Abstract
Lactobacillus reuteri mucus-binding protein (MUB) is a cell-surface protein that is involved in bacterial interaction with mucus and colonization of the digestive tract. The 353-kDa mature protein is representative of a broadly important class of adhesins that have remained relatively poorly characterized due to their large size and highly modular nature. MUB contains two different types of repeats (Mub1 and Mub2) present in six and eight copies, respectively, and shown to be responsible for the adherence to intestinal mucus. Here we report the 1.8-Å resolution crystal structure of a type 2 Mub repeat (184 amino acids) comprising two structurally related domains resembling the functional repeat found in a family of immunoglobulin (Ig)-binding proteins. The N-terminal domain bears striking structural similarity to the repeat unit of Protein L (PpL) from Peptostreptococcus magnus, suggesting binding in a non-immune Fab-dependent manner. A distorted PpL-like fold is also seen in the C-terminal domain. As with PpL, Mub repeats were able to interact in vitro with a large repertoire of mammalian Igs, including secretory IgA. This hitherto undetected activity is consistent with the current model that antibody responses against commensal flora are of broad specificity and low affinity.
Introduction
The human gastrointestinal tract (GIT)3 contains trillions of bacteria, representing hundreds of species and thousands of subspecies (1). They outnumber our own cells by a factor of 10 and contribute many physiological capabilities, including the provision of metabolic attributes not encoded in the human genome (2). A protective layer of mucus, consisting of a complex mixture of large, highly glycosylated proteins (mucins) (3), covers the epithelial cells of the intestine and offers an attachment site for the colonizing bacteria. These bacteria play important roles in maintaining normal gut function and in building resistance of the host to pathogenic micro-organisms (4, 5). Some may use mucins as their major carbon and energy source (6, 7).
Lactobacilli are Gram-positive microaerophilic bacteria naturally present in the dominant colonic microbiota and have been considered to be beneficial for human health (8). They are commonly used as probiotics, which are defined by the Food and Agriculture Organization/World Health Organization as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (9). As probiotic agents, lactobacilli can prevent or alleviate infectious diarrhea through their effects on the immune system and promote host resistance to colonization by pathogens (10, 11), and many have been shown to adhere to intestinal mucus (12–19). Confirmation of this lactobacillus-mucus association has not only been observed in vitro, but has also been validated by ex vivo/in vivo microscopic analysis of biopsy samples (20, 21). In most cases, lactobacilli adhesion to mucus has been proposed to be mediated by proteins (22–30). Compared with the present understanding of the adhesive mechanisms of human pathogenic bacteria, knowledge on the surface molecules mediating lactobacillus adhesion to the intestinal mucosa (i.e. epithelial cells, mucus layer, and/or extracellular matrices) and their corresponding receptors is less advanced.
The mucus adhesins from lactobacilli that have been identified and functionally characterized to date are the surface-associated mucus-binding protein (MUB) of Lactobacillus reuteri 1063 (23), the lectin-like mannose-specific adhesin of Lactobacillus plantarum WCFS1 (26), and the Mub of Lactobacillus acidophilus NCFM (25). These three mucus-binding proteins have a similar domain organization typical of cell-surface proteins of Gram-positive bacteria. At the N terminus is found a signal peptide targeting the protein for transport through the plasma membrane. An anchoring motif (LPXTG) that is recognized by a family of enzymes called sortases for covalent attachment of the transported protein to the peptidoglycan of the bacterial cell wall (31) is found at the C terminus. Interposed between these is the third and final domain containing a number of tandemly arranged mucus-binding repeats (Mub).
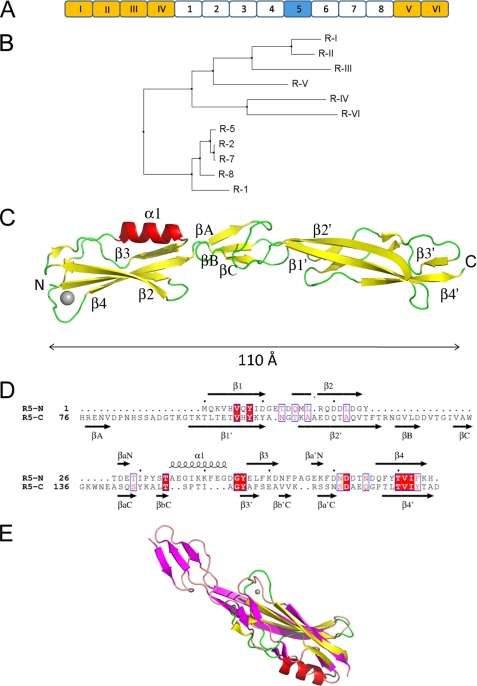
MUB from L. reuteri 1063 is predicted to have a 49-amino acid N-terminal secretion signal peptide, followed by a mature protein with a predicted molecular mass of 353 kDa. It is a highly repetitive protein containing two types of related amino acid repeats (Mub1 and Mub2) (Fig. 1, A and B), which have been shown to be responsible for the adherence to intestinal mucus. Six copies (RI through RVI) of a type 1 repeat (Mub1) are observed, ranging from 183 to 206 amino acids in length, and eight copies (R1 through R8) of a type 2 repeat (Mub2), all 184 amino acids long except for R1 with 186 amino acids, based on the Mub domain borders as described in a previous study (32), which differ slightly from the repeat sizes originally reported (23). These are organized in an interesting manner, with the Mub2 repeats inserted in between the Mub1 repeats RIV and RV (Fig. 1A). The six Mub1 repeats are rather diverse, whereas the Mub2 repeats show relatively low sequence variation (Fig. 1B). Mub repeat-containing proteins are most abundant in lactic acid bacteria (LAB), with the highest abundance in lactobacilli of the GIT, strongly suggesting that the Mub repeat is a functional unit specific to LAB that could fulfill an important function in host-microbe interactions.
FIGURE 1.
X-ray crystal structure of the mucus-binding protein Mub-R5 repeat. A, the organization of the modular repeat region of MUB from L. reuteri 1063. Repeats are labeled according to the nomenclature described in (Ref. 23). Type 1 repeats (RI to RVI) are colored gold and type 2 white except for the R5 repeat, which is highlighted in blue. B, neighbor joining tree as calculated by JALVIEW (82) for the non-redundant set of repeat sequences based on the percentage identity at each aligned position (note that repeats R2, R4, and R6 are identical as are R3 and R5). C, architecture of Mub-R5. α-Helices are colored red, and β-strands are yellow. The N and C termini of the protein are labeled, as are the major secondary structural elements. The single calcium ion bound to the N-terminal domain is represented as a gray sphere. D, structure-based sequence alignment of the B1 (R5-N) and B2 (R5-C) domains of Mub-R5. Secondary structural elements in both proteins are indicated and labeled. Identical residues are highlighted in red. E, an overlay of the structures of the B1 and B2 domains. The secondary structural elements of the B1 domain are shown in red (α-helices) and yellow (β-strands) and in magenta (β-strands) for the C-terminal B2 domain.
In this study, we report the first three-dimensional structure of a mucus binding repeat providing the first insights into a previously undetected Ig-binding activity for the repeat structural unit of MUB proteins.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Mub Repeats
L. reuteri 1063 was obtained from the American Type Culture Collection (strain ATCC 53608). Oligonucleotide primers for PCR amplification of DNA molecules encoding individual or multiple Mub repeats of the L. reuteri 1063 MUB protein were designed to anneal to specific Mub domain border regions, as defined in previous studies (32, 33) (supplemental Tables S1 and S2). Wild-type recombinant proteins were expressed from pETBlue-1 AccepTor (Novagen) in Escherichia coli Tuner™ (DE3/pLacI, Novagen). The L48M mutant of Mub-R5 was generated using the QuikChange® site-directed mutagenesis kit (Stratagene) with the gene-specific oligonucleotides listed in supplemental Table S2 and vector pETBlue-1:Mub-R5 as template. Wild-type Mub-R5 and mutant L48M proteins were labeled using the SelenoMet™ system (Athena Enzyme Systems™) and expressed in T&GE. coli B834 (DE3/pLacI) (Novagen). Recombinant Mub domains were purified from freeze-thaw or BugBuster® HT (Novagen)-soluble cell extracts by ion-exchange fast-protein liquid chromatography (Amersham Biosciences). See also the supplemental materials.
Biophysical Characterization of Recombinant Mub Repeats
Edman N-terminal protein sequencing was carried out by the Protein and Nucleic Acid Chemistry Facility (University of Cambridge, UK). ESI-MS of purified proteins was performed using a micrOTOF mass spectrometer (Bruker Daltonics Ltd.). Data were acquired in positive ionization mode at a capillary voltage of 4200 V and over a scan range of 250–3000 m/z. Trypsinized samples of proteins excised from SDS-PAGE gels were analyzed in an Ultraflex MALDI-ToF/ToF mass spectrometer (Bruker Daltonics Ltd.) or by liquid chromatography-tandem mass spectrometry in an LTQ-Orbitrap™ mass spectrometer (ThermoFisher), and MS data were searched against the relevant sequence databases using Mascot 2.1 and 2.2 search engines (Matrix Science Ltd.), respectively. CD spectroscopy was performed in a JASCO J-710 spectropolarimeter (Great Dunmow, Cambs, UK). A scan speed of 50 nm/min was used over a scan range of 260–185 nm with a bandwidth of 1.0 nm, a response time of 2.0 s, and a data pitch of 0.5 nm. The data were analyzed with JASCO Spectra Manager 32 v1.40.00a software, and CONTINLL (34) from the CDPro suite of programs was used to calculate the spectra and the proportion of each type of secondary structure (using IBasis reference set 3). Sedimentation equilibrium experiments were performed in a Beckman XLI analytical ultracentrifuge, equipped with absorbance optics, at 20 °C and speeds of 9,000, 15,000, and 20,000 rpm. Mub-R5 (20.0 μm), Mub-R6 (20.0 μm), Mub-RI (23.6 μm), and Mub-RI–III (8.0 μm) were prepared in PBS buffer (pH 7.4) each in a total volume of 110 μl prior to centrifugation, and samples were analyzed against PBS buffer blanks. Scans were recorded every 4 h to determine when proteins had reached equilibrium in the centrifuge, after which time five scans were recorded for each sample. The freeware program UltraScan II (Borries Demeler, University of Texas) was used to fit the obtained sedimentation equilibrium profiles to single molecular species. Complete details of all methods are given in the supplemental materials.
Protein Binding by Slot-blot Analysis
Recombinant Mubs were labeled with fluorescein isothiocyanate at pH 9.3 using an adapted standard protocol (see supplemental materials). Target proteins, including human secretory-IgA, IgG, and IgM (Sigma), human IgG-Fab/κ and IgG-Fc fragments (Bethyl Laboratories Inc., Montgomery, TX), and bovine serum albumin Fraction V (Sigma) in PBS buffer (pH 7.4) were vacuum-blotted onto an Immobilon-P polyvinylidene difluoride membrane (3.8 × 11.6 cm, 0.45 μm, Millipore) using a Hoefer PR600 24-slot apparatus. 1–20 μg of target protein was loaded per slot in a total volume of 100 μl. Blots were blocked for 18 h with gentle rocking at room temperature in 10 ml of Thermoblock protein-free blocking agent in PBS buffer (Thermo Scientific). All subsequent washing steps were carried out with 20 ml of PBS buffer containing 0.05% (v/v) Tween 20. Blocked membranes were incubated at room temperature with 10 ml of fluorescein-conjugated Mub proteins (200 μg/ml f-Mub, fluorescein/protein (F/P) ratio: 0.99–2.37) or fluorescein-conjugated protein L (18 μg/ml f-PpL; F/P ratio 0.63) in PBS buffer (±1 mm CaCl2) with gentle rocking in the dark for 20 h. Following excitation at 488 nm, fluorescein signals were detected at 530 nm in a Pharos FX™ Plus Molecular Imager (Bio-Rad) and quantified using Quantity One® v4.6.1 software (Bio-Rad). Background-subtracted signals were normalized to a probe F/P ratio of 1.0 and a probe concentration of 1 μm.
Crystallization and Crystal Structure Determination
Purified native Mub-R5 was concentrated to 2 mg/ml prior to crystallization. Single crystals were obtained by vapor diffusion at 4 °C using a precipitant solution containing 0.2 m ammonium formate and 22% (w/v) polyethylene glycol 3350. Crystals were cryoprotected by increasing the concentration of polyethylene glycol 3350 in the drops to 30% (v/v) and a native diffraction dataset was subsequently collected to 1.8-Å resolution at 100 K. These crystals were of space group P212121 and contained two copies of the protein in the asymmetric unit with a solvent content of 48% (v/v). Crystals of the SeMet-(L48M) mutant of Mub-R5 grew under similar conditions to those found for the native protein and were cryoprotected in an identical fashion. These crystals were found to be essentially isomorphous with those of the native protein, and the structure was solved by selenium SAD using heavy atom sites located by SOLVE (35). Initial phase estimates were improved with RESOLVE (36) and used to calculate an electron density map at 2.0-Å resolution. A preliminary molecular model was built comprising >80% of the polypeptide using ARP/wARP (37). This was completed by manual building using COOT (38) alternating with simulated annealing with PHENIX (39) and maximum likelihood refinement with REFMAC (40). The structure of the native protein was solved by molecular replacement using the structure of the SeMet mutant protein as search model. Refinement at 1.8-Å resolution resulted in a final structural model lacking only the C-terminal alanine residue in both independent molecular copies of Mub-R5 in the crystallographic asymmetric unit. Data collection and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
Dataset nomenclature is as follows, SeMet(L48M)Mub-R5: selenomethionyl derivative of the L48 M mutant of Mub-R5. Numbers in parentheses refer to data in the highest resolution bin.
| Dataset | Se-Met(L48M)Mub-R5 (SAD data collection) | Native Mub-R5 |
|---|---|---|
| Beamline | Diamond ID02 | ESRF BM14 |
| Space group | P212121 | P212121 |
| Cell parameters, a, b, c (Å) | 44.9, 45.7, 191.4 | 44.9, 45.8, 191.6 |
| Wavelength (Å) | 0.979 | 1.033 |
| Resolution (Å) | 60-2.0 (2.11-2.00) | 60-1.80 (1.90-1.80) |
| Rsyma (%) | 5.9 (9.6) | 6.0 (20.4) |
| Ranomb (%) | 5.2 (6.4) | |
| 〈I/σI〉 | 34.6 (25.0) | 13.8 (9.5) |
| Independent reflections | 27,636 (3,931) | 36,236 (4391) |
| Completeness (%) | 100.0 (100.0) | 96.0 (81.9) |
| Multiplicity | 14.0 (14.4) | 2.8 (1.9) |
| Anomalous completeness (%) | 100.0 (100.0) | |
| Anomalous multiplicity | 7.5 (7.5) | |
| Overall temperature factor (Å2) | 14.3 | 15.4 |
| FOMc (SAD phasing) | 0.82 (0.75) | |
| FOMc (solvent flattened) | 0.90 (0.82) | |
| Refinement statistics | ||
| Mub-R5 heterodimers per AUd | 2 | |
| Refined structure | ||
| Total atoms | 3,639 | |
| Water molecules | 755 | |
| Rcryste | 20.2 (26.9) | |
| Rfreef | 25.9 (30.6) | |
| Ramachandran analysisg (%) | ||
| Most favored | 98.9 | |
| Disallowed | 0.0 | |
| r.m.s.d. | ||
| Bonds (Å) | 0.009 | |
| Angles (°) | 1.16 | |
| Planes (Å) | 0.005 | |
| Mean atomic B-value (Å2) | 14.6 | |
a Rsym = Σ|Ii − 〈I〉|/Σ Ii, where 〈I〉 is the average of symmetry equivalent reflections, and the summation extends over all observations for all unique reflections.
b Ranom = Σ|〈I+〉 − 〈I−〉|/Σ|〈I+〉 + 〈I−〉| (reported for the selenium SAD dataset to a maximum resolution of 2.0 Å).
c Average figures of merit (FOM) after solvent flattening with RESOLVE (Terwilliger (36)).
d AU refers to the crystallographic asymmetric unit.
e Rcryst = Σ½|Fo| − |Fc½|/Σ|Fo|, where Fo and Fc are the measured and calculated structure factors, respectively.
f For Rfree the summations extends over a randomly selected subset (5%) of reflections excluded from all stages of refinement.
g Structure validation was performed using MOLPROBITY (83).
Protein Structure Analysis
Protein structure superposition was performed with DALI (41). Analysis of functional regions via evaluation of residue evolutionary conservation scores was performed with CONSURF (42) using sequence alignments generated using T-COFFEE (43) and visualized with ESPript (44).
RESULTS
Purification and Biophysical Characterization of Recombinant Mub Repeats
The recombinant single Mub repeats Mub-RI, Mub-R5, and Mub-R6 and the triple Mub repeat Mub-RI–III were expressed in soluble form in T&GE. coli and purified to homogeneity by ion-exchange chromatography (supplemental Fig. S1). The electrophoretic mobility of the proteins gave molecular weight estimates higher than that predicted from the amino acid sequences, similar to that observed with recombinant MucB2 domain from Streptococcus pneumoniae surface protein SP1492 (33). However, ESI-MS and MALDI-ToF-MS confirmed the integrity of the proteins, giving masses within 1 mass unit of the predicted sizes (supplemental Table S3). The pIs of all four proteins, as determined by isoelectric focusing, were 4.39 (Mub-R5), 4.33 (Mub-R6), 4.64 (Mub-RI), and 5.20 (Mub-RI–III), in agreement with the theoretical values. The N-terminal sequence of the triple domain Mub-RI–III, as determined by Edman sequencing, was MQEAAISFYD, in agreement with the amino acid sequence. Analytical ultracentrifugation of the four Mub proteins demonstrated that they were monomers under the conditions tested (data not shown). After the production of SeMet-labeled proteins, ESI-MS analysis revealed that SeMet incorporation was essentially complete (92–98%) at the two or three methionine residues present in the recombinant 185-amino acid wild-type Mub-R5 and L48M proteins, respectively (supplemental Table S3). CD spectra of unlabeled and SeMet-Mub-R5/L48M proteins were similar, consisting predominantly of β-structure (∼63%) with <0.5% α-structure (supplemental Fig. S2).
Crystallization and Crystal Structure Determination
Recombinant native Mub-R5 and the SeMet derivative of the (L48M)Mub-R5 mutant were crystallized, and a structure for the polypeptide component of the asymmetric unit of the L48M mutant was determined by selenium SAD phasing and refined at 2.0-Å resolution. The crystallographic R-factor of this interim model was 28.2% (R-free 33.3%). The structure of the native protein was then solved by molecular replacement using the mutant protein structure as a search model and refined to give an overall crystallographic R-factor of 20.2% (R-free 25.9%) at 1.8-Å resolution (Table 1). Incorporation of an additional methionine into the mutant led to only local and minor structural differences with the native protein (data not shown). The two copies of native Mub-R5 found in the crystallographic asymmetric unit consist of 184 residues (including the additional N-terminal methionine) and are similar, with an r.m.s.d. calculated for the Cα atoms to be 1.1 Å. All subsequent discussion refers to the refined native protein repeat structure, and the residue numbering system used is such that residue numbers 2–184 in the structure of the repeat correspond to 2063–2245 in the full-length MUB (supplemental Table S1).
The Crystal Structure of Mub-R5
The overall structure of Mub-R5 resembles a distorted cylinder ∼110 Å long and 25 Å in diameter at its widest point. It is made up of two discrete domains: an N-terminal domain composed of the first 75 residues and a C-terminal domain comprising residues 76–184 (Fig. 1C). The domain limits are essentially equivalent to those suggested in a previous study (32), and the C-terminal domain coincides with the MucBP domain definition from the Pfam data base (PF06458) (45). We will subsequently refer to the N- and C-terminal domains of Mub-R5 as the B1 and B2 domains, respectively.
The B1 domain possesses an ubiquitin-like β-grasp fold most similar to that found in the Ig-binding superfamily (46). This fold consists of two pairs of antiparallel β-strands forming a four-stranded mixed β-sheet connected by an α-helix. The strand order of the β-sheet is 2143. Interpretation of the residual electron density maps for the refined structure revealed a peak at 8.0 σ above the mean. The octahedral coordination of this site together with the coordination distances (supplemental Table S4) allowed us to identify this feature as originating from a bound calcium ion (47). This was subsequently confirmed by refinement. The residues involved in binding this ion are located at the N terminus of strand β1 (the side chain of Gln-2) and in the loop connecting β3 and β4 (the side chain of Asp60 and the main-chain carbonyl groups of Asp62 and Asn65). Two water molecules complete the coordination sphere of the metal (supplemental Fig. S3). This bound ion serves to stabilize the conformation of the polypeptide loop preceding strand β4 in the N-domain.
The B1 and B2 domains possess a degree of structural similarity. Superposition using DALI (41) gives an r.m.s.d. of 2.5 Å for 66 aligned residues of which 14% are identical (Z-score 4.5) (Fig. 1, D and E). However, despite its structural similarity to the B1 domain, the B2 domain does not possess a canonical β-grasp fold, because it lacks the connecting helix, α1, replacing it instead with a β-strand. The structurally related regions form a significant proportion of the molecule as a whole, excluding only residues 76–94 and 120–141. These residues form a small β-sheet involving strands βA, βB, and βC at the interface between B1 and B2 (Fig. 1C) and support the extended nature of the tertiary structure of the protein. This is achieved by bracing interactions involving contacts with the B1 domain through salt bridges (Asp127–His75 and Glu47–His76) and hydrophobic interactions (Val129–Tyr46). An interesting arrangement wherein the guanidinium group of Arg122 inserts between the parallel indole rings of the tryptophan residues at positions 135 and 138 also provides a cap to the hydrophobic core of the B2 domain. The result of these interactions is to stabilize an arrangement in which the B2 domain is rotated ∼90° relative to B1.
Structural Homology with Ig-binding Proteins
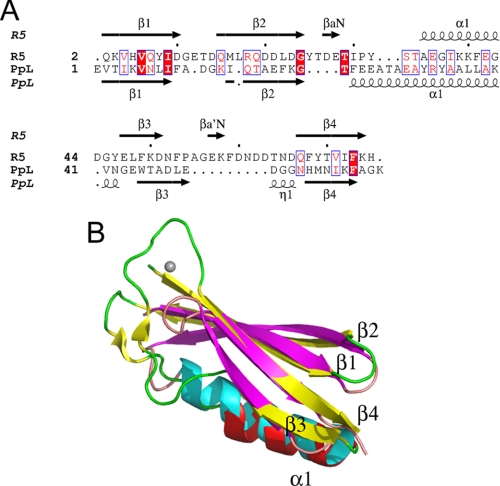
The closest structural homologue in the Protein Data Bank to the B1 domain of Mub-R5 is the B1-domain of Protein L (PpL) (48). The corresponding structural alignment has an r.m.s.d. of 2.9 Å over 57 aligned residues but shows only 5% sequence identity (Z-score, 5.1) (Fig. 2, A and B). Given its structural similarity to the B1 domain, it is not surprising that the B2 domain of Mub-R5 also shows structural homology, albeit weaker, to PpL (r.m.s.d. of 3.1 Å over 53 aligned residues and Z-score of 3.0). The sequence identity corresponding to this structural alignment is also low (13%). Protein L is a multidomain cell wall protein from Peptostreptococcus magnus, which belongs to a family of Ig-binding proteins (49), including Protein G from Streptococcus sp. (SpG). PpL binds to the VL domain of the κ chain of all Ig classes, whereas SpG binds predominantly to Fc but also has weaker Fab-binding activity (50). PpL is structurally similar to SpG. The major difference lies in a shorter loop in PpL between β3 and the connecting helix, α1. This loop is involved in Fc binding in SpG (51, 52). Its absence in PpL is thought to be related to the inability of PpL to bind to bind Fc (53).
FIGURE 2.
Structural similarity between the B1 domains of Mub-R5 and Protein L. A, a structure-based alignment of the sequences of the B1 domain of Mub-R5 (R5) and the B1 domain of Protein L (PpL). Secondary structural elements in both proteins are indicated and labeled. Identical residues are highlighted in red. A single turn of a 310 helix labeled η1 preceding strand β4 in the structure of Protein L is absent from the R5 domain. B, an overlay of the structures of the B1 domains of Mub-R5 and PpL. The secondary structure of the Mub-R5 B1 domain is shown in red (α-helices) and yellow (β-strands) with the coil in green. For Protein L, α-helices are colored cyan and β-strands are magenta with coil regions in pink. The calcium ion bound to the Mub-R5 B1 domain is shown as a gray sphere.
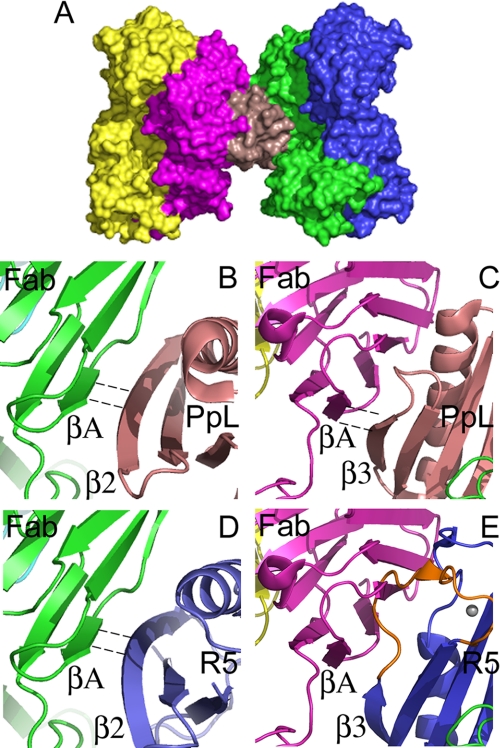
We used the x-ray crystal structures of the complex between PpL and a human antibody (PDB entry 1HEZ) to model the interaction of the B1 domain of the Mub-R5 repeat with IgG, by superimposition of Mub-R5 onto the coordinates of the bacterial proteins (Fig. 3A). The PpL-Fab complex has a 1:2 stoichiometry. In this complex a single PpL molecule is sandwiched between two Fab κ light chain domains, the two interfaces being characterized by β-zipper arrangement involving antiparallel and parallel hydrogen bonding arrangements involving the β2 and β3 strands, respectively (54, 55). In our structural model, the conserved β2 strand of Mub-R5 makes similar hydrogen bonds to the external VL A-strand (Fig. 3, B and C). However, as the β3 strand is displaced by two residues relative to the corresponding strand in PpL (see Fig. 2A), the model suggests that Mub-R5 may not form a similar parallel hydrogen bonding interaction with the Fab (Fig. 3, D and E). Furthermore, the polypeptide chain resulting from the insertion of nine amino acids following β3 in Mub-R5 clashes with the Ig light chain domain. Our simple modeling procedure therefore leads to the prediction that a ternary complex as observed for PpL is unlikely for the mucus-binding protein without major structural rearrangements.
FIGURE 3.
Modeling the interaction of the B1 domain of the Mub-R5 repeat with an IgG Fab domain. A, a molecular surface representation of the 1:2 ternary complex formed by a Protein L domain (PpL) with two human antibody Fab fragments (light red, PpL; green and magenta, Fab VL domains; and blue and yellow, Fab VH domains). Atomic coordinates were taken from PDB entry 1HEZ. B and C, close-up views of the molecular interfaces formed by the PpL domain in the 1:2 complex. The two interfaces have antiparallel (B) and parallel (C) hydrogen-bonding arrangements involving the β2 and β3 strands of the PpL domain, respectively, with the A strand of the VL domain. PpL is colored light red in both panels. Dashed lines indicate hydrogen-bonded β-strands. D and E, views of the model for an Mub-R5 B1 domain-Fab complex. Mub-R5 is colored blue in both panels. The conserved β2 strand of Mub-R5 makes similar antiparallel hydrogen bonds to PpL to the external VL A-strand (D). However, the polypeptide chain resulting from the insertion of nine amino acids in the loop (colored orange) following β3 in Mub-R5 relative to PpL clashes with the Ig light-chain domain (E) suggesting that a similar parallel hydrogen-bonding interaction with the Fab region may not occur with the mucus-binding protein repeat structure.
Structural Homology with Bacterial Adhesins
In addition to its homology with PpL, the B2 domain of Mub-R5 is also similar to a range of proteins with the structural classification of proteins prealbumin-like fold (46). Within this superfamily are found the transthyretin, IgG-rev fold of the Cna protein B-domain and starch-binding domain-like proteins. The greatest similarity detected (Z-score of 3.5 and r.m.s.d. of 2.6 Å over 82 aligned residues) was to the N2 domain of SpaB (GBS52), the minor pilin from the Gram-positive pathogen Streptococcus agalactiae (supplemental Fig. S4). SpaB is utilized by S. agalactiae to promote adhesion to pulmonary epithelial cells. The prealbumin fold is a variant of the Ig-like β-sandwich and is characterized by the presence of seven β-strands arranged in 3- and 4-stranded sheets. Of these, only the long β1′, β2′, and β4′ strands of the B2 domain of Mub-R5 (Fig. 1C), corresponding to the three-stranded sheet of SpaB, are well conserved. In this respect, it is useful to note that the core of a modified IgG-rev-like fold (such as observed in SpaB) can be generated from the β-grasp fold by deleting the α1-helix and replacing it with the edge EF strand pair found in the four-stranded antiparallel sheet of the pilin protein (56). This also occurs in the B2 domain of Mub-R5. Thus, although the Mub-R5 B1 domain is clearly structurally homologous to the Ig-binding domains of PpL, the similarity is less pronounced in the B2 domain, and we cannot discount the possibility of functional similarity to bacterial adhesins such as SpaB in this region.
The S. pneumoniae cell-surface protein SP1492 contains a 90-amino acid domain (MucB2) at its N terminus, which exhibits mucin and simple carbohydrate (mannose, sialic acid, and others)-binding activities (33). The sequence of the designated SP1492 mucin binding region shares 25% sequence identity with the B2 domain of Mub-R5. The majority of the residues strictly conserved across the B2 domains of all fourteen Mub1 and Mub2 repeats from L. reuteri 1063 are also found in SP1492. As such, it appears likely that they share a common fold. SP1492 has no region of amino acid sequence corresponding to the B1 domain of Mub-R5 suggesting that the B2 domain of the Mub repeat may be responsible for the mucin and/or carbohydrate-binding activity.
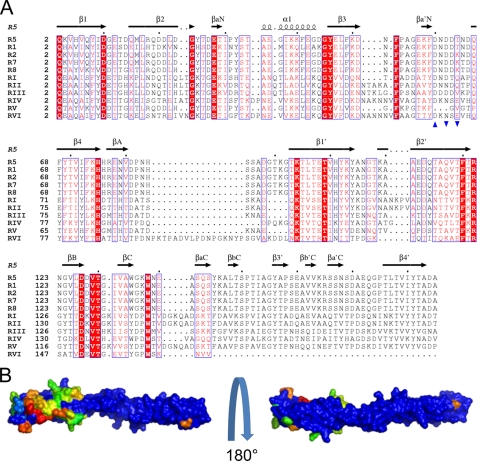
Inter-repeat Sequence Variability
Type 2 Mub repeats show relatively low sequence variation. As such, the presumption that these repeats share a common fold appears reasonable. CONSURF (42) analysis of the sequences of the eight type 2 repeats reveals strands β1 and β2 and the adjacent solvent-facing surface of helix α1 of the B1 domain to be the most variable in the repeat (Fig. 4, A and B). The similarity observed among the sequences of type 2 repeats is in contrast to that observed for type 1, where the six repeats have amino acid sequence identities ranging from 31 to 87%. Furthermore, the highest sequence identity between type 1 and 2 repeats is 48% (between RV and R8), but most Mub1–Mub2 identities are in the range of 30–40%. An interesting question, therefore, is whether the more divergent type 1 repeats will adopt the same fold as seen for the type 2 R5 repeat. Fig. 4A shows an alignment of the non-redundant type 1 and 2 repeat sequences. An immediate observation is the absence of a subset of the calcium-binding residues in R5 arising from a deletion in the loop joining β3 and β4 in RV. The implication is that this repeat does not possess a calcium binding site and so may be less ordered in this region. A number of further observations may be made. Firstly, insertions and deletions occur generally between secondary structural elements, and the hydrophobic cores of the B1 and B2 domains appear to be conserved. Secondly, the majority of the residues forming specific bracing interactions at the B1–B2 domain interface are also conserved. The conclusion is that the overall fold of type 1 repeats should resemble that of type 2. A number of residues are strictly conserved across all Mub repeats. A subset of these plays clear structural roles. Of the remainder, Tyr45 (and its structural mate in the C-domain, Tyr155), Thr98, and Pro150 are surface residues, have no clear role related to maintenance of structure, and may be involved in aspects of mucin binding.
FIGURE 4.
Conservation of repeat molecular surface properties. A, an alignment of the sequences of the non-redundant repeats from the L. reuteri mucus-binding protein. Secondary structural elements from the R5 repeat are shown. The residues contributing to the octahedral coordination sphere of the bound calcium ion in the Mub-R5 structure are indicated by blue triangles (pointing upward for residues with coordinating atoms in the residue side chain and downward for residues coordinating the metal via main-chain atoms). The starting residue numbers for each repeat in the full-length mucus-binding protein sequence can be found in the supplemental materials. B, views of a ConSurf (42) color-coded surface representation of Mub-R5 showing invariant and the semi-conserved residues of the type 2 repeats only according to the alignment shown in A. The normalized conservation scores calculated by ConSurf are a relative measure of evolutionary conservation at each residue position. The highest scores (9 on the ConSurf scale) represent the most conserved residue positions and are colored blue, and the least conserved are colored red. The molecular orientation on the left is approximately the same as in Fig. 1C. The region showing the least surface conservation includes the solvent-exposed surface of the N-domain β2 strand and the adjacent face of the α1 helix.
Mub Repeats Bind to Immunoglobulins
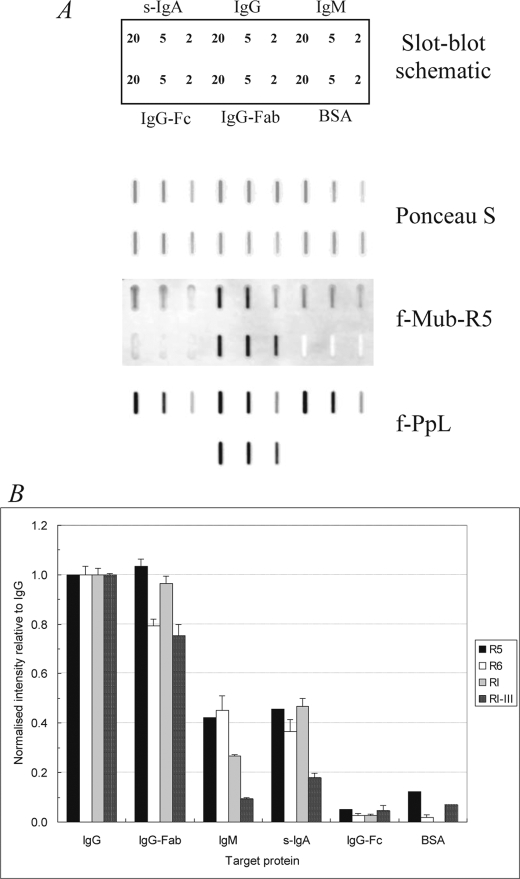
The binding of recombinant Mub repeats of type 2 (R5 and R6) and type 1 (RI and RI–III) to Igs was investigated against human secretory IgA, IgG, IgM, IgG-Fab/κ, and IgG-Fc. The Igs (and bovine serum albumin as a control) were slot-blotted and probed with fluorescein-Mub protein conjugates as well as f-PpL (Fig. 5A and supplemental Fig. S5). The binding profiles of the different Mub repeats were similar, but the relative fluorescence signal intensities to each Ig varied with the type and number of repeats (Fig. 5B and supplemental Fig. S5). The pattern of Mub-R5 binding to Igs was similar to that for PpL; Mub-R5 had affinity for IgG, IgM, and s-IgA and the IgG-Fab/κ fragment but not the heavy chain IgG-Fc fragment (Fig. 5, A and B). Mub-R6, which is 97% identical at the amino acid level to Mub-R5, bound to Igs in a similar fashion to Mub-R5 (Fig. 5B and supplemental Fig. S5). Specificity toward the IgG-Fab/κ fragment was also observed with Ig binding of type 1, Mub-RI, and Mub-RI–III, which are only 30–35% identical at the amino acid level to Mub-R5 and Mub-R6 (Fig. 5B and supplemental Fig. S5). However, after normalization for probe F/P ratio and molarity, the binding activities of the type 1 and 2 repeats to Igs appeared significantly lower when compared with full-length, four repeat-containing PpL (supplemental Table S5). The addition of Ca2+ had no effect on the binding activities of the Mub repeats to Igs (data not shown).
FIGURE 5.
Binding of Mub repeats and protein L to Igs immobilized on polyvinylidene difluoride membranes. A, slot blots of f-Mub-R5 (F/P ratio 1.05; 9.8 μm) and f-PpL (F/P ratio 0.63; 0.5 μm) bound to Igs, showing schematic of protein loading in micrograms per slot. Bovine serum albumin was also blotted as a negative control. Blots were scanned at the same laser intensity. B, quantification of Mub repeat binding from slot blots scanned at the same laser intensity using averaged fluorescence signals taken from three locations within each slot after background subtraction. The histogram displays averaged intensity signals for binding to 20 μg of blotted protein relative to those for IgG within each blot after normalization to a probe F/P ratio of 1.0 and a probe concentration of 1 μm (± S.D., n = 3).
DISCUSSION
In this study we have provided the first structural and functional evidence for the presence of proteins that exhibit non-antigenic binding to Igs at the surface of non-pathogenic bacteria. Mub-R5 is one of the 14 repeats present in the mucus-binding protein of L. reuteri. Similar Mub repeat-containing proteins are found predominantly, although not exclusively, in lactobacilli of the GIT and are very variable in size and sequence, making it difficult to determine precise domain boundaries. The Mub-R5 crystal structure, presented here, confirms the recent bioinformatics analysis of Mub domains from orthologous proteins, which predicted the presence of Mub type repeats of ∼100 to >200 amino acid residues (32). This is in contrast to the predicted size of the MucBP (Mucin-Binding Protein) domain from the Pfam data base (PF06458), which contains ∼50 amino acid residues. The tertiary structure of Mub-R5 reveals the presence of discrete N-terminal (B1, residues 1–75) and C-terminal (B2, residues 76–184) domains within the repeat, corresponding roughly to the MucB1 and MucB2 Mub sub-domains designated in a previous study (33). The structural homology between the domains is in accordance with the low but significant sequence homology at the amino acid level. However, in all Mub repeats from L. reuteri, the N-terminal (B1) domain is exclusively found in association with the C-terminal (B2) domain. Our structural data reveal that Mub-R5 exists in an extended conformation, spanning roughly 110 Å. The close-knit nature of the interactions between the B1 and B2 domains suggests that they may be limited in their relative movement. Furthermore, the high sequence homology observed among type 2 repeats and the absence of additional linking residues between these repeats in the protein sequence suggests that additional foot-to-head interactions between domains in adjacent repeats are likely. As such, an elongated structure for the mucus-binding protein is envisaged, at least in the region of the type 2 repeats, reminiscent of the structure of fibronectin-binding proteins at the surface of many pathogenic Gram-positive bacteria (57).
Our crystal structure of Mub-R5 allowed an unexpected prediction for the N-terminal domain. The fold of the Mub-R5 B1 domain is most similar to that of the PpL Ig-binding domain B1 (76 amino acids), as determined by NMR spectroscopy (58). It consists of a β-sheet formed from two pairs of anti-parallel β-strands and an α-helix that lies on top of the sheet. Several proteins that exhibit non-antigenic binding to Igs have been isolated from the surface of pathogenic Gram-positive bacteria such as Protein A from Staphylococcus aureus (59), Protein G of group C and G streptococci (60), and PpL of P. magnus (49). Structural analyses of these proteins have revealed that, although they share certain characteristics, including hydrophobic/charged tail domains anchoring them to cell membranes and C-terminal cell-wall-spanning motifs, these proteins contain multiple repeated domains (55–76 amino acids) that are divergent in nature (48, 61–63). These repeated domains are responsible for binding Igs, although they recognize different Ig regions. Proteins A and G bind to the CH2-CH3 interface of the Fc fragment of some classes of Ig, predominantly IgG (64, 65), whereas the Ig-binding domains of protein L bind exclusively to the framework region of the VL domain of κ light chains (κ-chains) (66, 67). Structural studies performed on PpL indicated that the residues involved in the interaction with the κ-chain are located along the β2-strand, the C-terminal end of the α-helix, and the loop between the α-helix and β3-strand (54, 68). From our model, the Mub-R5 B1 domain may interact similarly with the framework part of the light chain variable domain of Igs without contacting the hypervariable loops. This is corroborated by our binding studies, which indicate an interaction between Mub-R5 and the Fab region of IgG, albeit with a binding activity that is much weaker than that reported for PpL (50, 66, 69, 70). This can be explained on the basis of our model, which shows that, although the conserved β2 strand of Mub-R5 can form similar antiparallel hydrogen bonds to PpL to the external VL A-strand, the polypeptide chain resulting from the insertion of nine amino acids in the loop following β3 in Mub-R5 relative to PpL clashes with the Ig light chain domain. This suggests that a similar parallel β-zipper hydrogen-bonding interaction with the Fab may not occur with Mub-R5. Furthermore, MUB contains 14 Mub repeats, whereas PpL contains only four or five highly homologous, consecutive extracellular Ig-binding domains, depending on the bacterial strain from which it is isolated (63). Hence, it is expected that the avidity of binding to immunoglobulins of an individual Ig-binding domain may be lower than the full-length protein. For example, a single B1 PpL repeat has a 200-fold lower affinity than the full-length four-repeat PpL construct (48), suggesting the necessity for the presence of the full complement of repeats for optimum binding. In the present work, the binding of individual repeats (Mub-R5, Mub-R6, and Mub-RI) showed similar binding patterns to Igs but with variable affinities. The triple repeat Mub-RI–III did not show any synergistic effect on binding.
The lower Ig-binding activity reported in this study for L. reuteri Mubs may have significant biological implications. Secretory Igs such as IgA, IgM, and IgG that are present in mucosal surfaces potentially provide a first line of defense against microorganisms (71). Surface proteins that bind human Ig-Fc have been identified in many strains of Streptococcus pyogenes (group A streptococcus) and group B streptococcus, two important human pathogens (72). The presence of such molecules on the surface of Gram-negative bacteria, including T&GE. coli, has also been documented. These are the T&GE. coli Ig-binding proteins (73) and are proposed to afford an advantage to the bacterium through perturbation of Fc-dependent functions such as the interaction with phagocyte FcR, although firm evidence supporting this hypothesis has only been obtained for the IgA (74)- and IgG (75)-binding proteins from group A and B streptococci, and more recently from S. aureus (76). L. reuteri is an inhabitant of the human GIT (77). Unlike pathogens that are found within the body, having attached to the epithelial surface or penetrated, commensals live almost entirely within the intestinal lumen or within the mucus coat barrier. Proteins containing Mub repeats are most abundant in lactobacilli that are found mainly in the GIT, supporting the hypothesis that the repeat is primarily involved in adherence to intestinal mucus. The high variability in the number of Mub repeats in putative mucus-binding proteins suggests that these repeats are often duplicated or deleted in evolution. The genomes of bacteria that have a broader lifestyle and are less frequently encountered in the GIT, such as L. plantarum, encode a smaller number of these proteins (78). Compared with lactobacilli of the GIT, “domesticated” Lactococcus lactis strains live in a more restricted habitat (79), which could explain the presence of only a single Mub repeat-containing protein in this bacterium. Unlike the adaptive responses against pathogens that must be of high affinity and specificity, the antibody response to the commensal flora is expected to be of broad specificity and relatively low affinity, in agreement with our biochemical data. Furthermore, we have shown that the region of the repeat structure with the greatest sequence variation corresponds to that which modeling suggests may interact with the VL domain in Igs. The presence of low affinity antibodies to redundant surface epitopes of bacteria, or binding of IgA through bacterial lectin-mediated mechanisms, can probably be sufficient for the reinforced barrier effect of the mucus layer (80, 81).
Our observations that Mub proteins can bind to Igs as well as the reported binding to mucin add to the complexity of interactions that mediate the adhesion of commensal bacteria to the gut and to the mucus layer in particular. Mucus is continuously renewed, and therefore the ability to bind to intestinal surfaces is generally considered as a factor associated with probiotic bacteria. The attachment of bacteria to gastrointestinal surfaces extends their homing time in the gut and, as a consequence, may influence the host health by affecting the local microbial composition or by the stimulation of the gastrointestinal immune system. The large representation of Mub-containing proteins in LAB strains may thus be closely associated to their probiotic properties. This knowledge will be invaluable in selecting strains for fundamental research of the ecological role of lactobacilli in the GIT, for their use as probiotics in foods and supplements, and for pharmaceutical applications. In addition, this result may make possible further tests in mice to determine the physiological relevance of IgA- and mucus-mediated biofilm formation in the gut.
Supplementary Material
Acknowledgments
We thank Mark Philo and Andrew Watson (Institute of Food Research (IFR), Norwich) for protein ESI-MS and data analysis, Fran Mulholland (IFR, Norwich) for MALDI-ToF sample preparation and data analysis, Fiona Husband and Mike Ridout (IFR, Norwich) for assistance with CD analysis, and Tom Clarke (University of East Anglia, Norwich) for analytical ultracentrifugation. We acknowledge access to the facilities of synchrotron beamlines BM14 at the European Synchrotron Radiation Facility (Grenoble, France) and ID02 at Diamond Light Source Ltd. (Didcot, Oxfordshire, United Kingdom).
The work was supported by a Biotechnology and Biological Sciences Research Council core strategic grant.
The atomic coordinates and structure factors (code 3I57) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1–S5.
- GIT
- gastrointestinal tract
- CH
- constant domain of heavy chain
- ESI-MS
- electrospray ionization mass spectrometry
- f-
- fluorescein
- Fab
- variable antigen-binding superdomain of Ig
- Fc
- constant superdomain of Ig
- FcαR
- receptor specific for Fc-superdomain of Ig
- κ-chains
- κ light chains
- LAB
- lactic acid bacteria
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight
- MUB
- mucus-binding protein
- Mub-R
- mucus binding repeat
- PpL
- protein L from P. magnus
- r.m.s.d.
- root mean square deviation
- sIgA
- secretory form of IgA
- VL
- light-chain variable region
- PBS
- phosphate-buffered saline
- F/P
- fluorescein/protein ratio
- SeMet
- selenomethionyl
- SpG
- Protein G from Streptococcus sp.
- SAD
- single wavelength anomalous dispersion.
REFERENCES
- 1.Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. (2008) Science 320, 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E. (2006) Science 312, 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linden S. K., Sutton P., Karlsson N. G., Korolik V., McGuckin M. A. (2008) Nature Mucosal Immunol. 1, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper L. V., Gordon J. I. (2001) Science 292, 1115–1118 [DOI] [PubMed] [Google Scholar]
- 5.Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005) Science 307, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 6.Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. (2005) Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 7.Martens E. C., Chiang H. C., Gordon J. I. (2008) Cell Host Microbe 4, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter J. (2008) Appl. Environ. Microbiol. 74, 4985–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill H., Prasad J. (2008) Adv. Exp. Med. Biol. 606, 423–454 [DOI] [PubMed] [Google Scholar]
- 10.de Vrese M., Marteau P. R. (2007) J. Nutr. 137, 803S–811S [DOI] [PubMed] [Google Scholar]
- 11.Lebeer S., Vanderleyden J., De Keersmaecker S. C. (2008) Microbiol. Mol. Biol. Rev. 72, 728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirjavainen P. V., Ouwehand A. C., Isolauri E., Salminen S. J. (1998) FEMS Microbiol. Lett. 167, 185–189 [DOI] [PubMed] [Google Scholar]
- 13.Ouwehand A. C., Tuomola E. M., Tölkkö S., Salminen S. (2001) Int. J. Food. Microbiol. 64, 119–126 [DOI] [PubMed] [Google Scholar]
- 14.Lee Y. K., Puong K. Y., Ouwehand A. C., Salminen S. (2003) J. Med. Microbiol. 52, 925–930 [DOI] [PubMed] [Google Scholar]
- 15.Ma Y. L., Guo T., Xu Z. R., You P., Ma J. F. (2006) Lett. Appl. Microbiol. 42, 369–374 [DOI] [PubMed] [Google Scholar]
- 16.Vesterlund S., Karp M., Salminen S., Ouwehand A. C. (2006) Microbiology 152, 1819–1826 [DOI] [PubMed] [Google Scholar]
- 17.Collado M. C., Grześkowiak Ł., Salminen S. (2007) Curr. Microbiol. 55, 260–265 [DOI] [PubMed] [Google Scholar]
- 18.Li X. J., Yue L. Y., Guan X. F., Qiao S. Y. (2008) J. Appl. Microbiol. 104, 1082–1091 [DOI] [PubMed] [Google Scholar]
- 19.Muñoz-Provencio D., Llopis M., Antolín M., de Torres I., Guarner F., Pérez-Martínez G., Monedero V. (2009) Arch. Microbiol. 191, 153–161 [DOI] [PubMed] [Google Scholar]
- 20.Macfarlane S., Furrie E., Cummings J. H., Macfarlane G. T. (2004) Clin. Infect. Dis. 38, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 21.Macfarlane S., Dillon J. F. (2007) J. Appl. Microbiol. 102, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 22.Rojas M., Ascencio F., Conway P. L. (2002) Appl. Environ. Microbiol. 68, 2330–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos S., Jonsson H. (2002) Microbiology 148, 433–442 [DOI] [PubMed] [Google Scholar]
- 24.Granato D., Bergonzelli G. E., Pridmore R. D., Marvin L., Rouvet M., Corthésy-Theulaz I. E. (2004) Infect. Immun. 72, 2160–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck B. L., Altermann E., Svingerud T., Klaenhammer T. R. (2005) Appl. Environ. Microbiol. 71, 8344–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pretzer G., Snel J., Molenaar D., Wiersma A., Bron P. A., Lambert J., de Vos W. M., van der Meer R., Smits M. A., Kleerebezem M. (2005) J. Bacteriol. 187, 6128–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi Y., Okada S., Uchimura T., Satoh E. (2006) Biosci. Biotechnol. Biochem. 70, 1622–1628 [DOI] [PubMed] [Google Scholar]
- 28.Sun J., Le G. W., Shi Y. H., Su G. W. (2007) Lett. Appl. Microbiol. 44, 79–85 [DOI] [PubMed] [Google Scholar]
- 29.Tallon R., Arias S., Bressollier P., Urdaci M. C. (2007) J. Appl. Microbiol. 102, 442–451 [DOI] [PubMed] [Google Scholar]
- 30.Macías-Rodríguez M. E., Zagorec M., Ascencio F., Vázquez-Juárez R., Rojas M. (2009) J. Appl. Microbiol. 10.1111/i.1365-2672.2009.04368 [DOI] [PubMed] [Google Scholar]
- 31.Desvaux M., Dumas E., Chafsey I., Hébraud M. ( 2006) FEMS Microbiol. Lett. 256, 1– 15 [DOI] [PubMed] [Google Scholar]
- 32.Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. (2006) Microbiology 152, 273–280 [DOI] [PubMed] [Google Scholar]
- 33.Bumbaca D., Littlejohn J. E., Nayakanti H., Lucas A. H., Rigden D. J., Galperin M. Y., Jedrzejas M. J. (2007) Proteins 66, 547–558 [DOI] [PubMed] [Google Scholar]
- 34.van Stokkum I. H., Spoelder H. J., Bloemendal M., van Grondelle R., Groen F. C. (1990) Anal. Biochem. 191, 110–118 [DOI] [PubMed] [Google Scholar]
- 35.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger T. C. (2003) Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 37.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 44.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 45.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 47.Harding M. M. (1999) Acta Crystallogr. Sect. D 55, 1432–1443 [DOI] [PubMed] [Google Scholar]
- 48.Kastern W., Sjöbring U., Björck L. (1992) J. Biol. Chem. 267, 12820–12825 [PubMed] [Google Scholar]
- 49.Björck L. (1988) J. Immunol. 140, 1194–1197 [PubMed] [Google Scholar]
- 50.Beckingham J. A., Bottomley S. P., Hinton R., Sutton B. J., Gore M. G. (1999) Biochem. J. 340, 193–199 [PMC free article] [PubMed] [Google Scholar]
- 51.Derrick J. P., Wigley D. B. (1994) J. Mol. Biol. 243, 906–918 [DOI] [PubMed] [Google Scholar]
- 52.Sauer-Eriksson A. E., Kleywegt G. J., Uhlén M., Jones T. A. (1995) Structure 3, 265–278 [DOI] [PubMed] [Google Scholar]
- 53.Housden N. G., Harrison S., Roberts S. E., Beckingham J. A., Graille M., Stura E., Gore M. G. (2003) Biochem. Soc. Trans. 31, 716–718 [DOI] [PubMed] [Google Scholar]
- 54.Graille M., Stura E. A., Housden N. G., Beckingham J. A., Bottomley S. P., Beale D., Taussig M. J., Sutton B. J., Gore M. G., Charbonnier J. B. (2001) Structure 9, 679–687 [DOI] [PubMed] [Google Scholar]
- 55.Graille M., Harrison S., Crump M. P., Findlow S. C., Housden N. G., Muller B. H., Battail-Poirot N., Sibaï G., Sutton B. J., Taussig M. J., Jolivet-Reynaud C., Gore M. G., Stura E. A. (2002) J. Biol. Chem. 277, 47500–47506 [DOI] [PubMed] [Google Scholar]
- 56.Krishnan V., Gaspar A. H., Ye N., Mandlik A., Ton-That H., Narayana S. V. (2007) Structure 15, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarz-Linek U., Höök M., Potts J. R. (2006) Microbes Infect. 8, 2291–2298 [DOI] [PubMed] [Google Scholar]
- 58.Wikström M., Drakenberg T., Forsén S., Sjöbring U., Björck L. (1994) Biochemistry 33, 14011–14017 [DOI] [PubMed] [Google Scholar]
- 59.Forsgren A., Sjöquist J. (1966) J. Immunol. 97, 822–827 [PubMed] [Google Scholar]
- 60.Björck L., Kronvall G. (1984) J. Immunol. 133, 969–974 [PubMed] [Google Scholar]
- 61.Sjodahl J. (1977) Eur. J. Biochem. 73, 343–351 [DOI] [PubMed] [Google Scholar]
- 62.Guss B., Eliasson M., Olsson A., Uhlén M., Frej A. K., Jörnvall H., Flock J. I., Lindberg M. (1986) EMBO J. 5, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy J. P., Duggleby C. J., Atkinson M. A., Trowern A. R., Atkinson T., Goward C. R. (1994) Mol. Microbiol. 12, 911–920 [DOI] [PubMed] [Google Scholar]
- 64.Langone J. J. (1982) Adv. Immunol. 32, 157–252 [PubMed] [Google Scholar]
- 65.Goward C. R., Scawen M. D., Murphy J. P., Atkinson T. (1993) Trends Biochem. Sci. 18, 136–140 [DOI] [PubMed] [Google Scholar]
- 66.Nilson B. H., Solomon A., Björck L., Akerström B. (1992) J. Biol. Chem. 267, 2234–2239 [PubMed] [Google Scholar]
- 67.Enokizono J., Wikström M., Sjöbring U., Björck L., Forsén S., Arata Y., Kato K., Shimada I. (1997) J. Mol. Biol. 270, 8–13 [DOI] [PubMed] [Google Scholar]
- 68.Wikström M., Sjöbring U., Drakenberg T., Forsén S., Björck L. (1995) J. Mol. Biol. 250, 128–133 [DOI] [PubMed] [Google Scholar]
- 69.Beckingham J. A., Housden N. G., Muir N. M., Bottomley S. P., Gore M. G. (2001) Biochem. J. 353, 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svensson H. G., Wedemeyer W. J., Ekstrom J. L., Callender D. R., Kortemme T., Kim D. E., Sjöbring U., Baker D. (2004) Biochemistry 43, 2445–2457 [DOI] [PubMed] [Google Scholar]
- 71.Woof J. M., Mestecky J. (2005) Immunol. Rev. 206, 64–82 [DOI] [PubMed] [Google Scholar]
- 72.Kazeeva T. N., Shevelev A. B. (2009) Biochemistry 74, 12–21 [DOI] [PubMed] [Google Scholar]
- 73.Leo J. C., Goldman A. (2009) Mol. Immunol. 46, 1860–1866 [DOI] [PubMed] [Google Scholar]
- 74.Pleass R. J., Areschoug T., Lindahl G., Woof J. M. (2001) J. Biol. Chem. 276, 8197–8204 [DOI] [PubMed] [Google Scholar]
- 75.Agniswamy J., Nagiec M. J., Liu M., Schuck P., Musser J. M., Sun P. D. (2006) Structure 14, 225–235 [DOI] [PubMed] [Google Scholar]
- 76.Langley R., Wines B., Willoughby N., Basu I., Proft T., Fraser J. D. (2005) J. Immunol. 174, 2926–2933 [DOI] [PubMed] [Google Scholar]
- 77.Reuter G. (2001) Curr. Issues Intest. Microbiol. 2, 43–53 [PubMed] [Google Scholar]
- 78.Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O. P., Leer R., Tarchini R., Peters S. A., Sandbrink H. M., Fiers M. W., Stiekema W., Lankhorst R. M., Bron P. A., Hoffer S. M., Groot M. N., Kerkhoven R., de Vries M., Ursing B., de Vos W. M., Siezen R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolotin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S. D., Sorokin A. (2001) Genome Res. 11, 731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peterson D. A., McNulty N. P., Guruge J. L., Gordon J. I. (2007) Cell Host Microbe 2, 328–339 [DOI] [PubMed] [Google Scholar]
- 81.Macpherson A. J., McCoy K. D., Johansen F. E., Brandtzaeg P. (2008) Mucosal Immunol. 1, 11–22 [DOI] [PubMed] [Google Scholar]
- 82.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.