHD10S-1 vector (a gift from Dr. Gerard Grosveld). The stable cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1 μg/ml tetracycline, 0.5 μg/ml puromycin, and 1 mg/ml G418 (Invitrogen). HeLa and IMR-90 cell lines were purchased from American Type Culture Collection and cultured as recommended.
Histone Purification
Cultured cells were centrifuged in ice-cold PBS3 supplemented with 5 mm sodium butyrate. The cell pellets were resuspended in ice-cold Triton extraction buffer (PBS containing 0.5% (v/v) Triton X-100, 2 mm phenylmethylsulfonyl fluoride, 0.02% (v/v) NaN3, and protein inhibitor mixture) at a cell density of 5 × 106 cells/ml and incubated on a rotator. The nuclei were isolated and lysed in 0.4 n H2SO4 by incubation overnight at 4 °C. The supernatant of the centrifugation was added with trichloroacetic acid to reach a final concentration of 33% (v/v) so that histones were precipitated out. The core histones were further fractionated into H2A, H2B, H3, and H4 by reversed-phase HPLC (12).
Western Analysis of Histone H3K23 Propionylation
H3K23-propionylated peptide KQLATKprAARC and Lys23-acetylated peptide KQLATKacAARC were solid phase-synthesized on an ABI peptide synthesizer. The propionylated peptide was used to immunize rabbits for the preparation of polyclonal antibodies specific for H3K23 propionylation. Cross-reactivity to Lys23 acetylation was minimized by passing the antibody solution through an affinity column loaded with beads cross-linked with the acetylated Lys23 peptide. Peptide dot blotting analysis demonstrated that the purified H3K23 propionylation antibody had a 9-fold affinity over the H3K23-acetylated peptide and a 100-fold affinity over the unmodified peptide (supplemental Fig. 1). Western blot analysis was carried out using the propionylation-specific antibodies with total protein extracts of different cell lines.
Mass Spectrometry
Histones were digested overnight either with trypsin or Arg-C proteases (enzyme/protein of ∼1:100; Roche Applied Science). After digestion, the peptides were dried with reduced vacuum and then redissolved in 0.1% formic acid. The peptides were fractionated by reversed-phase HPLC on a microbore C18 column and subjected to MALDI-TOF analysis or directly subjected to LC-MS/MS analysis. MALDI-TOF mass spectrometric experiments were performed using a Bruker Autoflex instrument in the reflectron mode. For regular MALDI-TOF experiments, all spectra were externally calibrated using a peptide standard mixture provided by Bruker. A 1.0-μl aliquot of the peptide solution was mixed with 1.0 μl of α-cyano-4-hydroxycinnamic acid matrix (5 mg/ml in 50:50 acetonitrile/water and 0.1% formic acid; Sigma). Data from a set of 100∼200 laser shots were accumulated to give an acceptable spectrum. For high accuracy mass determination, the spectra of modified histone peptides were calibrated by the lock mass of a co-eluted known histone peptide. LC-MS/MS identification of histone modifications was carried out by nanoelectrospray on a Waters Q-TOF Ultima instrument that was coupled to a Waters capillary HPLC system. A 140-min gradient (mobile phase A, 0.1% formic acid; mobile phase B, 0.1% formic acid in acetonitrile) with a flow rate of 6 μl/min was run through a splitter. After splitting, a flow rate of ∼300 nl/min was run through a Waters nano C18 column (150 mm × 75 μm) to the nanoelectrospray source. A 1.8-kV capillary voltage and 45-V cone voltage were optimized for nanoelectrospray. The Q-TOF method utilized consisted of a survey scan mode in the mass range from 300 to 1500 Da during which up to three precursor ions were selected for parallel collision-activated dissociation (collision gas, argon) analysis using an automated data-dependent MS-to-MS/MS and MS/MS-to-MS switching function. Precursor ion selection was based on a normalized threshold of 10 counts/s, and ions of potentially modified peptides were preferentially selected for MS/MS fragmentation. MS/MS experiments used an m/z-dependent collision energy applied linearly from 20 to 42 V from m/z 300 to 1500. Product ion scans were measured in the range from m/z 50 to 2000. The Q-TOF instrument was calibrated with the MS/MS fragmentation ions of the peptide Glu-Fib, resulting in <20 ppm mass accuracy for precursor ions when a lock mass was applied and ±0.02 Da for product ions. Modifications of histones were manually determined with the aid of the Prospector program. Quantitative LC-MS/MS analyses of histone acetylation and propionylation were performed by derivatization of unmodified lysines using d6-acetic anhydride as described previously (13), and the quadruple resolution was set with a low window (low mass = 2 and high mass = 2) to select precursor ions at approximately ±8 Da for MS/MS fragmentation. The acetylation proportions of Lys18 and Lys23 were calculated by the intensity relative to the total peak value of diacetylated peptide (Lys18/Lys23), Lys18- or Lys23-monoacetylated peptide, and Lys18- and Lys23-unacetylated peptide. The propionylation level was calculated by the intensity of the Lys23 propionylation peak over the total intensity of acetylation and propionylation peaks.
Propionylation and Depropionylation Assays
In vitro propionylation and acetylation assays were carried out as described previously (9). Basically, 2.5 ng of human recombinant histone H3 (Millipore) was dissolved in 10 μl of acetylation buffer (50 mm Tris (pH 7.9), 10% glycerol, and 1 mm dithiothreitol). After the addition of 1 μl of [14C]acetyl-CoA or [14C]propionyl-CoA (55 mCi/mmol; American Radiolabeled Chemicals, Inc.) and 0.5 μg of p300 (Upstate), the solution was incubated at room temperature for a given time. The reaction mixture was then subjected to SDS-PAGE, followed by autoradiography and/or Coomassie Blue staining. For the propionylation/acetylation assay with nonradioactive propionyl-CoA or acetyl-CoA, the reaction mixture incubated for 1 h was subjected to enzymatic digestion with Arg-C, followed by MALDI-TOF and LC-MS/MS analyses. For the depropionylation assay, histone H3 was first incubated with p300 in the presence of [14C]propionyl-CoA (or [14C]acetyl-CoA as a control) for 1 h, and the reaction was stopped by heating at 60 °C for 5 min. 10 μl (100 μg/300 μl of solution) of human Sir2 proteins (sirtuins 1–3; BPS Bioscience) was then added to the solution together with 50 mm NAD+. The mixtures were incubated at room temperature for 30 min, and reactions were stopped by the addition of gel loading buffer. Samples were then subjected to SDS-PAGE analysis with Coomassie Blue staining and autoradiography. Meanwhile, a synthetic Lys23-propionylated H3 peptide was incubated with Sir2 proteins and NAD+ for 30 min, and the mixture was analyzed by MALDI-TOF mass spectrometry.
Monocytic Differentiation
U937 and HL-60 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. For induced differentiation, cells were washed and resuspended in fresh medium supplemented with 100 nm PMA (or Me2SO as a mock control). After 72 h of treatment, cells were harvested for analysis.
Flow Cytometric Analysis
Cells were harvested, washed with cold PBS, and counted. 1 × 106 cells were incubated with allophycocyanin-conjugated anti-CD11b antibody (BD Biosciences) on ice for 30 min in the dark. After washing twice with PBS, cells were resuspended in 200 μl of fluorescence-activated cell sorting buffer (PBS with 5 μg/ml propidium iodide and 2% fetal bovine serum) and subjected to FACSCalibur analysis.
RESULTS
Identification of Histone H3 Lysine 23 Propionylation in the U937 Cell Line
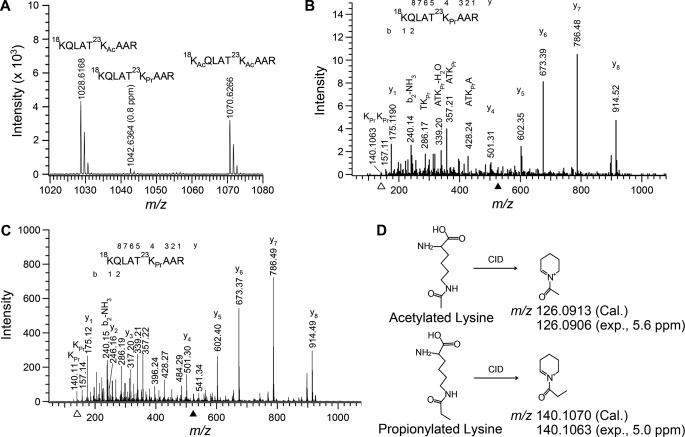
In the analysis of post-translational modifications of histones isolated from the U937 leukemia cell line, we identified a novel modification, propionylation at histone H3 lysine 23, by mass spectrometry. From LC-MS/MS analysis of histone H3 samples, we observed an unusual peak at m/z 521.82 (2+), which eluted between the peaks at m/z 514.82 (2+) and 535.82 (2+), which corresponded to peptides singly and doubly acetylated at Lys18 and/or Lys23, respectively (data not shown). We then performed HPLC peptide purification of Arg-C-digested histone H3 and conducted MALDI-TOF analysis of the pool of Lys18/Lys23-modified peptides. A peak at m/z 1042.6364 (0.8 ppm) was detected between the peaks at m/z 1028.6168 and 1070.6266, which corresponded to the singly and doubly acetylated peptides as stated above (Fig. 1A). The 14-Da mass increase of the singly acetylated form could result from an additional methylation on either the peptide or the acetyl group, namely propionylation. Fragmentation of the selected ion at m/z 521.82 (2+) by electrospray-LC-MS/MS indicated that Lys18 was free of modification but that Lys23 was attached with a moiety of 56 Da (Fig. 1B). The 56-Da change could be due either to a sum of one acetylation and one methylation or to one propionylation. There has been no precedent that methylation and acetylation can occur at the same amino group, although this possibility exists. Notably, a stable six-membered ring immonium ion at m/z 140.1063 that can be produced only from propionylated lysine was detected (Fig. 1B). Moreover, we analyzed a synthetic Lys23-propionylated peptide with a sequence identical to that of the Arg-C-digested peptide of histone H3 isolated from U937 cells. The MS/MS spectrum of this synthesized peptide was almost identical to that of the peptide from U937 cells (Fig. 1C). A lysine residue acetylated and methylated simultaneously cannot form an immonium ion at m/z 140.1070 (calculated monoisotopic mass) because nitrogen with a positive charge makes no more than four bonds (i.e. NH4+) theoretically (Fig. 1D). Therefore, the modification at Lys23 was conclusively demonstrated to be propionylation.
FIGURE 1.
Identification of histone H3 lysine 23 propionylation in U937 cells. A, MALDI-TOF spectrum of peptides containing modifications at Lys18 and Lys23. These peptides were purified by HPLC from Arg-C-digested histone H3 of U937 cells: 18KQLAT23KAcAAR (m/z), 1028.6168 (expected (exp.), 4.2 ppm) and 1028.6211 (calculated (Cal.)); 18KQLAT23KPrAAR (m/z), 1042.6364 (expected, 0.8 ppm) and 1042.6368 (calculated); and 18KAcQLAT23KAcAAR (m/z), 1070.6266 (expected, 4.8 ppm) and 1070.6317 (calculated). B, MS/MS spectrum of the doubly charged ion at m/z 521.8221 (solid arrowhead). Histone H3 prepared from U937 cells was digested with Arg-C prior to the LC-MS/MS analysis. The fragmentation pattern (mainly b ions and y ions) established the peptide sequence indicated. C, MS/MS spectrum of a synthetic peptide with Lys23 propionylation. Note that the spectrum of this authentic propionylated peptide is identical to that of the propionylated peptide identified in U937 cells (see B). D, illustration of distinctive immonium ions (open arrowheads in B and C) generated via collision-induced dissociation (CID) from an acetylated lysine (at m/z 126.0906; spectrum not shown) and from a propionylated lysine (at m/z 140.1063). The immonium ions were calibrated with the lock mass of the y1 (arginine) ion at m/z 175.1190.
H3K23 Propionylation Is Specific for U937 Leukemia Cells
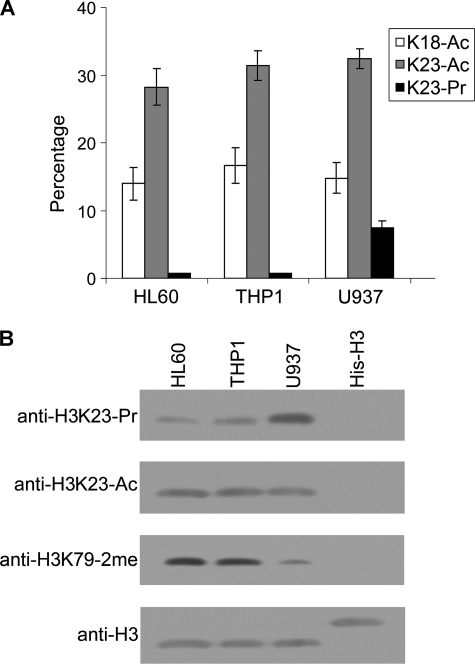
We then determined the relative abundance of propionylation at H3K23 in three leukemia cell lines. To this end, histone H3 was reacted with d6-acetic anhydride in d3-acetyl acid solution, followed by digestion with trypsin and analysis by LC-MS/MS. As shown in Fig. 2A, ∼7% of Lys23 was propionylated in the U937 cell line, whereas only a background level of propionylation was detected in two other leukemia cell lines, HL-60 and THP-1. No significant H3K23 propionylation was detected in two non-leukemia cell lines, HeLa and IMR-90 (supplemental Fig. 2). In addition, the propionylation did not seem to be linked to acetylation because no differences in acetylation were observed at Lys18 or Lys23 among the three leukemia cell lines (Fig. 2A). Western analysis with propionylated H3K23-specific antibody confirmed that Lys23 propionylation was more predominant in U937 cells (Fig. 2B). In contrast, the Lys23 acetylation levels were similar in the different cell lines. The global H3K79–2me hypomethylation is a characteristic of U937 cells, in which the CALM-AF10 fusion protein affects the association of the H3K79 methyltransferase hDOT1L with chromatin as described in our previous report (6).
FIGURE 2.
Quantification and Western blot confirmation of H3K23 propionylation in U937 cells. A, quantification of H3K23 propionylation in leukemia cells by LC-MS/MS analysis. Histone H3 isolated from each cell line was treated with d6-acetyl anhydride and digested with Arg-C prior to the mass analysis. The H3K23 propionylation level was significantly higher in U937 cells than in HL-60 or THP-1 cells, whereas no variation was seen for acetylation at Lys18 and Lys23 in the three cell lines compared. Experiments were performed three times, and the S.D. is shown by the error bars. Ac, acetylated; Pr, propionylated. B, Western detection of Lys23 propionylation using an antibody specific for Lys23 propionylation. Note that the H3K23 propionylation signal was significantly stronger in the U937 cell line, whereas the acetylation levels at the same site were similar in the three cell lines. Blotting with anti-histone H3 antibody served as a loading control. His6-tagged recombinant histone H3 lacking any modification was used as a negative control to validate the performance of the modification-specific antibodies.
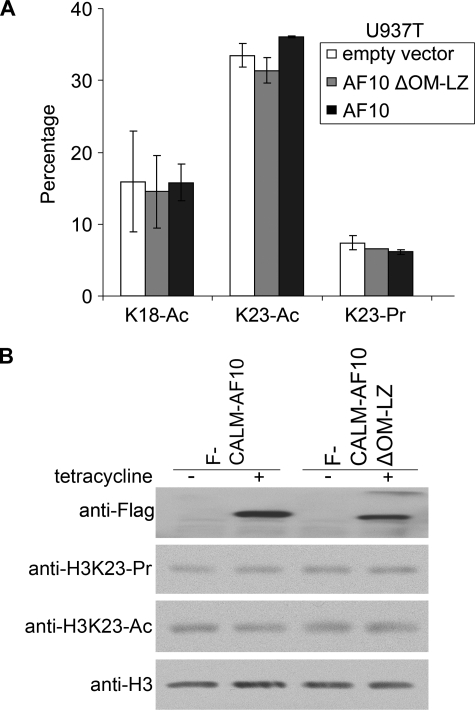
Down-regulation of AF10 or Ectopic Expression of CALM-AF10 Fusion Protein Does Not Increase Propionylation
We then examined whether the elevated propionylation is related to the decreased level of the AF10 transcription factor (14) due to the specific presence of the CALM-AF10 fusion gene in U937 cells. However, no change in H3K23 propionylation was observed in U937 cells upon ectopic expression of the wild-type AF10 protein or the mutant AF10 protein with a deletion of the hDOT1L-binding domain OM-LZ (Fig. 3A), which is crucial for AF10-mediated restoration of H3K79 in U937T cells (6). This result suggests that hyperpropionylation in U937 cells is not caused by down-regulation of AF10. To investigate the possible link between H3K23 hyperpropionylation and CALM-AF10 fusion, CALM-AF10 and its OM-LZ deletion mutant were induced to express in T-REx-293 stable cell lines with tetracycline. Consistent with our previous report (6), global hypomethylation of H3K79 was detected upon induction of CALM-AF10 but not the OM-LZ deletion mutant (data not shown). However, the H3K23 propionylation level did not change significantly in response to the induced expression of the CALM-AF10 proteins in the stable cell lines, and neither did the acetylation level (Fig. 3B). These data suggest that the H3K23 hyperpropionylation in U937 cells is unlikely associated with the expression of the CALM-AF10 fusion protein.
FIGURE 3.
H3K23 hyperpropionylation in U937 cells is not caused by down-regulation of AF10 or expression of the CALM-AF10 fusion protein. A, ectopic expression of AF10 in U937 cells did not reduce H3K23 propionylation. Quantitative LC-MS/MS analysis was performed to measure acetylation and propionylation of histone H3 isolated from the U937 cell lines ectopically expressing wild-type AF10 or mutant AF10 deleted of the hDOT1L-binding domain (OM-LZ). Experiments were performed three times, and the S.D. is shown by the error bars. Ac, acetylated; Pr, propionylated. B, inducible ectopic expression of CALM-AF10 in T-REx-293 cells did not increase H3K23 propionylation. CALM-AF10 or its mutant (OM-LZ deletion in AF10) was induced to express with tetracycline (+). Total cell extracts were examined by Western blot analysis using the antibodies indicated on the left. The uninduced cells (−) were included as negative controls.
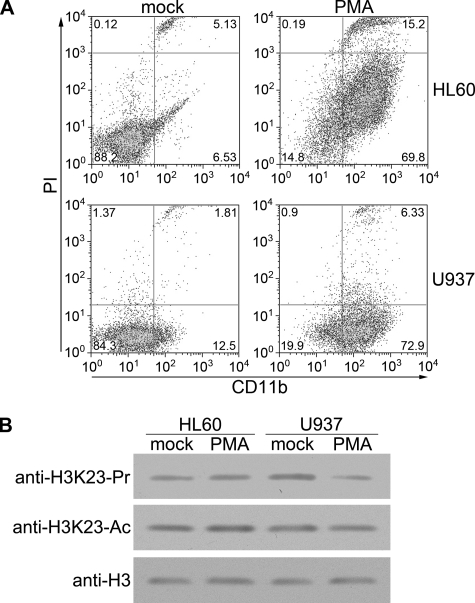
H3K23 Propionylation Decreases during Monocytic Differentiation of U937 Cells
Genome-wide reprogramming of histone modifications is closely associated with cell growth and differentiation in development (15, 16). To examine the potential role of H3K23 propionylation in cell differentiation, we mimicked monocytic differentiation in U937 cells with PMA induction. Following PMA treatment, U937 cells became adherent and, over the course of several days, differentiated into monocyte/macrophage-like cells. Differentiation of promonocytic U937 cells into monocytes/macrophages is characterized by the elevated expression of the cell-surface marker CD11b (17). We confirmed with fluorescence-activated cell sorting analysis that 72.9% of the U937 cells became CD11b-positive upon PMA treatment (Fig. 4A), which is consistent with a previous report (18). We then examined the H3K23 modification during monocytic differentiation of U937 cells and the control cell line HL-60 by Western blotting and LC-MS/MS. Interestingly, the propionylation level of H3K23 in U937 cells decreased markedly after PMA induction, whereas the acetylation level remained unchanged (Fig. 4B and supplemental Fig. 3). Furthermore, HL-60 cells did not show such a change during monocytic differentiation, suggesting that the hyperpropionylation of H3K23 in U937 cells is associated with cell differentiation.
FIGURE 4.
H3K23 propionylation decreases during differentiation of U937 cells. A, U937 and HL-60 cells were induced for monocytic differentiation. Cells were treated with PMA or Me2SO (mock) for 72 h, harvested, and analyzed by flow cytometry. CD11b was used as a cell-surface marker for monocytic differentiation. DNA was stained with propidium iodide (PI). B, shown are the results from Western blot analysis of H3K23 modifications in differentiated HL-60 and U937 cells. Upon induction with PMA for 72 h, cells were harvested, and the cell extracts were analyzed with the indicated antibodies. Ac, acetylated; Pr, propionylated.
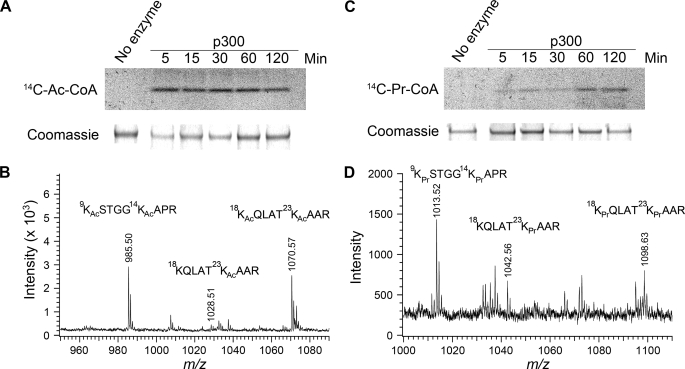
Histone Acetyltransferase p300 Propionylates Histone H3 in Vitro
Previous experiments have shown that the histone acetyltransferase p300 propionylates histone H4 at multiple sites and that PCAF propionylates lysine 14 of histone H3 (9, 10). To test whether the histone acetyltransferase p300 can also propionylate lysine 23 of histone H3, we set up an in vitro enzymatic assay with human recombinant histone H3 as a substrate. First, we performed the assay using [14C]propionyl-CoA as the propionyl donor and the acetylation cofactor [14C]acetyl-CoA as a positive control. As shown in Fig. 5(A and C), p300 propionylated histone H3 although with much slower kinetics compared with acetylation. Furthermore, we conducted the same propionylation assay using unlabeled propionyl-CoA, followed by analysis of the H3 substrate with MALDI-TOF and LC-MS/MS. As shown in Fig. 5 (B and D), p300 propionylated lysine 23 of histone H3 as well as three other residues (Lys9, Lys14, and Lys18) known to be acetylation sites.
FIGURE 5.
Histone acetyltransferase p300 can catalyze H3K23 propionylation in vitro. A, in vitro acetylation of human recombinant histone H3 by p300 in the presence of [14C]acetyl-CoA (14C-Ac-CoA). Upper panel, autoradiography; lower panel, Coomassie Blue staining of H3 upon incubation with p300. The time points at which samples were taken from the incubation are indicated. B, MALDI-TOF measurement of histone H3 upon incubation with p300 in the presence of acetyl-CoA. C, in vitro propionylation of human recombinant histone H3 by p300 in the presence of [14C]propionyl-CoA (14C-Pr-CoA). D, MALDI-TOF measurement of histone H3 upon incubation with p300 in the presence of propionyl-CoA. The acetyl and propionyl modifications at Lys9, Lys14, Lys18, and Lys23 were confirmed by LC-MS/MS sequencing analysis of Arg-C-digested peptides upon incubation with p300 (not shown).
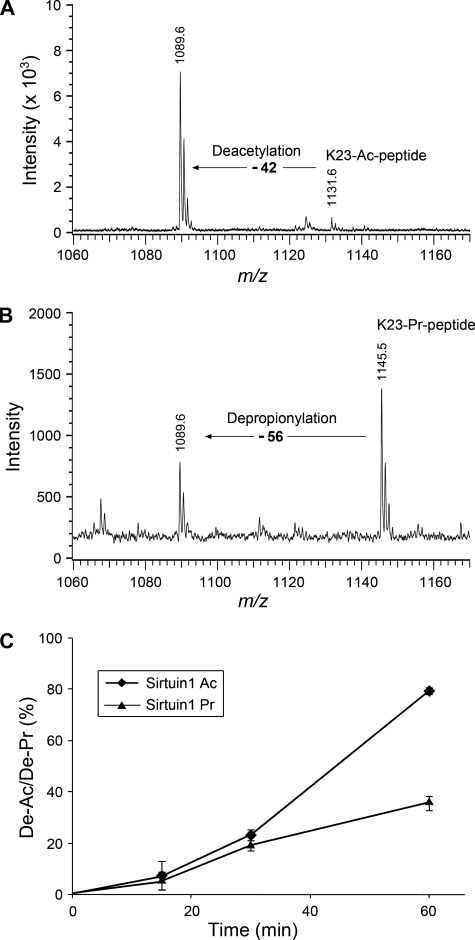
Sir2 Depropionylates Histone H3 Propionylation
Sir2 protein is an NAD+-dependent protein deacetylase (19). In humans, eight Sir2 homologs (sirtuins 1–8) have been identified. Garrity et al. (8) reported that both the bacterial and human Sir2 histone deacetylases can remove propionylation from the propionyl-CoA synthetase. Molecular modeling indicated that the hydrophobic pocket in the active site of Sir2 can readily accommodate the additional methyl of propionyl-lysine (20). Following this scenario, we asked whether Sir2 would depropionylate histone H3K23. To this end, we incubated a synthetic Lys23-propionylated peptide with sirtuin proteins in the presence of NAD+ in a time course for up to 1 h. Meanwhile, parallel experiments using synthetic Lys23-acetylated peptide as the substrate were performed to compare the enzyme activities between depropionylation and deacetylation. The peptides were then analyzed by MALDI-TOF mass spectrometry. For the input acetylated peptide (KQLATKacAARC, MH+ 1131.6), a new peak 42 Da smaller in mass appeared after incubation with sirtuin 1 (Fig. 6A), indicating the deacetylation activity of sirtuin 1. Similarly, for the propionylated peptide (KQLATKprAARC, MH+ 1145.5), a new peak with a decrease of 56 Da appeared (Fig. 6B), indicating the depropionylation activity of sirtuin 1. Sirtuins 2 and 3 could also catalyze depropionylation, although sirtuin 3 had a lower enzymatic activity than sirtuins 1 and 2 (supplemental Fig. 5). Notably, the sirtuin 1 depropionylation activity was about half of its deacetylation activity (Fig. 6C).
FIGURE 6.
Sir2 protein can depropionylate H3K23 in vitro. A, MALDI-TOF analysis of Lys23-acetylated peptides (K23-Ac-peptide) upon incubation with a Sir2 protein (sirtuin 1) for 1 h. The spectrum before the deacetylase treatment is not shown. B, MALDI-TOF analysis of Lys23-propionylated peptides (K23-Pr-peptide) upon incubation with sirtuin 1 for 1 h. C, time course of deacetylation/depropionylation (De-Ac/De-Pr) at Lys23 of the peptides by sirtuin 1. The deacetylation and depropionylation percentages were calculated by the following equations: Lys23 deacetylation = intensitypeak 1089.6/total intensitypeak 1089.6 & peak 1131.6; and Lys23 depropionylation = intensitypeak 1089.6/total intensitypeak 1089.6 & peak 1145.5. Experiments were performed three times, and the S.D. is shown by the error bars.
DISCUSSION
The regulation of gene expression is largely mediated by post-translational modification of histones in chromatin. Although the most abundant types of post-translational modifications (such as acetylation and methylation) that are known to play vital roles in transcriptional silencing and activation have been extensively studied, the existence of rare types of post-translational modifications in histones remains to be characterized. Our work presents the first evidence for the existence of propionylation at mammalian histone H3. We found that lysine 23 in the H3 N-terminal tail is specifically propionylated up to 7% in the U937 leukemia cell line. During the preparation of this manuscript, Zhang et al. (21) reported the detection of Lys23 propionylation in yeast histone H3. Therefore, propionylation at H3K23 constitutes a novel type of histone mark that is evolutionarily conserved from yeast to human.
Biological Significance of H3K23 Propionylation
Histone propionylation is expected to exert functions similar to the widespread acetylation due to the structural similarity of the propionyl and acetyl groups. This modification neutralizes the positive charge of a lysine residue, thus rendering an active local chromatin structure. The added propionyl group may also provide a mark for recognition by chromatin “readers” such as bromodomain-containing proteins that mediate further chromatin modification and transcriptional regulation. Propionyl-Lys23 can also exert its effect by inhibiting or stimulating the modification at the neighboring sites. The high abundance of histone H3K23 propionylation in U937 cells and its decrease upon differentiation may imply an important role of this modification in modulating cell differentiation. H3K23 propionylation might regulate transcription of genes crucial for cell growth and differentiation with a mechanism similar to acetylation. However, the genes controlled by propionylation might be different from the targeted genes regulated by other modifications because the dynamics of propionylation in U937 differentiation is different from that of Lys23 acetylation (Fig. 4B) and Lys79 methylation (data not shown). Because the propionylation levels became similar between U937 and other leukemia cell lines after differentiation, hyperpropionylation in U937 cells might be a stage-specific marker in hematopoiesis and leukemogenesis, which could serve as a diagnostic target for leukemia therapy.
The fact that histone propionylation is barely detectable in most mammalian cells suggests its highly restrictive occurrence in specific target genes under certain growth and metabolic conditions. In Salmonella, propionylation of the propionyl-CoA synthetase increases with increased levels of propionyl-CoA when the cells are grown on propionate medium (8). In animals, the concentration of the acyl-CoAs fluctuates in response to diet and other physiological conditions (22). Given the regulatory function of acyl donors, propionylation in chromatin is likely to reflect the cellular level of propionyl-CoA, an important intermediate generated from energy metabolism. In this connection, we speculate that genes involved in energy metabolism could be regulated by histone propionylation. The identification of target genes subject to propionylation would help elucidate the biological processes that this modification could modulate.
Regulatory Mechanism of H3K23 Propionylation
Our findings are in good agreement with previous reports that the addition and removal of propionyl groups can be catalyzed by the same set of enzymes as for acetylation and deacetylation (9, 10). Because the acetylation level in the U937 cell line is similar to that in other cell lines, we speculate that hyperpropionylation in U937 cells may result from the irregular accumulation of the propionyl donor molecule, propionyl-CoA. The accumulation of propionyl-CoA could arise from either up-regulation of propionyl-CoA synthetase or down-regulation of propionyl-CoA carboxylase, which removes propionyl-CoA by converting it into 2-methyl citrate. It would be of interest to examine the expression levels of these two enzymes directly involved in propionyl-CoA homeostasis in the related leukemia cells.
Several lines of evidence are in favor of the existence of distinct propionylation enzymes (or specific regulatory factors) other than acetyltransferase (deacetylase). H3K23 propionylation is found only in the U937 cell line but not in the other two related monocytic leukemia cell lines, HL-60 and THP-1. Despite the striking difference in propionylation levels, the acetylation levels at Lys23 are similar among these cell lines. Additionally, the histone acetyltransferase p300 propionylates four lysine residues (Lys9, Lys14, Lys18, and Lys23) of the H3 histone tail indiscriminately in vitro, and depropionylation by Sir2 is unlikely to be specific for the Lys23 site. A propionyltransferase that acts exclusively on H3K23 has not been found yet.
Consistent with the proposed relationship between a specific metabolic state and histone propionylation, the known genetic and epigenetic alterations in U937 cells do not seem to contribute to the prominent propionylation. The U937 cell line is characterized by the presence of a t(10;11)(p13;q14) chromosome translocation leading to the fusion of the CALM (clathrin assembly lymphoid myeloid leukemia) gene with the putative transcription factor gene AF10 (14). The CALM-AF10 fusion protein causes global hypomethylation at H3K79 due to the functional perturbation of the responsible histone methyltransferase hDOT1L by the fusion (6). Ectopic expression of CALM-AF10 in T-REx-293 cells results in H3K79 hypomethylation (6) but has no effect on Lys23 propionylation (Fig. 3A). Therefore, hyperpropionylation is not due to the expression of the leukemogenic fusion protein. Whether the differential propionylation in U937 leukemia cells can instead be attributed to a unique metabolic state associated with their monocytic differentiation stage is an intriguing possibility that warrants further investigation.
Supplementary Material
Acknowledgment
We thank Gerard Grosveld for the U937T cell line and pUHD10S-1 vector.
This work was supported by Grants 2005CB522400 and 2006CB943900 from the Ministry of Science and Technology of China and the National Science Foundation of China (to G. X.) and by the Biochemistry Department of Loma Linda University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- PBS
- phosphate-buffered saline
- HPLC
- high performance liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- LC-MS/MS
- liquid chromatography-coupled tandem mass spectrometry
- PMA
- phorbol 12-myristate 13-acetate
- hDOT1L
- human DOT1L.
REFERENCES
- 1.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 2.Rice K. L., Hormaeche I., Licht J. D. (2007) Oncogene 26, 6697–6714 [DOI] [PubMed] [Google Scholar]
- 3.Seligson D. B., Horvath S., Shi T., Yu H., Tze S., Grunstein M., Kurdistani S. K. (2005) Nature 435, 1262–1266 [DOI] [PubMed] [Google Scholar]
- 4.Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V. M., Su L., Xu G., Zhang Y. (2005) Cell 121, 167–178 [DOI] [PubMed] [Google Scholar]
- 5.Okada Y., Jiang Q., Lemieux M., Jeannotte L., Su L., Zhang Y. (2006) Nat. Cell Biol. 8, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y. H., Kakadia P. M., Chen Y., Li Y. Q., Deshpande A. J., Buske C., Zhang K. L., Zhang Y., Xu G. L., Bohlander S. K. (2009) Blood 114, 651–658 [DOI] [PubMed] [Google Scholar]
- 7.Gal-Yam E. N., Saito Y., Egger G., Jones P. A. (2008) Annu. Rev. Med. 59, 267–280 [DOI] [PubMed] [Google Scholar]
- 8.Garrity J., Gardner J. G., Hawse W., Wolberger C., Escalante-Semerena J. C. (2007) J. Biol. Chem. 282, 30239–30245 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leemhuis H., Packman L. C., Nightingale K. P., Hollfelder F. (2008) ChemBioChem 9, 499–503 [DOI] [PubMed] [Google Scholar]
- 11.Boer J., Bonten-Surtel J., Grosveld G. (1998) Mol. Cell. Biol. 18, 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang K., Tang H., Huang L., Blankenship J. W., Jones P. R., Xiang F., Yau P. M., Burlingame A. L. (2002) Anal. Biochem. 306, 259–269 [DOI] [PubMed] [Google Scholar]
- 13.Smith C. M., Gafken P. R., Zhang Z., Gottschling D. E., Smith J. B., Smith D. L. (2003) Anal. Biochem. 316, 23–33 [DOI] [PubMed] [Google Scholar]
- 14.Dreyling M. H., Martinez-Climent J. A., Zheng M., Mao J., Rowley J. D., Bohlander S. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4804–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U. C., Cesari F., Lee C., Almouzni G., Schneider R., Surani M. A. (2008) Nature 452, 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 17.Grattage L. P., McKenzie I. F., Hogarth P. M. (1992) Immunol. Cell Biol. 70, 97–105 [DOI] [PubMed] [Google Scholar]
- 18.Ragg S. J., Kaga S., Berg K. A., Ochi A. (1998) J. Immunol. 161, 1390–1398 [PubMed] [Google Scholar]
- 19.Schwer B., Verdin E. (2008) Cell Metab. 7, 104–112 [DOI] [PubMed] [Google Scholar]
- 20.Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 501–507 [DOI] [PubMed] [Google Scholar]
- 21.Zhang K., Chen Y., Zhang Z., Zhao Y. (2009) J. Proteome Res. 8, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King M. T., Reiss P. D. (1985) Anal. Biochem. 146, 173–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.