Abstract
It is well known that phosphoglucose isomerase/autocrine motility factor (AMF) promotes cell migration in an autocrine manner in various tumor cells. However, it remains unclear whether certain cytokines modulate the effects of AMF on tumor cell migration. Because interleukin (IL)-8, a proinflammatory cytokine, is produced by melanoma cells and has been correlated with melanoma migration, the migratory ability of melanoma cells induced by AMF may also involve induction of IL-8 expression. In the present study, we assessed whether AMF promotes melanoma cell migration through autocrine production of IL-8. We found that AMF stimulation increased IL-8 production through up-regulation of IL-8 mRNA transcription, especially in biologically early stage melanoma cells. AMF-induced migration of these cells was inhibited by a specific neutralizing antibody against IL-8. The IL-8 production induced by AMF was mediated by the ERK1/2 pathways. These findings suggest that melanoma migration induced by AMF is mediated by autocrine production of IL-8 as a novel downstream modulator of the AMF signaling pathway.
Introduction
Phosphoglucose isomerase is a housekeeping cytosolic enzyme that plays key roles in both the glycolysis and glyconeogenesis pathways (1). Phosphoglucose isomerase is a secretable protein that behaves as a potent mitogen/cytokine in the extracellular environment. Molecular cloning and sequencing have identified phosphoglucose isomerase as autocrine motility factor (AMF),2 which is a kind of tumor-secreted cytokine that stimulates tumor cell motility (2). The motility stimulation by AMF is mediated by its interaction with AMF receptor (AMFR) molecules on the surface of target cells (3). AMFR is a 78-kDa seven-transmembrane glycoprotein that belongs to the G protein-coupled receptor family (4). There are several reports that enhanced expression levels of both AMF and AMFR are correlated with the progression of malignant tumors (5, 6). Analysis of the signaling pathway for AMF in tumor cells has revealed that receptor stimulation by AMF leads to pertussis toxin-sensitive G protein activation (7). AMF stimulation induces up-regulation and activation of small Rho-like GTPases, both RhoA and Rac1, with no changes in Cdc42 in melanoma cells (8). Furthermore, both Jun N-terminal kinase 1 (JNK1) and JNK2, which are downstream proteins of RhoA, are increased by AMF stimulation (8). However, it remains unclear whether certain secreted mediators or presently unknown factors modulate the effects of AMF on tumor cell migration.
Interleukin (IL)-8, a member of the CXC chemokine family also known as CXCL-8, is a proinflammatory cytokine whose principal roles in infection and inflammation appear to involve the recruitment and activation of circulating and tissue neutrophils to sites of tissue damage (9). IL-8 is expressed in various tumor cells, such as melanoma cells (10–12), breast carcinoma cells (13), ovarian cancer cells (14), and pancreatic cancer cells (15), which are all cancers with high metastatic indexes. It has been shown that IL-8 stimulates haptotactic migration and regulates pathological angiogenesis, proliferation, and metastasis in a wide variety of cell types (16). A previous study showed that IL-8 stimulates tumor cell migration, especially in melanoma cells (17). Highly metastatic melanoma cells express higher steady-state levels of IL-8 mRNA transcripts than low metastatic melanoma cells (18). These previous studies demonstrated that IL-8 production is directly correlated with melanoma cell migration and the melanoma metastatic potential. Autocrine production of IL-8 is known to be induced by various stimuli, including proinflammatory cytokines, such as IL-1 or tumor necrosis factor (19, 20), bacterial or viral products (21, 22), and cellular stress (23, 24). Therefore, AMF stimulation may also induce IL-8 production in melanoma cells.
In the present study, we examined the possible involvement of IL-8 in the motility-promoting action of AMF in melanoma cells. We found that AMF stimulation induced up-regulation of IL-8 expression in a melanoma cell line, derived from an early stage tumor. This up-regulation of IL-8 expression induced by AMF played a critical role in the induction of cell motility in early stage melanoma cells. Activation of ERK MAPKs, but not JNK and p38, was necessary for AMF-mediated IL-8 protein induction. These results suggest that autocrine production of IL-8 induced by AMF may be a novel downstream modulator of AMF-induced migration in malignant tumor cells.
EXPERIMENTAL PROCEDURES
Cell Culture
Human malignant melanoma SBcl-2 (SB cell line, clone-2) cells were provided by Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA). SBcl-2 cells were established in culture from primary cutaneous melanoma and are a poorly tumorigenic and nonmetastatic line in nude mice (18, 25). 1205Lu cells (ATCC CRL-2812) were obtained from the American Type Culture Collection (Manassas, VA). 1205Lu cells are a metastatic variant of WM793 cells, established from primary melanoma lesions. 1205Lu cells were selected in nude mice for metastatic formation after subcutaneous inoculation of WM793 cells (26, 27). Melanoma cells were grown in 2% Tu medium comprising MCDB153 medium supplemented with 20% L15 medium, 2% heat-inactivated fetal bovine serum, and 5 μg/ml insulin unless otherwise stated. For growth in protein-free medium to evaluate the activities of specific growth factors, fetal bovine serum and insulin were omitted from the medium. The cultures were maintained in a humidified chamber under 95% air and 5% CO2 at 37 °C. Cells were harvested and passaged for experiments with 0.25% trypsin and 0.025% EDTA, and viability was monitored by trypan blue exclusion.
Antibodies and Reagents
The following medium components were used: MCDB153 medium (Sigma), L-15 medium (Invitrogen), insulin (Nacalai Tesque Inc., Kyoto, Japan), and fetal bovine serum (Sigma). The following primary antibodies were used: neutralizing anti-IL-8 (R&D Systems Inc.); anti-AMFR (Abgent, San Diego, CA); anti-phospho-ERK, anti-ERK, anti-phospho-JNK, anti-JNK, anti-phospho-p38, anti-p38, anti-phospho-Akt, anti-phospho-IκB, anti-IκB, anti-phospho-IKK, and anti-IKK (Cell Signaling Technology Inc., Beverly, MA). An anti-β-actin antibody (Sigma) served as a control. The selective kinase inhibitors used were PD98059, SP600125, wortmannin, and NFκB activation inhibitor (Calbiochem).
AMF Protein Purification
Recombinant AMF (rAMF) fusion protein was kindly provided from Dr. Avraham Raz (28). AMF protein was isolated as a glutathione S-transferase fusion protein. Briefly, Escherichia coli pGEX-6P-2 cells (GE Healthcare) containing the desired plasmid were grown to log phase, and protein expression was induced by adding 0.1 mmol/liter isopropyl-l-thio-β-d-galactopyranoside (Amersham Biosciences). After 4 h, the bacteria were centrifuged, and the pellet was sonicated in 1× phosphate-buffered saline. After solubilization of the protein with 1% Triton X-100, the extract was centrifuged, and the supernatant was incubated with a slurry of glutathione-Sepharose 4B (Amersham Biosciences) with gentle agitation to allow binding of the fusion protein to the slurry. AMF was separated from the glutathione S-transferase moiety by incubation with PreScission Protease (GE Healthcare) and isolated by centrifugation. For each procedure, the isolated rAMF protein solution was used after dialysis in a Slide-A-Lyzer dialysis system (Pierce).
ELISA
The IL-8 protein concentrations in cell culture supernatants were measured using an ELISA kit (BIOSOURCE International, Camarillo, CA), according to the manufacturer's protocol. All experiments were repeated three times.
Proliferation Assay
Cell proliferation was measured by an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide tetrazolium assay. Briefly, melanoma cells (2 × 103 cells/well) were cultured in 96-well microtiter plates in a total volume of 100 μl/well. After initial cell seeding, the cells were analyzed using a cell counting kit (Dojindo Laboratories, Tokyo, Japan). Briefly, 10 μl of the cell-counting solution was added and incubated under a humidified 5% CO2 atmosphere at 37 °C for 2 h. The formazan produced was dissolved in 1 n HCl (100 μl/well), and the absorbances of the wells at 450 nm were obtained using a microtiter plate reader (BD Biosciences). All experiments were performed in triplicate.
RNA Extraction, cDNA Synthesis, and PCR Amplification
Total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The quantity of isolated RNA was measured using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Template cDNAs were synthesized from 1 μg of total RNA using an Omniscript reverse transcriptase kit (Qiagen, Valencia, CA). Total RNA was reverse-transcribed with 4 units of Omniscript Reverse Transcriptase (60 min at 37 °C, 5 min at 93 °C) and placed on ice (29). RT-PCR was performed using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The amplification steps for β-actin and IL-8, respectively, were denaturation at 94 °C for 30 s or 1 min, annealing at 54 or 65 °C for 30 or 45 s, and extension at 72 °C for 30 s or 1 min. The amplifications were carried out for 27 cycles for IL-8 and 25 cycles for β-actin. Appropriate positive controls and negative controls (PCR mix without Taq DNA polymerase) were included with each batch of reactions. The primers for IL-8 and β-actin were used with reference to previously published assays (30–32). Specifically, the following primers were used: IL-8, sense (5′-ATGACTTCCAAGCTGGCCG-3′) and antisense (5′-CTCAGCCCTCTTCAAAAACTT-3′), product size 291 bp; β-actin, sense (5′-CTCCTCCTGAGCGCAAGTACTC-3′) and antisense (5′-GATGTGGATCAGCAAGCAGGA-3′), product size 91 bp.
Real-time RT-PCR
Real-time RT-PCR analyses were performed using an ABI Prism 7000 sequence detection system (Applied Biosystems). The standard reaction volume was 20 μl and contained 1× SYBR Green PCR Master Mix (Applied Biosystems), 1.0 μl of cDNA template, and a 1.0 μm concentration of both forward and reverse primers. The initial PCR denaturation step was performed for 5 min at 95 °C, followed by 40 cycles of 60 s at 95 °C (melting) and 60 s at 60 °C (annealing/elongation). All reactions were performed in duplicate. The data were normalized to an internal control gene, β-actin, to control for the amount of RNA in the preparation.
mRNA Stability
Melanoma cells were grown to 90% confluence in 60-mm dishes. After 4 h of incubation in PF medium with or without 500 ng/ml rAMF, 5 μg/ml actinomycin D (ActD) (Sigma) was added to block transcription. The cells were harvested at 0, 1, 2, 4, 6, and 8 h after the addition of ActD. RT-PCR and real-time RT-PCR were performed to analyze IL-8 mRNA expression.
SDS-PAGE and Western Blotting
Aliquots of cell lysates after various treatments were separated by SDS-PAGE with equal amounts of protein (20 μg) loaded in each lane. All protein samples were dissolved in SDS sample buffer (100 mm Tris-HCl, pH 8.8, 0.01% bromphenol blue, 36% glycerol, 4% SDS) containing 1 mm dithiothreitol, boiled for 5 min, and separated in 5–20% Tris-Tricine Ready Gels (Bio-Rad). The separated proteins were electrotransferred to Hybond-enhanced chemiluminescence nitrocellulose membranes (Amersham Biosciences). The membranes were blocked with 5% nonfat dry milk in PBS-T for 30 min at room temperature and then incubated with a primary antibody diluted 1:1000 in PBS-T overnight at 4 °C. Next, the membranes were treated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies, depending on the primary antibody used. After two washes with PBS-T for 30 min each, the membranes were treated with an enhanced chemiluminescence reagent (Amersham Biosciences) and exposed to Amersham Biosciences high performance chemiluminescence film to visualize the positive bands. The densities of the bands were determined using Adobe Photoshop (Apple Inc., Cupertino, CA) and normalized by the corresponding density of the β-actin band.
Migration Assay
Cell motility was measured using 48-well Biocoat Cell Culture Inserts (BD Biosciences Inc., Bedford, MA). Briefly, 5 μg/ml fibronectin in serum-free medium was placed in each lower chamber, which was separated from the upper chamber by a membrane with 8-μm pores. A single-cell suspension of 5 × 104 melanoma cells in serum-free medium containing different concentrations of rAMF was placed in each upper chamber. After 6 h of incubation at 37 °C, the cells were fixed with 70% ethanol and stained with hematoxylin and eosin. The cells that migrated through the pores to the lower surface of the filter were counted under a microscope. A total of 10 random fields were counted in triplicate assays.
Statistical Analysis
For continuous variables, the data were expressed as means ± S.E. The significance of differences between values was determined using Student's t test. All differences were deemed significant at p < 0.05. All statistical analyses were performed with the JMP software package, version 5.01 (SAS Institute Inc., Cary, NC).
RESULTS
rAMF Induces Cell Migration but Not Cell Proliferation in Two Melanoma Cell Lines (SBcl-2 and 1205Lu)
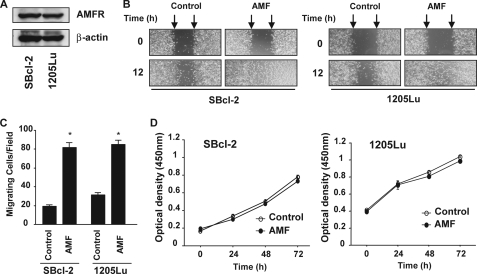
AMF is critical for the migration, invasion, metastasis, and anti-apoptotic effects of malignant tumor cells, and its multiple roles in tumor progression may be mediated by certain downstream pathways and effectors (2, 5, 6, 8, 33–35). A previous study reported that melanoma cells secrete and respond to AMF in an autocrine manner with increased motility (2). We investigated two melanoma cell lines in the present study, namely SBcl-2 cells derived from an early stage of malignant melanoma and 1205Lu cells derived from a metastatic site of malignant melanoma (36). We evaluated the expression of AMFR by Western blotting analysis. As shown in Fig. 1A, both SBcl-2 and 1205Lu cells expressed AMFR protein, suggesting that these cell lines are responsive to AMF in an autocrine manner. We purified human rAMF using a glutathione S-transferase fusion protein purification method involving T&GE. coli competent cells and tested the migratory activities. Cultured monolayers of fresh SBcl-2 and 1205Lu cells were wounded with a pipette tip and cultured for a further 12 h in medium with or without rAMF. Wound healing was significantly more rapid in the rAMF-exposed cells compared with the untreated control cells in both cell lines (Fig. 1B). We further confirmed the migratory activities using a modified Boyden chamber assay. Both SBcl-2 and 1205Lu cells were allowed to migrate through the 8-μm pores of polycarbonate filters during in vitro culture. As shown in Fig. 1C, rAMF induced ∼4-fold increases in the migration of the two melanoma cell lines. These findings demonstrate that purified rAMF induce cell motility in these cell lines.
FIGURE 1.
AMFR expression and biological activity of purified rAMF in melanoma cells. A, expression of AMFR in melanoma cells. Total cell extracts were separated by SDS-PAGE using 12% gels and probed with a polyclonal antibody against AMFR to detect its expression. B, representative examples of wounding experiments in SBcl-2 and 1205Lu cells cultured with or without rAMF (500 ng/ml). SBcl-2 and 1205Lu cells were wounded (time 0) and maintained for 12 h in conditioned medium with or without rAMF (500 ng/ml). The arrows point to the edges of the wounds. Note that the wound healing (measured after 12 h) is faster in rAMF-treated cells than in untreated cells in both cell lines. C, migration assays. The mean cell counts ± S.E. (n = 10) of cells that migrated through the pores to the lower surface are shown. SBcl-2 and 1205Lu cells with or without rAMF treatment were tested for migration using a modified Boyden chamber. D, proliferation assays. The mean optical densities ± S.E. (n = 3) of the melanoma cells are shown. SBcl-2 and 1205Lu cells were cultured on 96-well microtiter plates with or without rAMF (500 ng/ml). The reagent was injected after 0, 24, 48, or 72 h of culture, and the cells were incubated for a further 2 h. The optical densities were detected using a microplate reader.
AMF has been shown to enhance cell proliferation in NIH/fibroblast cell lines (37). Therefore, we investigated the effects of exogenous AMF on the cell proliferation of the melanoma cell lines in vitro. rAMF did not induce cell proliferation in either of the cell lines examined (Fig. 1D). These results indicate that rAMF is capable of inducing cell motility but not cell proliferation in both early stage and metastatic melanoma cells.
Regulation of IL-8 Expression by AMF
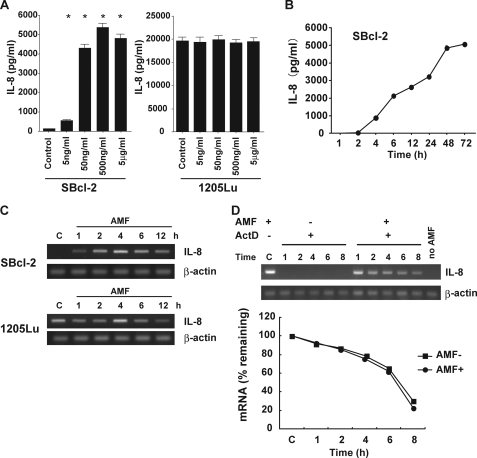
Previous studies have shown that IL-8 stimulates tumor cell migration, especially in melanoma cells (17). To examine whether rAMF induced IL-8 production in our melanoma cell lines, we measured the IL-8 protein levels produced by the melanoma cell lines after rAMF stimulation using an ELISA. In SBcl-2 cells, IL-8 production was significantly augmented after rAMF stimulation in a dose-dependent manner (Fig. 2A). On the other hand, 1205Lu cells exhibited constantly high IL-8 production compared with SBcl-2 cells, and further production of IL-8 was not induced by rAMF stimulation. After rAMF stimulation, the IL-8 protein level in SBcl-2 cells started to increase at 4 h and reached a plateau at 48 h (Fig. 2B). Next, we examined the IL-8 mRNA expression levels in the melanoma cell lines after rAMF stimulation. Melanoma cells were treated with rAMF for the indicated hours in vitro, and their total RNA was extracted for RT-PCR analysis. As shown in Fig. 2C, rAMF stimulation induced significantly high levels of IL-8 mRNA expression in SBcl-2 cells. On the other hand, 1205Lu cells constitutively expressed IL-8 protein before and after rAMF stimulation. These results are consistent with the IL-8 protein data and demonstrate that rAMF can increase IL-8 expression at both the mRNA and protein levels in early stage melanoma cells but not metastatic cells.
FIGURE 2.
AMF induces IL-8 protein production and IL-8 mRNA transcription. A, mean IL-8 protein concentrations ± S.E. (n = 3) detected by ELISA in cell culture media in response to stimulation with rAMF (500 ng/ml). SBcl-2 and 1205Lu cells were incubated with various concentrations of rAMF for 12 h. B, mean IL-8 protein concentrations ± S.E. (n = 3) detected by ELISA in cell culture media from untreated SBcl-2 cells and SBcl-2 cells stimulated with rAMF (500 ng/ml). SBcl-2 cells were incubated with or without rAMF and extracted at various times until 96 h. C, representative RT-PCR amplification of IL-8. SBcl-2 cells were stimulated with rAMF (500 ng/ml) and collected after 1, 2, 4, 6, and 12 h. C, control samples. D (top), IL-8 mRNA stability. SBcl-2 cells were exposed to rAMF for 4 h and then maintained in fresh medium containing 5 μg/ml ActD (−AMF +ActD) or 5 μg/ml ActD plus 500 ng/ml rAMF (+AMF +ActD). The cells were harvested at different time points. Total RNA was extracted, and the IL-8 and β-actin (control) mRNA levels were detected by RT-PCR. D (bottom), mean IL-8 mRNA ratios (n = 3) measured by real-time RT-PCR in cells undergoing the protocols described for the top. The mRNA levels were expressed as percentages relative to those measured after 4 h of rAMF treatment.
To examine the possibility that rAMF increases IL-8 mRNA stability in melanoma cells, SBcl-2 cells were treated with ActD, a transcription inhibitor, for 4 h after rAMF stimulation. The cells were harvested at the indicated hours after ActD addition and analyzed by RT-PCR. In the presence of both ActD alone and ActD plus rAMF, the ratios of the IL-8 mRNA levels did not differ for any of the culture periods (Fig. 2D). These results indicate that the IL-8 induction by AMF is mediated by acceleration of IL-8 mRNA transcription rather than by the promotion of IL-8 mRNA stability in early stage melanoma cells. We also investigated whether AMF stimulation changed the IL-8 receptor expression levels and found that the levels of these receptors remained unchanged after rAMF treatment in both cell lines (data not shown).
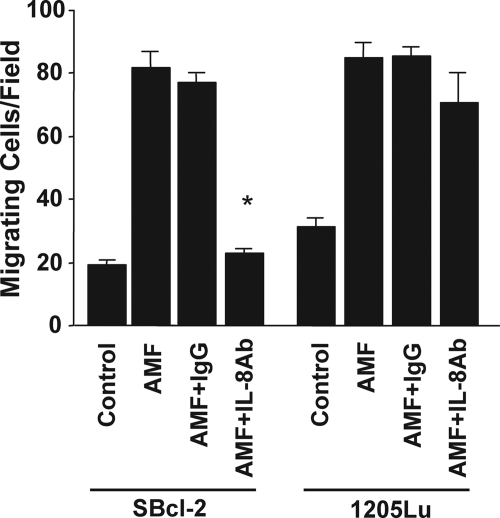
Responses of Melanoma Cells to AMF and Their Inhibition by an Anti-IL-8 Neutralizing Antibody
It is well known that IL-8 stimulates tumor cell migration, especially in melanoma cells (17). To examine whether IL-8 expression in response to rAMF stimulation induced migration in our melanoma cell lines, we performed a modified Boyden chamber assay using an anti-IL-8 neutralizing antibody. As shown in Fig. 3, the migratory ability of rAMF-treated SBcl-2 cells was abolished by treatment with the anti-IL-8 neutralizing antibody, indicating that IL-8 up-regulation by AMF is essential for the induction of migratory ability in early stage melanoma cells. In contrast, the anti-IL-8 neutralizing antibody treatment had no effect on the induction of migratory ability in 1205Lu cells after rAMF treatment, indicating that IL-8 production is dispensable for the induction of migratory ability in metastatic melanoma cells.
FIGURE 3.
AMF-induced IL-8 plays a significant role in melanoma migration. The mean cell counts ± S.E. (n = 10) of migrating SBcl-2 and 1205Lu cells measured using a modified Boyden chamber assay are shown. A neutralizing antibody against IL-8 (20 μg/ml) and a control IgG antibody were included to determine the functional contributions of individual molecules to cell migration. *, p < 0.05 versus rAMF and IgG control antibody-treated cells.
The Signaling Pathway for IL-8 Production Is ERK1/2 Pathway Activation Induced by AMF
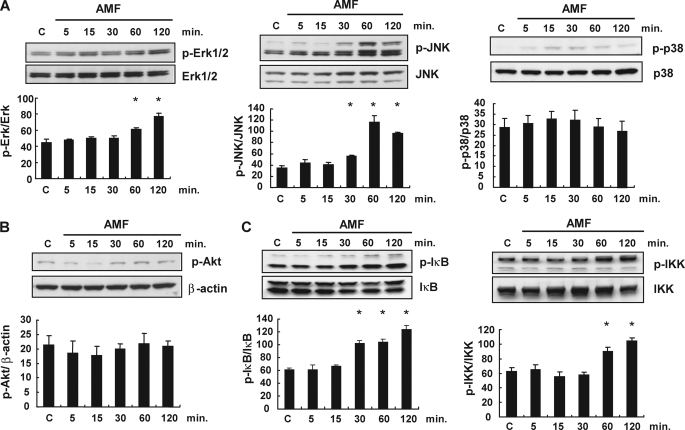
It has been shown that AMF exerts its effects through JNK activation in the MAPK pathway for cell migration and through the phosphatidylinositol 3-kinase pathway for angiogenesis of endothelial cells (8, 38). IL-8 is known to be produced through the MAPK, phosphatidylinositol 3-kinase, and NFκB pathways (39). We investigated the signaling pathway for IL-8 production by AMF in SBcl-2 melanoma cells, including the MAPK pathways. In a Western blotting analysis, both ERK1/2 and JNK phosphorylation were up-regulated in rAMF-treated melanoma cells by 2 h after stimulation (Fig. 4A). AMF stimulation also elicited significant increases in the IκB and IKK phosphorylation levels (Fig. 4C), indicating that NFκB signaling was also activated after rAMF stimulation. AMF stimulation did not activate p38 (Fig. 4A) or Akt (Fig. 4B) phosphorylation in the melanoma cells. These results suggest that rAMF induces activation of MAPK pathways, including the ERK, JNK, and NFκB pathways, in early stage melanoma cells.
FIGURE 4.
Basal and activation statuses of the MAPK, phosphatidylinositol 3-kinase, and NFκB pathways in SBcl-2 cells treated with AMF. SBcl-2 cells were stimulated with rAMF (500 ng/ml), and the cells were extracted after 5, 15, 30, 60, and 120 min. The levels of phosphorylated and total proteins were detected by Western blotting analysis. C, control samples. A, time-dependent changes in the expression levels of MAPKs, phospho-ERK (p-Erk), ERK, phospho-JNK (p-JNK), JNK, phospho-p38, and p38, in response to rAMF stimulation. Densitometric analyses of the data are shown under the Western blotting bands. *, p < 0.05 versus control cells. B, time-dependent changes in the expression levels of phospho-Akt and β-actin in response to rAMF stimulation. C, time-dependent changes in the expression levels of the NFκB pathway, phospho-IκB, IκB, phospho-IKK, and IKK, in response to rAMF stimulation. Densitometric analyses of the data are shown under the Western blotting bands. *, p < 0.05 versus control cells. Data represent the means ± S.E. of triplicate analyses.
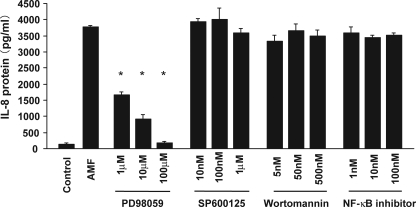
To examine which signaling pathway is critical for IL-8 induction by AMF in early stage melanoma cells, we pretreated SBcl-2 cells with inhibitors of various signaling molecules. Briefly, SBcl-2 cells were pretreated with a MAPK pathway inhibitor, stimulated with rAMF, and analyzed for their IL-8 production by ELISA. Pretreatment with a selective inhibitor of ERK1/2 (PD98059) significantly inhibited AMF-induced IL-8 production in a dose-dependent manner (Fig. 5). In contrast, melanoma cells pretreated with specific inhibitors of JNK (SP600125) or p38 (SB203580) did not alter their IL-8 protein induction by rAMF (Fig. 5) (data not shown). Pretreatment with a selective inhibitor of the NFκB pathway did not influence AMF-induced IL-8 up-regulation in the melanoma cells. Taken together, these findings suggest that ERK1/2 activation is essential for AMF-induced IL-8 up-regulation in melanoma cells.
FIGURE 5.
Roles of the ERK1/2, JNK, phosphatidylinositol 3-kinase, and NFκB pathways in AMF-induced IL-8 protein expression. Mean IL-8 protein concentrations ± S.E. (n = 3) in SBcl-2 cells measured after 12 h of culture without rAMF (Control), with rAMF alone (AMF), or with rAMF plus various concentrations of PD98059 (IC50 = 2 μm), SP600125 (IC50 = 100 nm), wortmannin (IC50 = 5 nm), and NFκB activation inhibitor (IC50 = 7–11 nm). Cells pretreated with the selective inhibitors were incubated for 30 min before rAMF injection. *, p < 0.05 versus AMF alone.
DISCUSSION
The main findings of the present study are as follows: 1) AMF induces a significant increase in IL-8 production in early stage melanoma cells; 2) the increase in IL-8 production is caused by an increase in the IL-8 transcription rate rather than enhanced IL-8 mRNA stability; 3) the ERK1/2 pathway enhances IL-8 transcription in response to AMF; 4) autocrine IL-8 effects are required for AMF-induced melanoma cell migration.
We report here, for the first time, that AMF induces melanoma cells to produce IL-8, which then acts in an autocrine manner to promote the migration of these cells. IL-8 is secreted by various tumor cells, including melanoma cells, in response to cytokines, bacterial or viral products, and cellular stress. Previous studies have reported that melanoma cells express the IL-8 receptors CXCR1/2, which are both G-protein-linked seven-transmembrane proteins, and that IL-8 signaling directly promotes cell migration of tumor cells, which may be relevant to tumor invasion and metastasis (17, 18). The motogenic effects of IL-8 are mediated independently of its effects on cell proliferation. Melanoma cells are also stimulated to migrate by certain cytokines, such as insulin-like growth factor-1. Insulin-like growth factor-1 is known as a strong mediator of tumor progression in melanoma cells (40–42), and the motogenic effects of these cytokines are mediated through autocrine production of IL-8 (43). Angiopoietin-1 is an angiogenic modulator of normal and pathologic cells and exerts its migratory, proliferative, and angiogenic effects in an autocrine system dependent on IL-8 production (44). Therefore, autocrine IL-8 production by normal and tumor cells frequently plays important roles in their migration. These findings support our observations that AMF-induced migration requires autocrine IL-8 production.
IL-8 gene expression is regulated through transcriptional activation and enhanced mRNA stability (39). With respect to IL-8 transcription, many reports have identified NFκB and AP-1 transcription factors as the main regulators of IL-8 promoter activity in response to a variety of stimuli. In the present study, AMF-induced IL-8 production was caused by up-regulation of IL-8 transcription rather than enhanced mRNA stability.
We also investigated the signaling pathway for IL-8 production. In Western blotting analyses, phospho-ERK1/2 were constantly activated, and AMF induced further phosphorylation of ERK1/2. In addition, we found that AMF triggered JNK pathway phosphorylation for IL-8 transcription but did not trigger p38 or Akt phosphorylation. Our findings of attenuation of IL-8 production by an ERK inhibitor suggest that the induction of IL-8 transcription is mediated by the ERK pathway. Metastatic melanoma cells with constant marked IL-8 production also exhibited inhibition of their IL-8 production in response to the ERK inhibitor (data not shown). Taken together, these findings suggest the possibility that these cellular responses, namely AMF-IL-8 signaling effects, accelerate tumor progression by leading to the acquisition of metastatic potential.
Regarding AMF signaling in cell migration, it has been reported that AMF stimulation induces RhoA and Rac1 activation, which in turn leads to activation of JNK activation, a downstream protein of RhoA (8). Our findings suggest that AMF to IL-8 signaling is a novel downstream signaling event of AMF for cell migration and that this signaling is mediated by the ERK pathway. In the present study, we showed that antibody-mediated blockade of IL-8 induction abolished AMF-induced cell migration, suggesting that a different pathway through autocrine production of IL-8 elicits the motogenic effects of AMF. Considering these facts, the induction of the migratory ability by AMF may have two pathways. One pathway indirectly stimulates cellular migration dependent on autocrine IL-8 production, and the other pathway directly stimulates cellular migration through the RhoA and Rac1 pathways. The interaction of these pathways remains unclear and should be elucidated in future investigations.
In the present study, differences in the responses to AMF were detected between early stage and metastatic melanoma cells. Although AMF only induced IL-8 production in the early stage melanoma cells, the metastatic melanoma cells exhibited marked IL-8 production that remained unchanged by AMF stimulation. In fact, the amount of IL-8 production in the metastatic melanoma cells was about 4-fold higher than that in the early stage melanoma cells. We estimated that the metastatic melanoma cells had higher levels of IL-8 receptor expression than the early stage melanoma cells, although we found that the IL-8 receptor mRNA levels did not differ between the two cell lines by RT-PCR analysis (data not shown). In the metastatic melanoma cells, overexpression of IL-8 may cause anergy against IL-8 induction signaling by AMF. Considering that AMF-induced migration was not inhibited by an anti-IL-8 neutralizing antibody in the metastatic cells, once the melanoma cells have become metastatic, AMF signals may directly stimulate cell migration rather than acting through autocrine IL-8 production. We propose the hypothesis that autocrine AMF stimulation induces cellular migration dependent on autocrine IL-8 production and that this induction enhances the malignant potential (migration or invasion), thereby leading to metastasis. This signaling for AMF-induced migration through IL-8 autocrine production may represent a new target for cancer therapies toward preventing cancer metastatic progression.
In the present study, we have shown for the first time that AMF induces significant IL-8 production in melanoma cells, especially in early stage melanoma cells, as a result of increased IL-8 transcription. These effects are mediated through activation of the ERK1/2 pathway. Finally, enhanced IL-8 production plays an important role in mediating AMF-induced melanoma cell migration, and we propose that these cellular responses accelerate the progression to acquisition of malignant potential in malignant tumors.
Acknowledgments
We thank Dr. Meenhard Herlyn for supplying the SBcl-2 melanoma cell line, and we thank Dr. Hitoshi Kimura for his special efforts in this research work.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and the Support Program for Improving Graduate School Education from MEXT.
- AMF
- autocrine motility factor
- AMFR
- AMF receptor
- rAMF
- recombinant AMF
- JNK
- Jun N-terminal kinase
- ERK
- extracellular signal-regulated kinase
- IL
- interleukin
- ELISA
- enzyme-linked immunosorbent assay
- RT
- reverse transcription
- ActD
- actinomycin D
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MAPK
- mitogen-activated protein kinase.
REFERENCES
- 1.Achari A., Marshall S. E., Muirhead H., Palmieri R. H., Noltmann E. A. (1981)Philos. Trans. R. Soc. Lond. B Biol. Sci. 293,145–157 [DOI] [PubMed] [Google Scholar]
- 2.Liotta L. A., Mandler R., Murano G., Katz D. A., Gordon R. K., Chiang P. K., Schiffmann E. (1986)Proc. Natl. Acad. Sci. U.S.A. 83,3302–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silletti S., Watanabe H., Hogan V., Nabi I. R., Raz A. (1991)Cancer Res. 51,3507–3511 [PubMed] [Google Scholar]
- 4.Shimizu K., Tani M., Watanabe H., Nagamachi Y., Niinaka Y., Shiroishi T., Ohwada S., Raz A., Yokota J. (1999)FEBS Lett. 456,295–300 [DOI] [PubMed] [Google Scholar]
- 5.Timar J., Trikha M., Szekeres K., Bazaz R., Tovari J., Silletti S., Raz A., Honn K. V. (1996)Cancer Res. 56,1902–1908 [PubMed] [Google Scholar]
- 6.Watanabe H., Takehana K., Date M., Shinozaki T., Raz A. (1996)Cancer Res. 56,2960–2963 [PubMed] [Google Scholar]
- 7.Kohn E. C., Liotta L. A., Schiffmann E. (1990)Biochem. Biophys. Res. Commun. 166,757–764 [DOI] [PubMed] [Google Scholar]
- 8.Tsutsumi S., Gupta S. K., Hogan V., Collard J. G., Raz A. (2002)Cancer Res. 62,4484–4490 [PubMed] [Google Scholar]
- 9.Luscinskas F. W., Kiely J. M., Ding H., Obin M. S., Hébert C. A., Baker J. B., Gimbrone M. A., Jr. (1992)J. Immunol. 149,2163–2171 [PubMed] [Google Scholar]
- 10.Norgauer J., Metzner B., Schraufstätter I. (1996)J. Immunol. 156,1132–1137 [PubMed] [Google Scholar]
- 11.Colombo M. P., Maccalli C., Mattei S., Melani C., Radrizzani M., Parmiani G. (1992)Melanoma Res. 2,181–189 [DOI] [PubMed] [Google Scholar]
- 12.Schadendorf D., Möller A., Algermissen B., Worm M., Sticherling M., Czarnetzki B. M. (1993)J. Immunol. 151,2667–2675 [PubMed] [Google Scholar]
- 13.Freund A., Chauveau C., Brouillet J. P., Lucas A., Lacroix M., Licznar A., Vignon F., Lazennec G. (2003)Oncogene 22,256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatakrishnan G., Salgia R., Groopman J. E. (2000)J. Biol. Chem. 275,6868–6875 [DOI] [PubMed] [Google Scholar]
- 15.Wigmore S. J., Fearon K. C., Maingay J. P., Lai P. B., Ross J. A. (1997)Am. J. Physiol. 273,E720–E726 [DOI] [PubMed] [Google Scholar]
- 16.Brat D. J., Bellail A. C., Van Meir E. G. (2005)Neuro. Oncol. 7,122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J. M., Taraboletti G., Matsushima K., Van Damme J., Mantovani A. (1990)Biochem. Biophys. Res. Commun. 169,165–170 [DOI] [PubMed] [Google Scholar]
- 18.Singh R. K., Gutman M., Radinsky R., Bucana C. D., Fidler I. J. (1994)Cancer Res. 54,3242–3247 [PubMed] [Google Scholar]
- 19.Baggiolini M., Clark-Lewis I. (1992)FEBS Lett. 307,97–101 [DOI] [PubMed] [Google Scholar]
- 20.Kasahara T., Mukaida N., Yamashita K., Yagisawa H., Akahoshi T., Matsushima K. (1991)Immunology 74,60–67 [PMC free article] [PubMed] [Google Scholar]
- 21.Aihara M., Tsuchimoto D., Takizawa H., Azuma A., Wakebe H., Ohmoto Y., Imagawa K., Kikuchi M., Mukaida N., Matsushima K. (1997)Infect. Immun. 65,3218–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbie S., Chen L. M., Davis R. J., Galán J. E. (1997)J. Immunol. 159,5550–5559 [PubMed] [Google Scholar]
- 23.DeForge L. E., Preston A. M., Takeuchi E., Kenney J., Boxer L. A., Remick D. G. (1993)J. Biol. Chem. 268,25568–25576 [PubMed] [Google Scholar]
- 24.Sonoda Y., Kasahara T., Yamaguchi Y., Kuno K., Matsushima K., Mukaida N. (1997)J. Biol. Chem. 272,15366–15372 [DOI] [PubMed] [Google Scholar]
- 25.Verschraegen C. F., Giovanella B. C., Mendoza J. T., Kozielski A. J., Stehlin J. S., Jr. (1991)Anticancer Res. 11,529–535 [PubMed] [Google Scholar]
- 26.Satyamoorthy K., DeJesus E., Linnenbach A. J., Kraj B., Kornreich D. L., Rendle S., Elder D. E., Herlyn M. (1997)Melanoma Res. 7,Suppl. 2,S35– S42 [PubMed] [Google Scholar]
- 27.Juhasz I., Albelda S. M., Elder D. E., Murphy G. F., Adachi K., Herlyn D., Valyi-Nagy I. T., Herlyn M. (1993)Am J. Pathol. 143,528–537 [PMC free article] [PubMed] [Google Scholar]
- 28.Haga A., Niinaka Y., Raz A. (2000)Biochim. Biophys. Acta 1480,235–244 [DOI] [PubMed] [Google Scholar]
- 29.Neudauer C. L., McCarthy J. B. (2003)Exp. Cell Res. 286,128–137 [DOI] [PubMed] [Google Scholar]
- 30.Shimoda K., Begum N. A., Shibuta K., Mori M., Bonkovsky H. L., Banner B. F., Barnard G. F. (1998)Hepatology 28,108–115 [DOI] [PubMed] [Google Scholar]
- 31.Furuya M., Nishiyama M., Kimura S., Suyama T., Naya Y., Ito H., Nikaido T., Ishikura H. (2004)J. Pathol. 203,551–558 [DOI] [PubMed] [Google Scholar]
- 32.Tikhonov I., Doroshenko T., Chaly Y., Smolnikova V., Pauza C. D., Voitenok N. (2001)Clin. Exp. Immunol. 125,414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haga A., Funasaka T., Niinaka Y., Raz A., Nagase H. (2003)Int. J. Cancer 107,707–714 [DOI] [PubMed] [Google Scholar]
- 34.Funasaka T., Hu H., Yanagawa T., Hogan V., Raz A. (2007)Cancer Res. 67,4236–4243 [DOI] [PubMed] [Google Scholar]
- 35.Tsutsumi S., Yanagawa T., Shimura T., Kuwano H., Raz A. (2004)Clin. Cancer Res. 10,7775–7784 [DOI] [PubMed] [Google Scholar]
- 36.Schaider H., Oka M., Bogenrieder T., Nesbit M., Satyamoorthy K., Berking C., Matsushima K., Herlyn M. (2003)Int. J. Cancer 103,335–343 [DOI] [PubMed] [Google Scholar]
- 37.Tsutsumi S., Yanagawa T., Shimura T., Fukumori T., Hogan V., Kuwano H., Raz A. (2003)J. Biol. Chem. 278,32165–32172 [DOI] [PubMed] [Google Scholar]
- 38.Funasaka T., Haga A., Raz A., Nagase H. (2002)Int. J. Cancer 101,217–223 [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002)J. Leukocyte Biol. 72,847–855 [PubMed] [Google Scholar]
- 40.Herlyn M., Thurin J., Balaban G., Bennicelli J. L., Herlyn D., Elder D. E., Bondi E., Guerry D., Nowell P., Clark W. H. (1985)Cancer Res. 45,5670–5676 [PubMed] [Google Scholar]
- 41.Stracke M. L., Kohn E. C., Aznavoorian S. A., Wilson L. L., Salomon D., Krutzsch H. C., Liotta L. A., Schiffmann E. (1988)Biochem. Biophys. Res. Commun. 153,1076–1083 [DOI] [PubMed] [Google Scholar]
- 42.Stracke M. L., Engel J. D., Wilson L. W., Rechler M. M., Liotta L. A., Schiffmann E. (1989)J. Biol. Chem. 264,21544–21549 [PubMed] [Google Scholar]
- 43.Satyamoorthy K., Li G., Vaidya B., Kalabis J., Herlyn M. (2002)Cell Growth Differ. 13,87–93 [PubMed] [Google Scholar]
- 44.Abdel-Malak N. A., Srikant C. B., Kristof A. S., Magder S. A., Di Battista J. A., Hussain S. N. (2008)Blood 111,4145–4154 [DOI] [PubMed] [Google Scholar]