Abstract
Isothiocyanates are a class of phytochemicals with widely reported anti-cancer and anti-inflammatory activity. However, knowledge of their activity at a molecular level is limited. The objective of this study was to identify biological targets of phenethyl isothiocyanate (PEITC) using an affinity purification approach. An analogue of PEITC was synthesized to enable conjugation to a solid-phase resin. The pleiotropic cytokine macrophage migration inhibitory factor (MIF) was the major protein captured from cell lysates. Site-directed mutagenesis and mass spectrometry showed that PEITC covalently modified the N-terminal proline residue of MIF. This resulted in complete loss of catalytic tautomerase activity and disruption of protein conformation, as determined by impaired recognition by a monoclonal antibody directed to the region that receptors and interacting proteins bind to MIF. The conformational change was supported by in silico modeling. Monoclonal antibody binding to plasma MIF was disrupted in humans consuming watercress, a major dietary source of PEITC. The isothiocyanates have significant potential for development as MIF inhibitors, and this activity may contribute to the biological properties of these phytochemicals.
Introduction
Isothiocyanates are a class of phytochemicals with recognized anti-cancer activity. They can act in a chemopreventive capacity via inhibition of carcinogen-activating phase I enzymes (1) and induction of phase II detoxification enzymes (2). Isothiocyanates are also active in the post-initiation phase of tumorigenesis and are, therefore, proposed to have chemotherapeutic potential (3, 4). Isothiocyanate-mediated disruption of cancer progression is achieved by a variety of mechanisms including modulation of cell growth (5), inhibition of angiogenesis (6), suppression of metastasis (7), and induction of apoptosis (8, 9). Isothiocyanates can also modulate inflammatory pathways via inhibition of the transcription factor nuclear factor κB (10).
The electrophilic carbon residue in the isothiocyanate moiety (-N C
C S) is capable of reacting with biological nucleophiles such as cysteine in proteins and the tripeptide glutathione (11, 12). Binding of isothiocyanates to Kelch-like ECH-associated protein 1 (Keap1) (13), transient receptor potential channels (14), MEKK1 protein kinase (15), and tubulin (16) has been demonstrated to occur via covalent modification of cysteine. Reaction with amines to form stable thiourea derivatives can also occur. However, this is generally considered to be a less favorable reaction at physiological pH (11).
S) is capable of reacting with biological nucleophiles such as cysteine in proteins and the tripeptide glutathione (11, 12). Binding of isothiocyanates to Kelch-like ECH-associated protein 1 (Keap1) (13), transient receptor potential channels (14), MEKK1 protein kinase (15), and tubulin (16) has been demonstrated to occur via covalent modification of cysteine. Reaction with amines to form stable thiourea derivatives can also occur. However, this is generally considered to be a less favorable reaction at physiological pH (11).
To elucidate the major cellular targets of biologically active isothiocyanates, we have utilized an affinity-based target identification approach. An amine linker was added to phenethyl isothiocyanate (PEITC)3 without compromising cytotoxicity, and the molecule was immobilized to a solid phase resin. The pleiotropic cytokine macrophage migration inhibitory factor (MIF) was identified as a major biological target of PEITC. Using mass spectrometry and site-directed mutagenesis, we identified the N-terminal proline of MIF as the target residue and have shown that conjugation disrupts the catalytic tautomerase activity of MIF and conformational integrity of the protein in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Synthesis of Amino-PEITC
Amino-PEITC (2-(3-(2-aminoethyl)phenyl)ethyl isothiocyanate) was prepared from the diamine precursor (1,3-bis(2-aminoethyl)benzene) (17) by selective protection as a mono-t-Boc derivative (18) followed by reaction with thiophosgene (19) and deprotection. 1,3-Bis(2-aminoethyl)benzene was obtained by reduction of 1,3-bis(cyanomethyl)benzene (20) with a nickel-modified borohydride reagent (21). NMR spectra were obtained on a Varian Unity 500 spectrometer.
A solution of the bistrifluoroacetate salt of 1,3-bis(2-aminoethyl)benzene (17) (291 mg, 0.74 mmol) and triethylamine (205 μl, 1.48 mmol) in dry methanol (15 ml) was stirred at room temperature for 10 min, then a solution of di-t-butyl dicarbonate (41.3 mg, 0.19 mmol) in methanol (10 ml) was added dropwise over 5 min, and the solution was stirred at room temperature for 24 h. The solvents were removed in vacuo, and the yellow residue was washed successively with diethyl ether, ethyl acetate, and dichloromethane (5 ml). The combined organic extracts were washed with saturated aqueous sodium bicarbonate and evaporated to give the mono t-Boc amine (67 mg) as a yellow oil. 1H NMR (500 MHz, chloroform-d1) δ 7.24 (1H, t, J = 7.5 Hz, ArH), 7.1-7.0 (3H, m, ArH), 3.35 (2H, m, CH2NHCO), 2.96 (2H, t, J = 7 Hz, CH2NH2), 2.78 (2H, broad t, J = 6.5 Hz, CH2CH2NHCO), 2.73 (2H, t, J = 7 Hz, CH2CH2NH2), 1.43 (9H, s, C(CH3)3) ppm.
The crude t-Boc amine (67 mg) was dissolved in dichloromethane (6 ml) and partitioned with saturated aqueous sodium bicarbonate (3 ml) at 0 °C for 5 min. Thiophosgene (19.3 μl, 0.25 mmol) was then added to the organic layer, and the mixture was stirred at 0 °C for 20 min. The dichloromethane was then separated and evaporated in vacuo to give a yellow oil (69 mg) which was filtered through a column of silica gel (2 g) with 1:1 petroleum ether/diethyl ether and evaporated to give the t-Boc isothiocyanate as a clear oil (56 mg). 1H NMR (500 MHz, chloroform-d1) δ 7.28 (1H, t, J = 7.5 Hz, ArH), 7.12-7.0 (3H, m, ArH), 3.72 (2H, t, J = 6.5 Hz, CH2NCS), 3.38 (2H,m, CH2NHCO), 2.97 (2H, t, J = 7Hz, CH2CH2NCS), 2.81 (2H, m, CH2CH2NHCO), 1.44 (9H, s, C(CH3)3) ppm.
The crude t-Boc isothiocyanate (56 mg) was dissolved in dichloromethane (3 ml) and stirred with trifluoroacetic acid (0.5 ml) for 3 h. Evaporation of the solvents at room temperature in vacuo gave the product, as the trifluoroacetate salt, in the form of an orange oil (53 mg). One peak detected at 270 nm on a Jasco PU 980 HPLC system with a reverse phase HPLC column (Prodigy 5-μm C18 column, 250 × 4 mm, Phenomenex, San José, CA) eluted with 50% acetonitrile, 50% water containing 0.1% trifluoroacetic acid at a rate of 1 ml/min. 1H NMR (500 MHz, chloroform-d1) δ 7.6 (3H, broad, NH3), 7.33 (1H, t, J = 7.5 Hz, ArH), 7.14 (2H, broad t, J = 8 Hz, ArH), 7.09 (1H, broad s, ArH), 3.72 (2H, t, J = 6.5 Hz, CH2NCS), 3.30 (2H,m, CH2NH3), 3.0 (4H, m, CH2CH2NCS + CH2CH2NH3) ppm. IR (film) υmax 2186, 2113 (N C
C S). 1780 (C
S). 1780 (C O) cm−1. Electrospray ionisation mass spectrometry (Bruker Daltonics MicroTOF, positive ion) mass to charge ratio found that 207.0946 C11H15N2S requires 207.0950.
O) cm−1. Electrospray ionisation mass spectrometry (Bruker Daltonics MicroTOF, positive ion) mass to charge ratio found that 207.0946 C11H15N2S requires 207.0950.
Cell Culture
The Jurkat T-lymphocyte cell line was obtained from American Type Culture Collection (Manassas, VA) and was maintained in RPMI 1640 containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were grown at 37 °C in a humidified atmosphere with 5% CO2.
Cytotoxicity Assay
Plasma membrane integrity was monitored using propidium iodide staining. After a 24-h treatment with isothiocyanates propidium iodide (5 μg) was added to cells, and samples were allowed to incubate in the dark for 10 min before cell fluorescence was measured using a FC500 MPL Flow Cytometry system (Beckman Coulter Inc., Fullerton, CA).
Preparation of Affi-PEITC
100 μl of Affi-Gel® 10 activated immunoaffinity support (Bio-Rad) was thoroughly washed with 0.1 m NaHCO3 and resuspended in 500 μl of the same solution. The Affi-Gel suspension was reacted with 7 μl of amino-PEITC (100 mg/ml in DMSO) for 1 h at room temperature with constant rotation. The remaining reactive ester groups were blocked by the addition of 50 μl of 1 m ethanolamine, pH 8.0. The suspension was then incubated for an additional 1 h at room temperature with constant rotation. An unreactive resin (blocked Affi-Gel) was prepared by incubating 100 μl of AffiGel-10 in 500 μl of 0.1 m NaHCO3 containing 50 μl of 1 m ethanolamine, pH 8.0, for 2 h with constant rotation. The resulting Affi-PEITC and blocked Affi-Gel preparations were thoroughly washed with binding buffer (0.1 m KCl, 20 mm HEPES, pH 7.6, 0.1 m EDTA, 0.1% Nonidet P-40, 0.25 mm phenylmethylsulfonyl fluoride) before use.
Identification of Intracellular Targets of PEITC
Jurkat cells were collected and lysed in a buffer consisting of 25 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 10 mm MgCl2, 1 mm EDTA, 10% glycerol and Complete™ protease inhibitors (Roche Diagnostics). Protein concentration was determined using a Dc protein assay (Bio-Rad) and adjusted to 2 mg/ml with additional buffer. Affi-PEITC and blocked Affi-Gel were incubated with 1 ml of cell lysate for 1 h at room temperature with constant rotation. After incubation, the resins were thoroughly washed with binding buffer. Bound protein was eluted by boiling the resin in the presence of 100 μl of reducing sample buffer (62.5 mm Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.025% bromphenol blue, and 700 mm β-mercaptoethanol). The resin was pelleted, and the resulting supernatant was resolved by SDS-PAGE. Total protein was stained with SYPRO® Ruby (Invitrogen) and visualized using a Molecular Imager® FX (Bio-Rad). Bands of interest were excised from the gel and digested, and the resulting peptide fragments were analyzed by MALDI-TOF mass spectrometry (Centre for Protein Research, University of Otago, Dunedin, New Zealand).
Mass Spectrometry of Purified MIF
Recombinant human MIF and MIF mutant proteins were expressed, purified, and renatured from pET11b vector as previously described (22). 10 μg of recombinant human MIF and mutant MIF proteins were reacted with a 10-fold molar excess of PEITC (Sigma) for 20 min. Samples were passed through spin columns pre-equilibrated with water and then diluted 1:1 with acetonitrile containing 0.1% formic acid. Mass spectrometry was performed using an LCQ™ DECA XPplus ion trap instrument (ThermoFinnigan, San Jose, CA). Samples were directly infused using a Hamilton syringe at a flow rate of 5 μl/min. A full scan for the mass range 100–2000 m/z was monitored. Data were collected for 1 min before deconvolution using BioworksBrowser 3.1 SR1 (ThermoFinnigan, San Jose, CA).
Immunoblot Analysis of MIF
Samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween 20 (TBST20). Blots were probed with goat anti-human MIF antibody (0.2 μg/ml) or mouse anti-human MIF antibody (clone 12302, 2 μg/ml) (R&D Systems, Minneapolis, MN) in TBST20 containing 2% skim milk. Proteins were visualized using horseradish peroxidase-conjugated secondary antibodies and the ECL™ system (GE Healthcare). Images were obtained using a ChemiDoc™ XRS system (Bio-Rad).
Recombinant Human MIF (rhMIF) Tautomerase Activity Assay
A l-dopachrome methyl ester solution was prepared just before use by combining 72 μl of a sodium periodate stock (20 mm) with 108 μl of l-3,4-dihydroxyphenylalanine methyl ester solution (4 mm) in 1620 μl of sodium phosphate buffer (10 mm sodium phosphate, 1 mm EDTA, pH 6.2). rhMIF was diluted to 1 μm with sodium phosphate buffer, and 20-μl aliquots were transferred to a 96-well plate. Appropriate dilutions of PEITC were added and incubated at room temperature before 180 μl of dopachrome solution was added to all wells, and the change in absorbance at 475 nm due to dopachrome tautomerization was monitored for 2 min. A blank containing sodium phosphate buffer was included in all experiments.
Cellular MIF Tautomerase Activity Assay
After treatment, 1.5 × 106 Jurkat cells were collected and resuspended in 60 μl of lysis buffer consisting of 40 mm HEPES, pH 7.4, containing 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1.6 mg/ml Complete™ protease inhibitors, and 1% CHAPS (Roche Diagnostics). Insoluble material was removed by centrifugation at 15,000 × g for 4 min. Cell extracts (25 μl) were transferred to a 96-well plate in duplicate. 200 μl of dopachrome solution was added to each well, and the change in absorbance at 475 nm was monitored for 2 min. A blank containing lysis buffer was included in all experiments.
The Effect of Isothiocyanate Exposure on Binding of MIF to Affi-PEITC
Jurkat cells were treated with PEITC for 30 min before lysis in a buffer containing 25 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 10 mm MgCl2, 1 mm EDTA, 10% glycerol and Complete™ protease inhibitors. Protein concentration was determined and adjusted to 2 mg/ml with additional buffer. Affi-PEITC or blocked Affi-Gel was incubated with 1 ml of cell lysate preparations for 1 h at room temperature with constant rotation. After incubation the resins were thoroughly washed with binding buffer. Bound protein was eluted by boiling the resin in reducing sample buffer. The resin was pelleted, and the resulting supernatant was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Membranes were blocked with 5% skim milk in TBST20. Blots were probed with goat anti-human MIF antibody (0.2 μg/ml). Proteins were visualized as previously described, and Quantity One® software (Bio-Rad) was utilized to quantify MIF binding to Affi-PEITC.
Molecular Modeling Studies
Isothiocyanates were built within sybyl8.0.3 using sketcher and minimized using MMFF forcefield with 1000 iterations of conjugate gradient method. Modified isothiocyanate compounds were docked into an active site of the homotrimer of MIF covalently bound to inhibitor 4-iodo-6-phenylpyrimidine (PDB code 3B9S) using GOLD4.0.1 using the covalent bond constraint.
Immunoreactivity of rhMIF and Jurkat Cell MIF by Quantitative ELISA
rhMIF was reacted with PEITC for 10 min before detectable MIF levels were determined by a commercial ELISA (R&D Systems) according to the manufacturer's instructions. 106 Jurkat cells (106 cells/ml) were resuspended in fresh media and incubated for 4 h at 37 °C before the addition of 15 μm PEITC for 1 h. A control sample was prepared by resuspending Jurkat cells in media and incubating for 5 h at 37 °C. After treatment, cells were pelleted by centrifugation at 10,000 × g for 1 min. Media was collected and stored before cells were lysed in 1 ml of water and clarified by centrifugation at 15,000 × g for 4 min. Detectable levels of both extracellular and intracellular MIF were determined by commercial ELISA according to manufacturer's instructions.
Immunoreactivity of MIF by Immunoblotting
1 × 106 Jurkat cells were treated with PEITC for 1 h before cells were lysed in a buffer consisting of 40 mm HEPES, pH 7.4, containing 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1.6 mg/ml Complete™ protease inhibitors, and 1% CHAPS (Roche Diagnostics). Insoluble material was removed by centrifugation at 15,000 × g for 4 min. Samples were diluted in reducing sample buffer and resolved by SDS-PAGE before immunoblot analysis as described above.
Ex Vivo Immunoreactivity of MIF after Ingestion of Watercress
Blood was obtained from healthy human volunteers under approval of the Upper South A Regional Ethics Committee. Volunteers consumed 50 g of fresh watercress, and blood was drawn into heparinized tubes just before and 1 and 2 h after ingestion. Plasma was prepared by immediate centrifugation of the blood samples at 1000 × g for 10 min and stored at −80 °C. Plasma MIF levels were determined by a commercial ELISA according to the manufacturer's instructions.
Quantitative Determination of Plasma Isothiocyanate/Dithiocarbamate Levels
Plasma isothiocyanate and dithiocarbamate derivatives were determined by a cyclocondensation reaction as previously described (23). Fractions were analyzed using a Waters 2690 HPLC system (Waters, Milford, MA) with a reverse phase HPLC column (Luna 5-μm C18 column, 250 × 4.6 mm, Phenomenex) and eluted with 80% methanol, 20% water at a rate of 0.6 ml/min. The 1,3-benzodithiole-2-thione was eluted at ∼12 min, and the peak was detected and integrated by a Photodiode array detector (Waters Model 996) at 365 nm. The instrument was calibrated with pure 1,3-benzodithiole-2-thione, and plasma samples spiked with known concentrations of isothiocyanates were included to ensure completion of the cyclocondensation reaction.
RESULTS
Identification of Protein Targets of PEITC
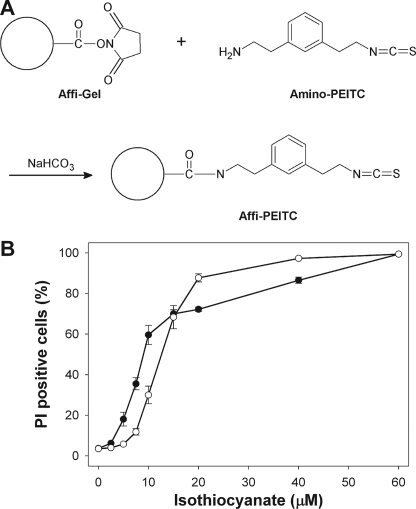
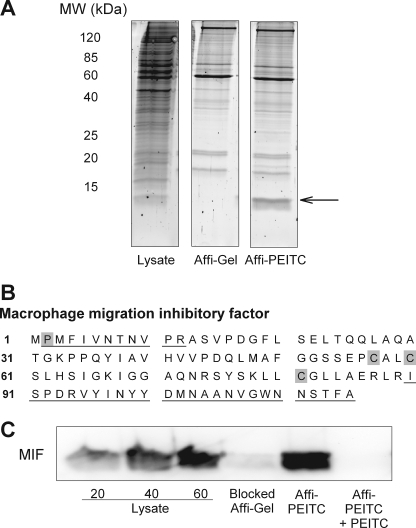
A novel analogue of PEITC, amino-PEITC, containing a meta-aminoethyl group was synthesized to enable coupling to Affi-Gel®, a solid-phase resin containing reactive N-hydroxysuccinimide ester groups (Fig. 1A). Amino PEITC retained cytotoxicity, albeit with slightly reduced potency compared with PEITC (Fig. 1B). Amino-PEITC was conjugated to Affi-Gel®, and lysates from Jurkat T-lymphoma cells were incubated with the resin. Proteins that bound to the matrix were eluted and separated by SDS-PAGE. A prominent band with an apparent molecular mass of 12 kDa was consistently captured from the lysates (Fig. 2A). This band was only faint in the whole cell lysate, indicating considerable concentration during capture, and it was absent when lysates were incubated with an unreactive resin (blocked Affi-Gel), indicating that the interaction was not due to nonspecific binding to the matrix itself. MALDI-TOF mass spectrometry analysis of the excised band identified the protein as the 12.5-kDa pro-inflammatory and pro-tumorigenic cytokine MIF (Fig. 2B). As confirmation, eluates were resolved by SDS-PAGE and immunoblotted with a polyclonal anti-MIF antibody. Whereas antibody binding was absent in the elution fraction from blocked Affi-Gel resin (Fig. 2C), MIF was clearly shown to associate with the Affi-PEITC resin. Pretreatment of lysates with PEITC before incubation with Affi-PEITC prevented the capture of MIF, indicating that isothiocyanate modification was sufficient to inhibit subsequent binding of protein to the resin. Pretreatment of lysates with the isothiocyanate sulforaphane before incubation with Affi-PEITC also prevented the capture of MIF (data not shown).
FIGURE 1.
A, amino-PEITC, a derivative of PEITC, was synthesized and conjugated to a solid phase resin to generate the Affi-PEITC probe. B, cytotoxicity of amino-PEITC and PEITC is shown. Jurkat cells were exposed to amino-PEITC or PEITC for 24 h. Loss of cell viability was assessed by flow cytometric determination of the uptake of propidium iodide (PI). Values are the mean ± S.E. from four experiments.
FIGURE 2.
A, Affi-PEITC or a non-reactive Affi-Gel resin equivalent was incubated with a Jurkat cell lysate. Protein bound to the resins was eluted and resolved by SDS-PAGE. The highlighted protein band was excised from the gel and identified by MALDI-TOF mass spectrometry to be the 12.5-kDa cytokine MIF. B, matched peptides (underlined) were identified by MALDI-TOF mass spectrometry. A 33% coverage of the MIF sequence was positively identified. Cysteine residues and the N-terminal proline are highlighted. C, immunoblot detection of MIF binding to Affi-is PEITC is shown. Binding of a polyclonal anti-human MIF antibody to increasing amounts of a Jurkat cell lysate (lanes 1–3), an eluate recovered from blocked Affi-Gel (lane 4), an eluate from Affi-PEITC (lane 5), and an eluate recovered from Affi-PEITC when the lysate was preincubated with PEITC (lane 6) is shown. All gels are representative of results from at least three experiments.
Covalent Modification of MIF by PEITC
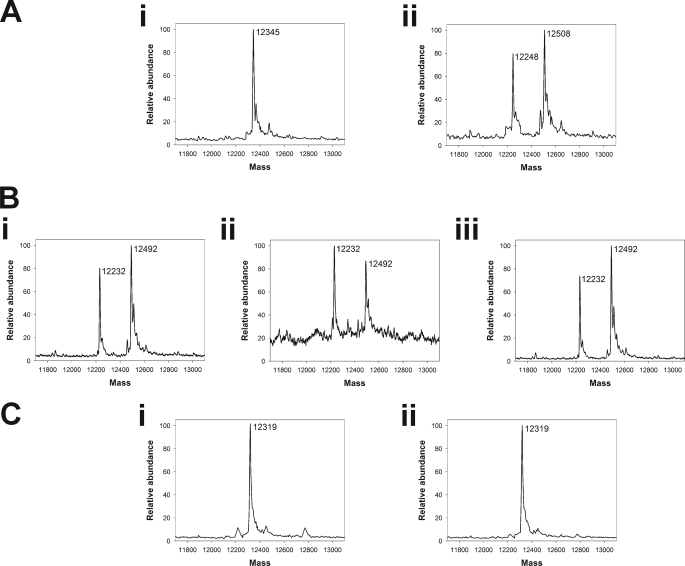
To assess the stoichiometry and site of isothiocyanate binding to MIF, rhMIF was incubated with PEITC, and adduct formation was monitored by mass spectrometry. Coincubation of rhMIF with PEITC resulted in a species with a mass corresponding to the addition of one PEITC (Fig. 3A). MIF has three cysteine residues (Fig. 2B), but rhMIF proteins with C57S, C60S, and C81S mutations were still able to bind PEITC (Fig. 3B), suggesting an alternate site of modification. In all of the isothiocyanate-treated samples, a second peak was present with a mass 97 Da less than the parent protein. We proposed that this resulted from loss of proline and tested the ability of PEITC to bind rhMIF in which the N-terminal proline was mutated to alanine (P2A). P2A rhMIF did not bind PEITC, and the truncated product was absent (Fig. 3C), suggesting that in native MIF the N-terminal proline is cleaved in an Edman degradation reaction. The cleavage itself is likely to be an in vitro artifact driven by acidification in the buffer used for positive ion mass spectrometry. Indeed, in the absence of acid, a single peak with a mass equivalent to PEITC-modified rhMIF was observed (see the supplemental figure).
FIGURE 3.
rhMIF was reacted with a 10-fold excess of PEITC for 10 min before analysis of PEITC-adduct formation by mass spectrometry. A, shown is wild type rhMIF before (i) and after (ii) incubation with PEITC. A peak with a mass of 12,508 Da, corresponding to MIF (12,345 Da) with the addition of 1 PEITC (163 Da) resulted. In addition, a peak with a mass of 12,248 Da was also apparent. B, shown is rhMIF with a C60S mutation (i), a C57S mutation (ii) or a C81S mutation (iii) (12329 Da) after incubation with PEITC. In all cases, a peak with a mass of 12,492 Da, corresponding to cysteine mutant MIF with addition of one PEITC, resulted. In addition, a peak with a mass of 12,232 Da was also formed. C, shown is P2A rhMIF before (i) and after (ii) incubation with PEITC. Only the mass of the parent protein (12,319 Da) was observed.
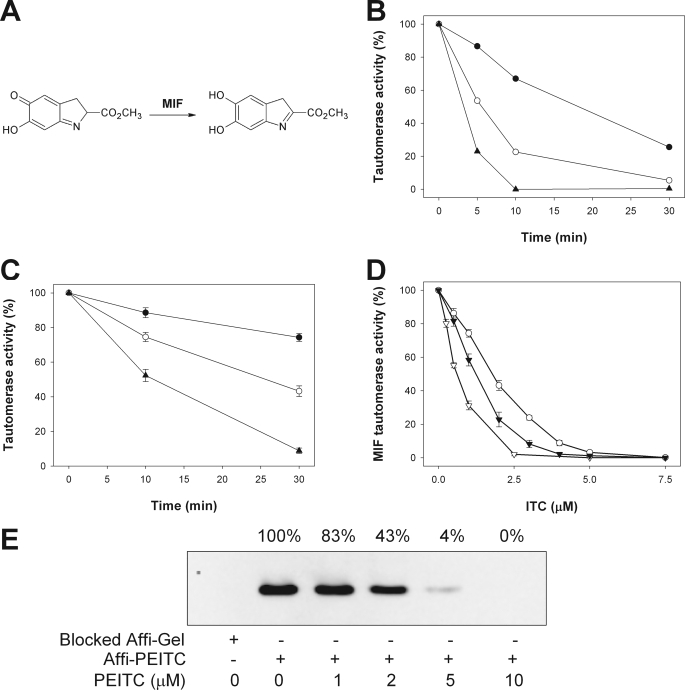
The Effect of Isothiocyanate Adduct Formation on MIF Tautomerase Activity
MIF possesses a catalytic tautomerase activity and can convert the methyl ester of l-dopachrome to an indole derivative (Fig. 4A). The N-terminal proline lies within the tautomerase active site and plays a critical role as a catalytic residue in the tautomerase reaction (24). Coincubation of rhMIF with PEITC resulted in a dose- and time-dependent loss of MIF tautomerase activity (Fig. 4B). PEITC also inhibited MIF tautomerase activity in Jurkat cells in a time- and concentration-dependent manner with a half-maximal inhibitory concentration (IC50) of 2 μm after a 30-min treatment (Fig. 4C). Additional naturally occurring isothiocyanates were shown to inhibit cellular MIF tautomerase activity with IC50 values of 1.3 μm for sulforaphane and 0.6 μm for benzyl isothiocyanate (Fig. 4D). When cells were preincubated with PEITC before affinity capture, a dose-dependent inhibition of MIF binding was observed (Fig. 4E). The concentrations at which capture was disrupted were very similar to the concentrations required to inhibit tautomerase activity. Consistent with the purified system, these results indicate that PEITC directly modifies cellular MIF and inhibits its tautomerase activity.
FIGURE 4.
A, MIF catalyzes the tautomerization of the chromogenic substrate dopachrome methyl ester to a colorless indole derivative. B, the time course of the inhibition of rhMIF tautomerase activity by 10 μm (●), 20 μm (○), and 30 μm (▴) PEITC is shown. Results are representative of three experiments. C, time course of the inhibition of cellular MIF tautomerase activity by 1 μm (●), 2 μm (○), and 4 μm (▴) PEITC is shown. Values are the mean ± S.E. of three experiments. D, concentration dependence of the inhibition of cellular MIF tautomerase activity after a 30-min exposure to PEITC (○), sulforaphane (▾), or benzyl isothiocyanate (▿) is shown. Values are the mean ± S.E. of at least three experiments. E, Jurkat cells were treated with increasing concentrations of PEITC for 30 min. Cell lysates were prepared, and the ability of MIF to bind Affi-PEITC was measured. A representative gel is shown.
Isothiocyanate Binding and the Conformational Integrity of MIF
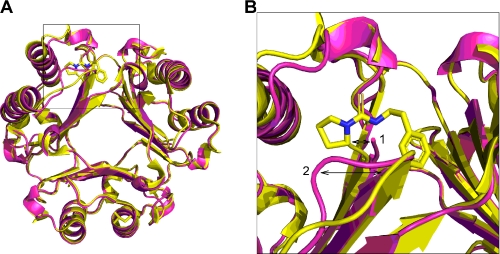
Although the biological significance of inhibiting MIF tautomerase activity is controversial, a previous study recognized that covalent modification of the N-terminal proline residue can disrupt the integrity of epitope(s) critical to the biological activity of MIF (25). In silico modeling of PEITC at the tautomerase active site highlighted a conformational change with the catalytic proline shifting 2 Å compared with that seen in the unmodified structure (Fig. 5). In addition, lysine 32, which sits above the active site, shifted by 1.6 Å. This is a direct consequence of the formation of a thiourea adduct with the N-terminal proline where the active site must accommodate the planar thiourea group (R2NC SNR) including the planar (sp2 hybridization) proline nitrogen.
SNR) including the planar (sp2 hybridization) proline nitrogen.
FIGURE 5.
In silico model of PEITC bound to the tautomerase active site of MIF via covalent modification of the N-terminal proline residue. The MIF homotrimer is shown with PEITC (yellow) docked in one of the tautomerase active sites. MIF without PEITC bound is shown in magenta (PDB code 3B9S). Conformational shifts of the catalytic proline by 2 Å (arrow 1) and lysine 32 by 1.6 Å (arrow 2) are highlighted. A zoom image of the highlighted region in A is shown in B.
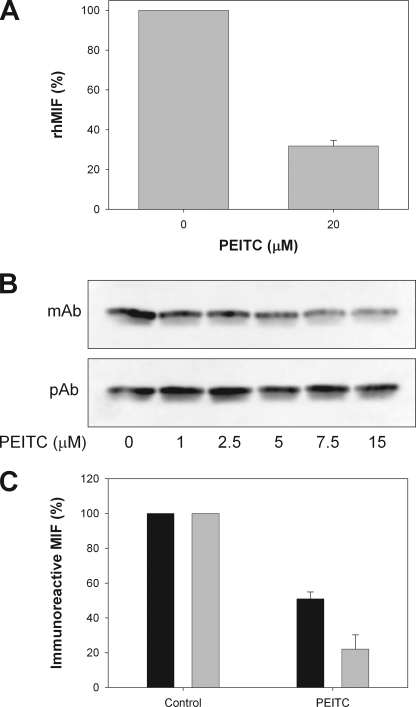
Covalent modification of MIF by acetaminophen metabolites has previously been shown to affect immunorecognition of MIF by a monoclonal antibody (25). In the same study loss of immunorecognition was associated with impaired MIF biological activity. A short incubation of rhMIF with PEITC reduced the immunoreactivity of MIF (Fig. 6A) as measured by an ELISA using a neutralizing monoclonal anti-MIF antibody (clone 12302). Loss of antibody recognition is consistent with isothiocyanate modification disrupting the conformational integrity of MIF epitope(s) as predicted by in silico modeling. The immunoreactivity of cellular MIF was also compromised when Jurkat cells were treated with PEITC as observed by immunoblotting with the monoclonal anti-MIF antibody (Fig. 6B). No change in total MIF protein levels, as determined by binding of a polyclonal anti-MIF antibody, was observed. Disruption of MIF immunoreactivity was confirmed by ELISA with both intracellular and secreted (extracellular) MIF being susceptible to isothiocyanate modification (Fig. 6C).
FIGURE 6.
A, shown is immunoreactivity of rhMIF after a 10-min exposure to 20 μm PEITC as measured by a commercial ELISA. Values are the mean ± S.E. of three experiments. B, immunobinding of MIF monoclonal (mAb) and polyclonal (pAb) antibodies to extracts prepared from Jurkat cells treated with increasing concentrations of PEITC for 60 min. Images are representative of results from three experiments. C, cells were treated with 15 μm PEITC for 1 h before detectable levels of extracellular (black bars) and intracellular MIF (gray bars) were measured using a commercial ELISA. Values are the mean ± S.E. from three experiments.
MIF as an in Vivo Target of Isothiocyanates
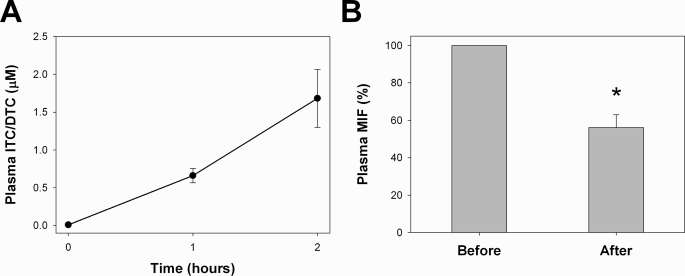
To determine whether MIF is a biologically relevant target of dietary isothiocyanates, a pilot study was designed to measure the immunoreactivity of plasma MIF in three human volunteers after ingestion of watercress (Nasturtium officinale), a rich dietary source of PEITC (27). Consumption of 50 g (wet weight) of watercress led to a time-dependent increase in plasma levels of isothiocyanates and their corresponding thiocarbonyl derivatives (dithiocarbamates) and a concomitant decrease in immunoreactive plasma MIF (Fig. 7). Two hours after ingestion, plasma immunoreactive MIF had decreased by ∼45%, indicating that isothiocyanates can target MIF in complex biological systems and at physiologically achievable concentrations.
FIGURE 7.
Three volunteers consumed 50 g of watercress. Blood was drawn just before eating (0), 1 h after eating (1), and 2 h (2) after eating. Plasma fractions were prepared immediately after venipuncture. A, plasma levels of isothiocyanates (ITC) and their corresponding dithiocarbamates (DTC) were measured at all time points in two of the volunteers using a cyclocondensation reaction with 1,2-benzenedithione. Values are the mean ± range. B, plasma MIF levels were measured by a commercial ELISA just before eating and 2 h after ingestion of watercress. Values are the mean ± S.E. The asterisk indicates a significant (p = 0.001) difference in the immunoreactivity of plasma MIF before and after watercress consumption as determined by a paired t test (SigmaStat, SPSS Inc., Chicago, IL).
DISCUSSION
Modification of proteins is recognized as a key mechanism underlying the biological activity of isothiocyanates, but knowledge of the major intracellular binding targets of these phytochemicals is lacking. We have used an affinity-based approach to identify targets of PEITC. A derivative of PEITC, amino-PEITC, was synthesized and retained biological activity. When coupled to a solid phase matrix, amino-PEITC permitted the identification of MIF as the most prominent intracellular binding target of isothiocyanates. PEITC was shown to bind MIF via a proline residue at the N terminus of the protein. Modification resulted in inhibition of MIF tautomerase activity and also triggered conformational changes.
MIF was originally identified as a lymphocyte-derived factor with cytokine-like activity (28, 29). MIF has since been shown to be constitutively expressed in numerous tissues and cell types (30) and have a variety of biological activities, including pivotal roles in the regulation of immune and inflammatory responses and promotion of tumorigenesis (31). Elevated levels of MIF have been observed in a number of disease states including cardiovascular disease (32), arthritis (33), diabetes (34), sepsis (35), and many cancer types (36, 37). Furthermore, genetic ablation of MIF has been shown to attenuate various disease states in murine models (38–40). Although details surrounding the mechanisms of MIF action are still in question, the clinical significance of MIF expression is such that targeted approaches to inhibit the biological activities of MIF are currently in development (41, 42). At least some of the biological activities of MIF are mediated by binding to, and downstream signaling from the extracellular receptors CD74, CXCR2, and CXCR4 (43, 44). However, MIF also possesses two distinct catalytic activities, thiol protein oxidoreductase activity (45) and the ability to catalyze a tautomerase reaction (46, 47). Whether or not either of these catalytic reactions is essential for MIF biological activity remains controversial, although some of the proinflammatory activities of MIF are impaired by mutations that affect enzymatic activity (22, 46, 48).
MIF has three cysteine residues, two of which (Cys-57 and Cys-60) are critical to the thiol protein oxidoreductase activity of MIF, whereas a third cysteine residue (Cys-81) is believed to play a role in maintenance of protein conformation (50). When we initially identified MIF as an isothiocyanate target, these cysteines were considered prime candidates for direct modification based on the known reactivity of isothiocyanates with thiols. Using mutant rhMIF in which the cysteine residues were isosterically mutated to serine, we have shown that PEITC does not directly react with any of these thiols. Instead, the N-terminal proline was identified as the binding site of PEITC. The sensitivity of the proline to attack by isothiocyanates can partly be explained by its unusually low pKa of 5.6, a value that is almost four pH units less than the pKa of free proline (48). The increased nucleophilicity of the N-terminal proline allows it to function as a general base catalyst in the tautomerase reaction (51) and favors reactivity with electrophiles such as isothiocyanates.
Not surprisingly, isothiocyanate binding to this proline inhibited MIF tautomerase activity. Because of the lack of an identified physiological substrate, whether tautomerase activity is a biologically relevant function of MIF is a matter of debate. On the negative side, P2 mutants retain MIF-like activity in assays of its proinflammatory action (52). However, others argue that the tautomerase active site may be critical for permitting and regulating interactions of MIF with receptors or binding proteins, including CD74 (41). Multiple sequence alignment analysis has revealed that many of the invariant residues in 11 MIF homologues cluster around the N-terminal proline, indicating an evolutionary pressure to preserve this site (48). Indeed, there is evidence that the interaction of MIF with CD74 occurs near the tautomerase active site and that inhibition of tautomerase activity correlates with inhibition of CD74 binding (41). We have shown that PEITC disrupts monoclonal antibody binding to MIF, clearly indicating that crucial epitopes are disturbed upon binding of the isothiocyanate. Results from the in vivo study indicate that ingested isothiocyanates can also bind MIF in the complex milieu of human plasma. As MIF is both proinflammatory and protumorigenic, we propose that its inhibition in plasma could contribute to the strong correlation between increased dietary intake of isothiocyanates and decreased risk of many types of cancer including lung, breast, and prostate cancers (53).
The probe used in this study proved to be effective at capturing a major target of isothiocyanates. Previously characterized target proteins, including Keap1 (13, 54), MEKK1 protein kinase (15), and tubulin (16) interact with isothiocyanates via formation of covalent adducts with cysteine. In contrast, our study indicates formation of a thiourea derivative with the N-terminal proline of MIF. Furthermore, it shows that this reaction must be favorable in biological systems. Modification of amines by isothiocyanates has been previously observed on the N-terminal glycine and phenylalanine residues in insulin, the N-terminal aspartic acid residue in epidermal growth factor (26), and a lysine residue in the transient receptor potential-A1 channel (49). However, modification of amines has not previously been shown to occur either in cells or in vivo. Although the current protocol favored the identification of irreversibly bound targets, detection of reversible modifications such as dithiocarbamate formation with cysteine might be possible utilizing the current approach with minor modifications.
In summary, this study has led to the discovery of MIF as a major target of isothiocyanates. This provides a foundation for the development of novel MIF inhibitors to be used in the treatment of cancer and inflammatory diseases and identifies a previously uncharacterized mechanism through which naturally occurring isothiocyanates can influence biological systems.
Supplementary Material
Acknowledgments
We thank Dr. Louise Paton and Dr. Peter Nagy for assistance with mass spectrometry and the National Research Centre for Growth and Development for funding this equipment. We also thank Dr. Rufus Turner for performing venipunctures and for assistance with HPLC.
This work was supported by the Canterbury-West Coast Division and the National Division of the Cancer Society of New Zealand. Support also came from the Maurice and Phyllis Paykel Trust, the University of Otago Oxidative Stress Theme, and German Research Council (Deutsche Forschungsgemeinschaft) Grant BE 1977/4-1/FOR809 (to J. B.).

The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
- PEITC
- phenethyl isothiocyanate
- MIF
- macrophage migration inhibitory factor
- rhMIF
- recombinant human MIF
- HPLC
- high pressure liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Nakajima M., Yoshida R., Shimada N., Yamazaki H., Yokoi T. (2001) Drug Metab. Dispos. 29, 1110–1113 [PubMed] [Google Scholar]
- 2.Zhang Y., Talalay P., Cho C. G., Posner G. H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2399–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiao J. W., Wu H., Ramaswamy G., Conaway C. C., Chung F. L., Wang L., Liu D. L. (2004) Carcinogenesis 25, 1403–1408 [DOI] [PubMed] [Google Scholar]
- 4.Pham N. A., Jacobberger J. W., Schimmer A. D., Cao P., Gronda M., Hedley D. W. (2004) Mol. Cancer Ther. 3, 1239–1248 [PubMed] [Google Scholar]
- 5.Gamet-Payrastre L., Li P., Lumeau S., Cassar G., Dupont M. A., Chevolleau S., Gasc N., Tulliez J., Tercé F. (2000) Cancer Res. 60, 1426–1433 [PubMed] [Google Scholar]
- 6.Xiao D., Singh S. V. (2007) Cancer Res. 67, 2239–2246 [DOI] [PubMed] [Google Scholar]
- 7.Hwang E. S., Lee H. J. (2006) J. Nutr. Biochem. 17, 837–846 [DOI] [PubMed] [Google Scholar]
- 8.Yu R., Mandlekar S., Harvey K. J., Ucker D. S., Kong A. N. T. (1998) Cancer Res. 58, 402–408 [PubMed] [Google Scholar]
- 9.Thomson S. J., Brown K. K., Pullar J. M., Hampton M. B. (2006) Cancer Res. 66, 6772–6777 [DOI] [PubMed] [Google Scholar]
- 10.Heiss E., Herhaus C., Klimo K., Bartsch H., Gerhäuser C. (2001) J. Biol. Chem. 276, 32008–32015 [DOI] [PubMed] [Google Scholar]
- 11.Drobnica L., Kristian P., Augustin J. (1977) The Chemistry of the NCS Group, pp. 1003–1221, John Wiley & Sons, Inc., New York [Google Scholar]
- 12.Zhang Y. S. (2000) Carcinogenesis 21, 1175–1182 [PubMed] [Google Scholar]
- 13.Hong F., Freeman M. L., Liebler D. C. (2005) Chem. Res. Toxicol. 18, 1917–1926 [DOI] [PubMed] [Google Scholar]
- 14.Macpherson L. J., Dubin A. E., Evans M. J., Marr F., Schultz P. G., Cravatt B. F., Patapoutian A. (2007) Nature 445, 541–545 [DOI] [PubMed] [Google Scholar]
- 15.Cross J. V., Foss F. W., Rady J. M., Macdonald T. L., Templeton D. J. (2007) BMC Cancer 7, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi L., Xiao Z., Hood B. L., Dakshanamurthy S., Wang X., Govind S., Conrads T. P., Veenstra T. D., Chung F. L. (2008) J. Biol. Chem. 283, 22136–22146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggli P., Prijs B. (1945) Helv. Chim. Acta 28, 674–690 [PubMed] [Google Scholar]
- 18.Callahan J. F., Ashton-Shue D., Bryan H. G., Bryan W. M., Heckman G. D., Kinter L. B., McDonald J. E., Moore M. L., Schmidt D. B., Silvestri J. S., Stassen F. L., Sulat L., Yim N. C. F., Huffman W. F. (1989) J. Med. Chem. 32, 391–396 [DOI] [PubMed] [Google Scholar]
- 19.Taylor M. S., Jacobsen E. N. (2004) J. Am. Chem. Soc. 126, 10558–10559 [DOI] [PubMed] [Google Scholar]
- 20.Dewey T. M., Du Bois J., Raymond K. N. (1993) Inorg. Chem. 32, 1729–1738 [Google Scholar]
- 21.Caddick S., Haynes A. K. D. K., Judd D. B., Williams M. R. (2000) Tetrahedron Lett. 41, 3513–3516 [Google Scholar]
- 22.Kleemann R., Rorsman H., Rosengren E., Mischke R., Mai N. T., Bernhagen J. (2000) Eur. J. Biochem. 267, 7183–7193 [DOI] [PubMed] [Google Scholar]
- 23.Ye L., Dinkova-Kostova A. T., Wade K. L., Zhang Y., Shapiro T. A., Talalay P. (2002) Clin. Chim. Acta 316, 43–53 [DOI] [PubMed] [Google Scholar]
- 24.Bendrat K., Al-Abed Y., Callaway D. J., Peng T., Calandra T., Metz C. N., Bucala R. (1997) Biochemistry 36, 15356–15362 [DOI] [PubMed] [Google Scholar]
- 25.Senter P. D., Al-Abed Y., Metz C. N., Benigni F., Mitchell R. A., Chesney J., Han J., Gartner C. G., Nelson S. D., Todaro G. J., Bucala R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traka M., Gasper A. V., Melchini A., Bacon J. R., Needs P. W., Frost V., Chantry A., Jones A. M., Ortori C. A., Barrett D. A., Ball R. Y., Mills R. D., Mithen R. F. (2008) PLoS ONE 3, e2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palaniswamy U. R., McAvoy R. J., Bible B. B., Stuart J. D. ( 2003) J. Agric. Food Chem. 51, 5504– 5509 [DOI] [PubMed] [Google Scholar]
- 28.Bloom B. R., Bennett B. (1966) Science 153, 80–82 [DOI] [PubMed] [Google Scholar]
- 29.Weiser W. Y., Temple P. A., Witek-Giannotti J. S., Remold H. G., Clark S. C., David J. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7522–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernhagen J., Calandra T., Bucala R. (1998) J. Mol. Med. 76, 151–161 [DOI] [PubMed] [Google Scholar]
- 31.Bach J. P., Rinn B., Meyer B., Dodel R., Bacher M. (2008) Oncology 75, 127–133 [DOI] [PubMed] [Google Scholar]
- 32.Burger-Kentischer A., Goebel H., Seiler R., Fraedrich G., Schaefer H. E., Dimmeler S., Kleemann R., Bernhagen J., Ihling C. (2002) Circulation 105, 1561–1566 [DOI] [PubMed] [Google Scholar]
- 33.Morand E. F., Leech M., Weedon H., Metz C., Bucala R., Smith M. D. (2002) Rheumatology 41, 558–562 [DOI] [PubMed] [Google Scholar]
- 34.Herder C., Kolb H., Koenig W., Haastert B., Müller-Scholze S., Rathmann W., Holle R., Thorand B., Wichmann H. E. (2006) Diabetes Care 29, 368–371 [DOI] [PubMed] [Google Scholar]
- 35.Emonts M., Sweep F. C., Grebenchtchikov N., Geurts-Moespot A., Knaup M., Chanson A. L., Erard V., Renner P., Hermans P. W., Hazelzet J. A., Calandra T. (2007) Clin Infect Dis 44, 1321–1328 [DOI] [PubMed] [Google Scholar]
- 36.Tomiyasu M., Yoshino I., Suemitsu R., Okamoto T., Sugimachi K. (2002) Clin Cancer Res. 8, 3755–3760 [PubMed] [Google Scholar]
- 37.Lee H., Rhee H., Kang H. J., Kim H. S., Min B. S., Kim N. K., Kim H. (2008) Am. J. Clin. Pathol. 129, 772–779 [DOI] [PubMed] [Google Scholar]
- 38.Bozza M., Satoskar A. R., Lin G., Lu B., Humbles A. A., Gerard C., David J. R. (1999) J. Exp. Med. 189, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizue Y., Ghani S., Leng L., McDonald C., Kong P., Baugh J., Lane S. J., Craft J., Nishihira J., Donnelly S. C., Zhu Z., Bucala R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14410–14415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos L. L., Dacumos A., Yamana J., Sharma L., Morand E. F. (2008) Clin. Exp. Immunol. 152, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cournia Z., Leng L., Gandavadi S., Du X., Bucala R., Jorgensen W. L. (2009) J. Med. Chem. 52, 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winner M., Meier J., Zierow S., Rendon B. E., Crichlow G. V., Riggs R., Bucala R., Leng L., Smith N., Lolis E., Trent J. O., Mitchell R. A. (2008) Cancer Res. 68, 7253–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng L., Metz C. N., Fang Y., Xu J., Donnelly S., Baugh J., Delohery T., Chen Y., Mitchell R. A., Bucala R. (2003) J. Exp. Med. 197, 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernhagen J., Krohn R., Lue H., Gregory J. L., Zernecke A., Koenen R. R., Dewor M., Georgiev I., Schober A., Leng L., Kooistra T., Fingerle-Rowson G., Ghezzi P., Kleemann R., McColl S. R., Bucala R., Hickey M. J., Weber C. (2007) Nat. Med. 13, 587–596 [DOI] [PubMed] [Google Scholar]
- 45.Kleemann R., Kapurniotu A., Frank R. W., Gessner A., Mischke R., Flieger O., Jüttner S., Brunner H., Bernhagen J. (1998) J. Mol. Biol. 280, 85–102 [DOI] [PubMed] [Google Scholar]
- 46.Rosengren E., Bucala R., Aman P., Jacobsson L., Odh G., Metz C. N., Rorsman H. (1996) Mol. Med. 2, 143–149 [PMC free article] [PubMed] [Google Scholar]
- 47.Rosengren E., Aman P., Thelin S., Hansson C., Ahlfors S., Björk P., Jacobsson L., Rorsman H. (1997) FEBS Lett. 417, 85–88 [DOI] [PubMed] [Google Scholar]
- 48.Swope M., Sun H. W., Blake P. R., Lolis E. (1998) EMBO J. 17, 3534–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinman A., Chuang H. H., Bautista D. M., Julius D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19564–19568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleemann R., Kapurniotu A., Mischke R., Held J., Bernhagen J. (1999) Eur. J. Biochem. 261, 753–766 [DOI] [PubMed] [Google Scholar]
- 51.Stamps S. L., Fitzgerald M. C., Whitman C. P. (1998) Biochemistry 37, 10195–10202 [DOI] [PubMed] [Google Scholar]
- 52.Hermanowski-Vosatka A., Mundt S. S., Ayala J. M., Goyal S., Hanlon W. A., Czerwinski R. M., Wright S. D., Whitman C. P. (1999) Biochemistry 38, 12841–12849 [DOI] [PubMed] [Google Scholar]
- 53.Higdon J. V., Delage B., Williams D. E., Dashwood R. H. (2007) Pharmacol. Res. 55, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.