Abstract
Heat shock proteins of 70 kDa (Hsp70s) and their J domain-containing Hsp40 cofactors are highly conserved chaperone pairs that facilitate a large number of cellular processes. The observation that each Hsp70 partners with many J domain-containing proteins (JDPs) has led to the hypothesis that Hsp70 function is dictated by cognate JDPs. If this is true, one might expect highly divergent Hsp70-JDP pairs to be unable to function in vivo. However, we discovered that, when a yeast cytosolic JDP, Ydj1, was targeted to the mammalian endoplasmic reticulum (ER), it interacted with the ER-lumenal Hsp70, BiP, and bound to BiP substrates. Conversely, when a mammalian ER-lumenal JDP, ERdj3, was directed to the yeast cytosol, it rescued the temperature-sensitive growth phenotype of yeast-containing mutant alleles in two cytosolic JDPs, HLJ1 and YDJ1, and activated the ATP hydrolysis rate of Ssa1, the yeast cytosolic Hsp70 that partners with Hlj1 and Ydj1. Surprisingly, ERdj3 mutants that were compromised for substrate binding were unable to rescue the hlj1ydj1 growth defect even though they stimulated the ATPase activity of Ssa1. Yet, J domain mutants of ERdj3 that were defective for interaction with Ssa1 restored the growth of hlj1ydj1 yeast. Taken together, these data suggest that the substrate binding properties of certain JDPs, not simply the formation of unique Hsp70-JDP pairs, are critical to specify in vivo function.
Introduction
Hsp70s3 constitute a highly conserved family of molecular chaperones that are found in all organisms and in all cellular organelles. Due to their ability to bind to unfolded regions on nascent polypeptides or unassembled subunits of heteromeric complexes in a nucleotide-dependent manner, these chaperones play critical roles in diverse cellular processes (1, 2). Two distinct sets of cofactors tightly monitor Hsp70 action by regulating ATPase activity (3, 4): J domain-containing proteins (JDPs) of the Hsp40/DnaJ family and nucleotide exchange factors. The highly conserved ∼70-amino acid J domain of JDPs contacts the nucleotide-binding domain of Hsp70 and enhances ATPase activity by inducing a conformational change (5, 6). This leads to enhanced binding of Hsp70s to substrates. Moreover, some JDPs directly bind to unfolded regions on substrate proteins through their substrate-binding domain and deliver the unfolded protein to the ATP-bound form of their Hsp70 partner (7, 8), whereas others contain atypical domains that specify exclusive functions (9, 10). The nucleotide exchange factors, on the other hand, release bound ADP, which triggers ATP rebinding and subsequent substrate release from the Hsp70.
The JDPs can be classified into three groups (11, 12): (i) type I JDPs are most similar to DnaJ and contain a J domain followed by a glycine/phenylalanine-rich region and a cysteine-rich region with four repeats of a CXXCXGXG-type zinc finger; (ii) type II JDPs lack the cysteine-rich region and are unable to coordinate Zn2+; and (iii) type III JDPs only have the J domain in common with DnaJ. Notably, the number of JDPs exceeds the number of Hsp70s and nucleotide exchange factors in most organisms/organelles. In fact, a single Hsp70 can interact with different JDPs to form unique Hsp70-JDP pairs that participate in specific cellular functions (12, 13). Despite the high conservation of the J domain, JDPs are not necessarily interchangeable between organelles or organisms (11, 14), further suggesting that there is specificity in the interaction between Hsp70s and JDPs. Specificity might be essential for substrate recognition and delivery, for the targeting of Hsp70 to distinct cellular locations, and/or for catalyzing protein folding in distinct chemical environments within organelles.
To begin to address this hypothesis, a recent study tested the ability of 13 different JDPs to interact with resident Hsp70s in the yeast cytoplasm (13). In many cases, the ability of a JDP to stimulate the ATPase activity of a particular Hsp70 was sufficient to constitute a functional pair. However, distinct features of JDPs, such as the ability to interact with substrates, might also be required for specific functions (7, 15–17). To better resolve this apparent discrepancy, we have compared the abilities of two type I JDPs, a mammalian, ER-lumenal protein (ERdj3), and a yeast cytoplasmic protein (Ydj1) to functionally substitute for each other. Our data, combined with those of a previous study (13), suggest that both the binding of JDPs to substrates and their association with Hsp70s are essential to support cell viability and chaperone-dependent function.
EXPERIMENTAL PROCEDURES
Preparation of Ydj1, Hlj1, ERdj3, and ERdj4 Constructs
To target Ydj1 to the mammalian ER, a signal sequence (ss) was engineered onto the N terminus of Ydj1 using pAV4 (18) as a template and the following PCR primers (lowercase letters represent the inserted signal sequence, and the underlined letters indicate a BamHI site): 5′ primer: CGGGATCCatggctccgcagaacctgagcaccttttgcctgttgctgctatacctcatcggggcggtgattgccGTTAAAGAAACTAAGTTTTACGATATTCTAGGTGTTCC, and 3′ primer: CGGGATCCTCATTGAGATGCACATTGAACACCTTC. The PCR product was digested with BamHI and inserted into 3HA-DSL, a mammalian expression vector that was modified from the original pSG5 vector (Neupogen) by the addition of several unique restriction enzyme recognition sequences to the multiple cloning site and a triple-HA encoding sequence at the 3′-end of the multiple cloning site. After determining the direction of insertion, the translation “STOP” codon in ssYdj1-3HA-DSL was destroyed using the QuikChange site-directed mutagenesis kit (Stratagene) with the following primers: 5′ primer: GAAGGTGTTCAATGTGCATCTCAAGGATCCCCGGAATTCCTCGAG, and 3′ primer: CTCGAGGAATTCCGGGGATCCTTGAGATGCACATTGAACACCTTC. Next, a ribosomal-binding Kozak sequence (underlined, see below) was inserted immediately upstream of the translation start site of ssYdj1 using the QuikChange site-directed mutagenesis kit with the following primers: 5′ primer: GTTTAAACGGATCCACCCGGGACAGAGGAACCATGGCTCCGCAGAAC and 3′ primer: GTTCTGCGGAGCCATGGTTCCTCTGTCCCGGGTGGATCCGTTTAAAC. A second construct, ssYdj1C406S, was made in which the farnesylation acceptor site in ssYdj1 was mutated (denoted by the underlined sequence) with the primer pair: 5′ primer: GGTGGCGAAGGTGTTCAAAGTGCATCTCAAGGATCCCCG and 3′ primer: CGGGGATCCTTGAGATGCACTTTGAACACCTTCGCCACC. To target Hlj1 to the mammalian ER, the signal sequence from ERdj3 was engineered onto the N terminus of Hlj1 using yeast genomic DNA as a template and the following PCR primers (lowercase letters represent the inserted signal sequence, and the underlined letters indicate EcoRV and BglII sites in the 5′ and 3′ primers, respectively): 5′ primer: CGGATATCACCCGGGACAGAGGAACCatggctccgcagaacctgagcaccttttgcctgttgctgctatacctcatcggggcggtgattgccTCTTTCACTGAGGATCAAGAAAAA, and 3′ primer: GGAGATCTACTAAACAGGTAATCTTTAATCATAGGAAGAACAAT. The PCR product was digested with EcoRV and BglII and inserted into 3HA-DSL.
To express ERdj3 in yeast, two constructs were produced under the control of the PGPD promoter in the multicopy pGPD426 vector (19) as follows. (i) Full-length human ERdj3 containing its endogenous ER-targeting was amplified by PCR from the ERdj3-3HA-DSL vector (20) with the following primer pair (the underlined letters represent the EcoRI and BamHI sites introduced for cloning purposes): 5′ primer: CGGAATTCGGACCCGGGAC, and 3′ primer: CGGGATCCATATCCTTGCAGTCCATTGTATACCTTCTG. The resulting PCR product was digested with EcoRI and BamHI and inserted into pGPD426. (ii) For cytosolic expression, CaaX-ERdj3 was generated using the following primer pair with ERdj3-3HA-DSL serving as the template (the underlined letters on the 5′ primer represent the beginning of the coding sequence for the mature ERdj3 protein without its signal sequence, while the underlined sequence on the 3′ primer represents the farnesylation sequence; the lowercase letters represent the restriction sites used for cloning): 5′ primer: CGggatccGGAACCATGGGACGAGATTTCTATAAGATCTTGGGG, and 3′ primer: CCCaagcttTCATTGAGATGCACATTGCAGTCCATTGTATACCTTCTGC. The resulting PCR product lacked the N-terminal signal sequence and contained a “CASQ” farnesylation sequence in lieu of the final two C-terminal amino acids (GY) in ERdj3. Next, the PCR product was digested with BamHI and HindIII and inserted into pGPD426. All CaaX-ERdj3 mutants were generated by QuikChange site-directed mutagenesis using primer pairs previously described (20).
Similarly, for expression of ERdj4 in the yeast ER, full-length ERdj4 was amplified from the pIRES2-EGFP-ERdj4 vector (a kind gift from Timothy Weaver, University of Cincinnati) using the primer pair (the underlined letters represent the EcoRI and HindIII sites introduced for cloning purposes into the 5′ and 3′ primers, respectively): 5′ primer: CGGAATCCATGGCTACTCCCCAGTCAATTTTCATCTTTGCA, and 3′ primer: GGAAGCTTCTACTGTCCTGAACAGTCAGTGTATGTAGTAAC. The resulting product was digested with EcoRI and HindIII enzymes and inserted into pGPD426. A cytosolic form of ERdj4 was constructed by amplifying ERdj4 with the primer pair (the underlined letters on the 5′ primer represent the beginning of the coding sequence for the cytosolic form of ERdj4 protein without its signal sequence, while the underlined sequence on the 3′ primer represents the farnesylation sequence; the lowercase letters represent the restriction sites used for cloning): 5′ primer: CGgaatccTTAATTCTGGCCTCAAAAAGCTACTATGATATCTT, and 3′ primer: GGaagcttTCATTTGAGATGCACACTGTCCTGAACAGTCAGTGTATGTAGTAA, digested with the EcoRI and HindIII enzymes, and inserted into the pGPD426 vector.
Expression and Detection of Ydj1 and Hlj1 in Mammalian Cells
Cells were transfected with the indicated vectors using the FuGENE 6 transfection reagent (Roche Applied Science). A vector that encodes Chinese hamster BiP has previously been described (21). For immunofluorescence (22), transfected cells grown on coverslips were fixed and stained with an anti-HA antibody to detect ssYdj1 followed by fluorescein isothiocyanate-labeled secondary antibody. Grp94, an abundant ER lumenal protein, served as an ER marker and was detected with an anti-Grp94 antiserum (22) followed by a TRITC-conjugated secondary antibody.
To detect interactions between JDPs and either BiP or substrate, 48 h post-transfection, cells were labeled with Tran35S-label (Amersham Biosciences) for 3 h, and cell lysates were prepared (20). To stabilize protein complexes, cells were treated with 150 μg/ml 3,3′-dithiobispropionic acid N-hydroxysuccinimide ester for 1 h on ice, quenched with 100 μl of 1 m glycine, and lysed in a non-ionic lysing buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% deoxycholic acid, and 0.5% Nonidet P-40). Solubilized proteins were incubated with the indicated antisera and precipitated with Protein A-Sepharose beads. The immunoprecipitated complexes were subjected to denaturing gel electrophoresis, and after enhancing with Amplify (Amersham Biosciences Bioscience), the signals were detected by chemiluminescence.
Protein Expression, Purification, and ATPase Assays
Hexahistidine-tagged recombinant wild-type and mutant human ERdj3 (20) and hamster BiP (23) proteins were expressed in Escherichia coli M15 cells and purified under non-denaturing conditions using nickel-nitrilotriacetic acid-agarose (Qiagen QIAexpress System) as described. The following proteins were purified using previously established protocols: Ssa1p (24), Ydj1 (25), glutathione S-transferase-tagged J domain of Hlj1 (26), hexahistidine-tagged Kar2 (24), and the glutathione S-transferase-tagged J domain of Sec63 (27). Steady-state ATPase assays using the indicated molar ratios of the JDP to Hsp70 were performed as described (24, 25).
Rescue of the Slow Growth Phenotype of Mutant Yeast Strains
The following Saccharomyces cerevisiae yeast strains were used for complementation studies: SCJ1JEM1 (MATα lys2-801 leu2-3,112 his3-Δ200 trp1-Δ901 ura3-52 suc2-Δ9) and scj1Δ jem1Δ (MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 scj1Δ::TRP1 jem1Δ::LEU2) (28); YDJ1 (MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100) and ydj1Δ (MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 ydj1-2::HIS3) (29); and HLJ1YDJ1 (MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100) and hlj1Δydj1-151 (MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hlj1Δ::TRP1 ydj1-2::HIS3 LEU2::ydj1-151) (26). The yeast strains were grown to logarithmic phase at 26 °C in yeast extract-peptone-dextrose medium containing 2% glucose and transformed with the indicated plasmids using lithium acetate (30). The resulting transformants were isolated and grown to logarithmic phase at 26 °C in selective synthetic complete medium containing 2% glucose. 10-fold serial dilutions were spotted onto solid medium and cultured at the indicated temperatures for 2 days. Where indicated, NaCl was included in the medium at a final concentration of 0.4 m, and sorbitol was included at a final concentration of 1 m.
Detection of ERdj3 and ERdj4 in Yeast
Wild-type cells transformed with an empty vector or plasmids expressing ERdj3, ERdj3-CaaX, ERdj4, or ERdj4-CaaX were grown to logarithmic phase (A600 of 0.8–1.0) in selective synthetic complete medium containing 2% glucose. A total of ∼200 A600 equivalents of cells were harvested, and ER-derived microsomal membranes were prepared as previously described (31). For protein detection, ∼20 μg of membranes was resolved by SDS-PAGE, and immunoblots were analyzed using a polyclonal anti-ERdj3 antiserum (32) or monoclonal anti-ERdj4 antibody (33), as indicated..
Assays for ER-associated Degradation
The gene encoding HA-tagged Ste6p* was subcloned from pSM1911 (2μ URA3 PPGK ste6–166::HA) (34) into pRS425 (2μ LEU2) (35) to generate the p425-Ste6p*-HA plasmid. To enable selection of p425-Ste6p*-HA in the hlj1Δydj1-151 strain, the LEU2 gene was replaced with the NAT cassette, which confers resistance to the antibiotic nourseothricin, using PCR-mediated gene disruption (36). Cycloheximide chase assays to measure the degradation efficiency of Ste6p* were performed as described before (31) at 37 °C.
Chaperone-mediated Refolding of Firefly Luciferase
Luciferase refolding assays were performed as described (37). The luminescence was measured in an analytical luminescence laboratory luminometer (BD Pharmingen, San Diego, CA). The molar amounts of Hsp70s and JDPs present in the reaction are indicated in the figure legends.
RESULTS
Ydj1 Expressed in the Mammalian ER Functions as an Hsp70 Cofactor
To better understand the restrictions guiding the formation of functional Hsp70-JDP pairs, we first asked whether Ydj1, a cytosolic yeast JDP, could interact with BiP/Grp78, the mammalian ER-lumenal Hsp70. Ydj1 is a type I JDP and bears an overall sequence identity of 37% to ERdj3, a cognate BiP JDP (Fig. 1 and supplemental Fig. S1). For this set of experiments, COS cells were transfected with plasmids expressing BiP along with either a C-terminally HA-tagged form of ERdj3 as a positive control, or with two different Ydj1 constructs engineered with an N-terminal ER signal sequence for targeting to the ER and a C-terminal HA tag for detection. In one construct, the Ydj1 farnesylation site was removed (ssYdj1C406S), and in the other, it was retained (ssYdj1). In yeast, farnesylation enables Ydj1 to associate with the cytosolic surface of the ER and is essential for the function of Ydj1 at elevated temperatures (18).
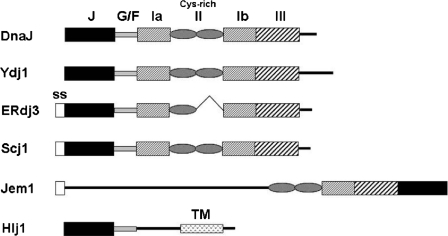
FIGURE 1.
A schematic representation of select JDPs. DnaJ, the founding member of this family of chaperones, is a bacterial type I JDP. Ydj1, ERdj3, and Scj1 are type I JDPs found in the yeast cytosol, mammalian ER, and yeast ER, respectively. Hlj1 is a type II JDP found in the yeast cytosol, whereas Jem1 is a type III JDP found in the yeast ER with an atypical domain organization. The domains are indicated by: J, J domain; G/F, glycine/phenylalanine-rich region; Ia, II, Ib, and III, various subdomains of the substrate-binding domain based on Ydj1 crystal structure (65); Cys-rich, cysteine-rich region containing the Zn2+-finger motifs; ss, signal sequence; and TM, transmembrane domain. Note that domains are not drawn to scale.
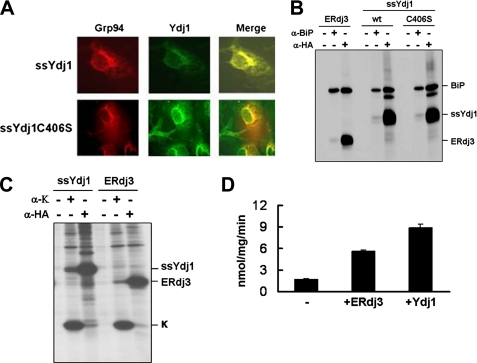
Post-transfection, the ER localization of the two Ydj1 isoforms was confirmed using indirect immunofluorescence (Fig. 2A). Next, the transfected cells were metabolically labeled, treated with a cross-linking agent to stabilize multiprotein complexes, and lysed. Proteins were immunoprecipitated with antiserum against BiP, or a monoclonal antibody against HA, to detect Ydj1. We found that the two Ydj1 isoforms efficiently associated with BiP at levels similar to those observed for ERdj3 (Fig. 2B). Interaction was detected regardless of whether the complex was isolated with anti-BiP or anti-HA antibodies, although the anti-HA antibody was better at detecting this association than the anti-BiP antibody. This is due to the fact that anti-HA immunoprecipitates nearly all of the HA-tagged forms of ERdj3 or Ydj1, whereas the polyclonal anti-BiP isolates only a fraction of BiP (data not shown). Furthermore, when COS cells were co-transfected with plasmids expressing Ydj1 and immunoglobulin κ light chain, which is normally a BiP and ERdj3 substrate (32), Ydj1 interacted with κ light chain to the same extent as ERdj3 (Fig. 2C). Taken together, these data suggested that Ydj1 attains its native conformation in the mammalian ER and is active. We also found that Ydj1 robustly stimulated the ATP hydrolysis activity of BiP to an even greater degree than ERdj3 (Fig. 2D). Finally, we discovered that purified Ydj1 enhanced the BiP-mediated refolding of denatured firefly luciferase as proficiently as ERdj3 (supplemental Fig. S2). Thus, Ydj1 can function as a BiP cofactor when targeted to the mammalian ER. This result was intriguing, because we had previously shown that Ydj1 does not bind to or functionally interact with the yeast ER-lumenal Hsp70, Kar2, in vitro (24).
FIGURE 2.
Ydj1 interacts with both BiP and κ light chain in the mammalian ER. A, COS cells transfected with HA-tagged ssYdj1 or ssYdj1-C306S were grown on coverslips, fixed and stained with an anti-HA antibody to detect the Ydj1 isoforms. Grp94 served as an ER marker. B, COS cells were co-transfected with cDNAs encoding BiP and the indicated HA-tagged ERdj3 or Ydj1 constructs. Metabolically labeled, 3,3′-dithiobispropionic acid N-hydroxysuccinimide ester cross-linked cell lysates were immunoprecipitated with anti-HA or anti-BiP antibodies, or Protein A-Sepharose alone. Isolated proteins were separated by denaturing gel electrophoresis. C, COS cells were co-transfected with cDNAs encoding κ light chain and either ERdj3 or ssYdj1. Cell lysates were immunoprecipitated with anti-κ or anti-HA antibodies, or Protein A-Sepharose alone. Samples were analyzed as described in B. D, the ATPase activity of BiP was measured in the absence (−) or presence of either ERdj3 or Ydj1. Reactions contained 1 μg of BiP and an 8-fold molar excess of ERdj3 or Ydj1. ATPase activity is expressed as nanomoles of ATP hydrolyzed per milligram of protein per minute. Data represent the means of a minimum of three independent experiments ± S.E.
To determine whether Ydj1 was unique in its ability to function in the mammalian ER, we expressed Hlj1, another yeast cytosolic JDP, in COS cells and examined its ability to bind to BiP or a BiP substrate. We found that Hlj1 associated poorly with BiP in these cells and was unable to detect any association with immunoglobulin γ heavy chains (supplemental Fig. S3). These data argue that not all JDPs can serve as a cofactor for BiP in vivo.
Overexpression of Cytosolically Localized ERdj3 Rescues the Temperature-sensitive Growth Defect of hlj1Δydj1-151 Yeast
In yeast, Scj1 and Jem1 are two ER-localized JDPs (28, 38) that interact with Kar2, the yeast BiP homolog, and are required for multiple ER functions, including protein folding (39) and ER-associated degradation (ERAD) (40). Scj1 is a soluble type I JDP (Fig. 1) whose structural organization is similar to ERdj3, although its overall sequence is only 30% identical to that of ERdj3 (supplemental Fig. S1). Jem1 is a membrane-associated type III JDP (Fig. 1), and, owing to its non-canonical domain arrangement, the sequence comparison of Jem1 and ERdj3 was limited to that of the J domains. This analysis revealed that Jem1 and ERdj3 share 35% sequence identity within their J domains (supplemental Fig. S1). Loss of both Scj1 and Jem1 (scj1Δjem1Δ) reduces the degradation efficiency of soluble ERAD substrates (40) and induces the unfolded protein response (28, 39). In addition, the scj1Δjem1Δ strain exhibits a slow growth phenotype at elevated temperatures (28).
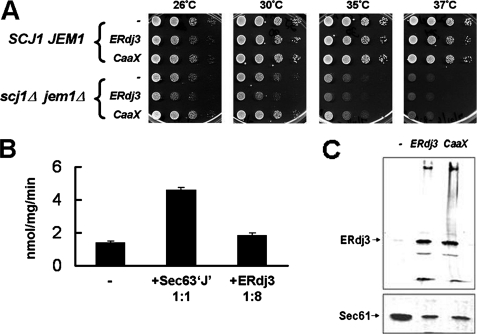
Because ERdj3 is ER-localized and contains intramolecular disulfide bonds (41), we initially tested whether this JDP could substitute for Scj1 and Jem1 in the yeast ER. Two ERdj3 constructs were created for strong, constitutive expression from the PGPD promoter (19). One contains an ER-targeting signal sequence (wild-type ERdj3), whereas the other lacks this sequence but possesses a CASQ farnesylation sequence at the C terminus that attaches the protein to the cytosolic side of the ER membrane (CaaX-ERdj3); the farnesylation sequence is the same as that found in Ydj1 (18). When the constructs were introduced into wild-type SCJ1JEM1 yeast, they did not affect growth, indicating that overexpression of these heterologous proteins is not toxic (Fig. 3A). As expected, when cytosolically targeted CaaX-ERdj3 was overexpressed in the scj1Δjem1Δ strain, it did not restore growth at elevated temperatures, however, neither did ERdj3 (Fig. 3B). To determine whether the lack of an effect on scj1Δjem1Δ growth was due to ineffective Kar2-ERdj3 interaction, we measured the ability of ERdj3 to stimulate the ATPase activity of Kar2. As shown in Fig. 3B, ERdj3 only weakly stimulated the ATP hydrolysis rate of Kar2, as compared with Sec63, a cognate Kar2 JDP that is essential for nascent polypeptide translocation across the ER membrane (27, 42, 43). Moreover, the lack of an effect was not due to ERdj3 being unstable in yeast cells, because we were able to detect its expression by Western blotting (Fig. 3C). Taken together, these observations suggest that ERdj3 is unable to functionally interact with Kar2 and hence cannot substitute for Scj1 and Jem1 in vivo.
FIGURE 3.
ER-expressed ERdj3 is unable to rescue the growth phenotype of the scj1Δjem1Δ strain. A, 10-fold serial dilutions were performed for wild-type (SCJ1JEM1) and mutant (scj1Δjem1Δ) yeast strains containing an empty vector (−), a vector for the expression of an ER-targeted form (ERdj3), or a vector encoding a cytosolically localized form (CaaX) of full-length ERdj3. Cells were plated onto selective medium and incubated for 2 days at the indicated temperatures. B, the ATPase activity of Kar2 was measured alone (−) or in the presence of equimolar amounts of the J domain of Sec63 or an 8-fold molar excess of ERdj3 as described in the legend to Fig. 2D. Data represent the means of a minimum of three independent experiments ± S.E. C, microsomal membranes were prepared from wild-type yeast strains that were transformed with an empty vector (−) or vectors encoding either ERdj3 or CaaX-ERdj3. ERdj3 was detected using anti-ERdj3 antisera and primarily migrates at its predicted molecular mass of 42 kDa. Sec61, as detected using anti-Sec61 antiserum (66), served as a loading control.
Based on our observation that a yeast cytosolic JDP could function in the mammalian ER (Fig. 2), we next asked if ERdj3 was able to function in the yeast cytosol. The yeast cytosol contains 13 JDPs of which the best characterized is Ydj1. YDJ1 deletion results in slow growth at 30 °C and inviability at elevated temperatures (44). However, the slow growth of the ydj1Δ strain can be rescued by the overexpression of at least five other cytosolic JDPs (13). Interestingly, expression of the J domain alone of these five JDPs is sufficient to substitute for Ydj1 at 30 °C, suggesting that the J domain-mediated activation of Hsp70 ATPase activity is critical to support the growth of ydj1Δ yeast (13).
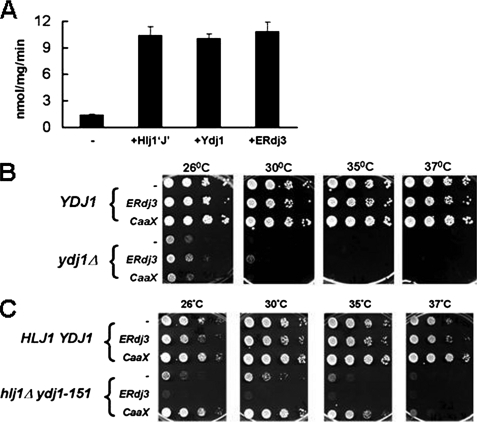
To determine whether ERdj3 could substitute for Ydj1, we first asked whether ERdj3 could stimulate the ATPase activity of Ssa1, an essential cytosolic Hsp70 that interacts with Ydj1 to execute key cellular functions (14, 26, 34, 45–49). We discovered that ERdj3 proficiently stimulated the ATP hydrolysis rate of Ssa1, to a similar extent as Ydj1 or Hlj1, another J domain containing Ssa1 cofactor (Fig. 4A; see below). Next, we performed serial dilution analyses on wild-type YDJ1 and mutant ydj1Δ yeast strains transformed with plasmids expressing either ERdj3 or CaaX-ERdj3. ERdj3 did not rescue the slow growth phenotype of ydj1Δ yeast, which was anticipated due to its expression in the ER lumen rather than on the cytosolic face of the ER. However, contrary to our expectations, neither did cytosolically targeted CaaX-ERdj3 (Fig. 4B). Furthermore, varying the levels of CaaX-ERdj3 expression through the use of several different promoters that drive varying levels of transcription did not enhance its ability to rescue the slow growth phenotype of the ydj1Δ strain (data not shown). These data suggest that, despite its ability to be expressed in yeast (Fig. 3C) and to stimulate the ATPase activity of Ssa1 in vitro (Fig. 4A), cytosolic expression of ERdj3 is not sufficient to overcome the strong impairment of cellular homeostasis that results from deleting the YDJ1 locus.
FIGURE 4.
Cytosolic expression of ERdj3 rescues the temperature-sensitive growth phenotype of the hlj1Δydj1-151 strain. A, the ATPase activity of Ssa1 was measured either alone (−) or in the presence of Ydj1, Hlj1, or ERdj3 as described in the legend to Fig. 2D. The molar ratio of Ssa1 to the JDPs was 1:2. Data represent the means of a minimum of three independent experiments ± S.E. B and C, either an empty vector (−), or a vector containing an ER-targeted form (ERdj3) or a vector engineered to produce an ER-tethered, cytosolically localized form (CaaX) of ERdj3 was transformed into (B) wild-type (YDJ1) and mutant (ydj1Δ) yeast strains, or (C) wild-type (HLJ1YDJ1) and mutant (hlj1Δydj1-151) yeast strains. The resulting transformants were serially diluted onto selective medium and incubated as described in the legend to Fig. 3A.
We therefore decided to utilize an alternate yeast strain, hlj1Δydj1-151, which contains a temperature-sensitive allele of YDJ1 and lacks another ER-associated JDP with a cytosolic J domain, Hlj1 (26). Hlj1 is a type II JDP and bears 27% overall sequence identity to ERdj3 (Figs. 1 and supplemental Fig. S1). The hlj1Δydj1-151 yeast strain exhibits a tight temperature-sensitive growth phenotype but not the slow growth displayed by ydj1Δ yeast; this strain also exhibits defects in the ERAD of select integral membrane proteins, such as a mutant form of the yeast a-factor transporter, Ste6p (i.e. Ste6p*) (34), and the cystic fibrosis transmembrane conductance regulator (26). Plasmids engineered for the expression of ERdj3 and CaaX-ERdj3 were transformed into HLJ1YDJ1 and hlj1Δydj1-151 yeast strains, and indirect immunofluorescence microscopy was performed to confirm that the proteins were expressed (supplemental Fig. S4). Next, serial dilution analyses were performed to analyze the growth of the transformed strains at various temperatures. In contrast to the results using ydj1Δ yeast, we observed that cytosolic CaaX-ERdj3, but not ER-targeted ERdj3, restored the growth of hlj1Δydj1-151 cells at temperatures up to 37 °C (Fig. 4C). In fact, the ER-lumenally expressed ERdj3 actually exacerbated the growth defect of the hlj1Δydj1-151 strain (Fig. 4C), probably due to its drastic effect on ER morphology (supplemental Fig. S4). These results indicate that CaaX-ERdj3 functions as a cochaperone for Ssa1 in vivo and can compensate for Hlj1 and Ydj1, but only in the presence of a partially functional copy of Ydj1.
Interestingly, we found that the expression of ERdj4, another mammalian ER JDP, in either the yeast cytoplasm (via a CaaX motif) or in the yeast ER (via its endogenous signal sequence), was unable to complement the growth defects of the ydj1Δ, hlj1Δydj1-151, or scj1Δjem1Δ yeast strains, despite robust ERdj4 expression from the PGPD promoter (data not shown). In accordance with other data (see above), this result suggests that not all eukaryotic JDPs are able to substitute for one another.
The Substrate Binding Domain of ERdj3 Is Required to Rescue the Slow Growth Phenotype of hlj1Δ ydj1-151 Yeast
Previous studies showed that substrate binding is essential for Ydj1 to optimally function in vivo and in vitro (15–17), and Ydj1 and ERdj3 have similar substrate-binding domains (supplemental Fig. S1). We recently demonstrated that the binding of ERdj3 to substrates requires three features: (i) the presence of domain II (Fig. 1), (ii) the pocket formed by hydrophobic amino acids in domain I, and (iii) dimerization, which occurs through interactions in the C-terminal region and requires the presence of phenylalanine 326 (20).
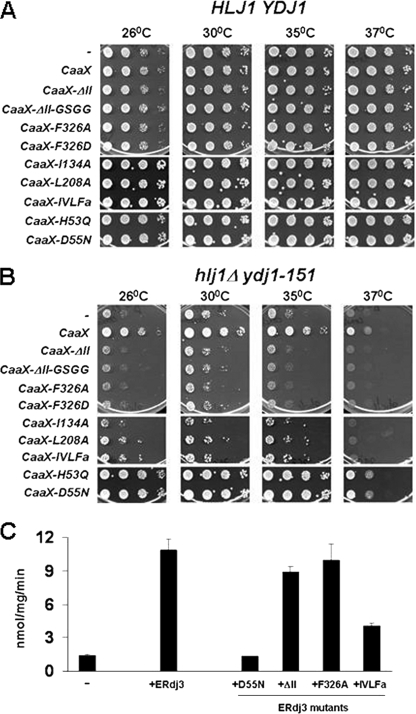
Having established a new system to assay the function of ERdj3, we next wished to determine whether substrate interaction is required for CaaX-ERdj3 to rescue the growth of hlj1Δydj1-151 yeast. We therefore expressed seven CaaX-ERdj3 substrate-binding mutants in this strain. These included mutants in which domain II, spanning amino acids 160–200, was deleted (ΔII) or was replaced by a “GSGG” linker (ΔII-GSGG), two mutants in which dimerization was inhibited (F326A and F326D), single amino acid substitution mutants in domain I (I134A and L208A), and a mutant in which four hydrophobic residues that affect substrate binding were altered (IVLFa). In addition, we expressed two versions of CaaX-ERdj3 that are defective for Hsp70 interaction, H53Q and D55N (32, 50). These mutations lie in the invariant HPD motif of the J domain (51).
Expression of the mutant CaaX-ERdj3 proteins in wild-type HLJ1YDJ1 yeast did not affect cell growth (Fig. 5A), indicating that these proteins do not exert a dominant negative effect. Moreover, none of the substrate-binding mutants rescued the temperature-sensitive phenotype of the hlj1Δydj1-151 strain, even though CaaX-ERdj3 was again able to improve growth up to 37 °C (Fig. 5B). The failure of the substrate-binding mutants to rescue the growth phenotype was not due to their inability to stimulate the ATPase activity of Ssa1, because a member of each mutant class stimulated Ssa1 ATP hydrolysis to levels comparable to wild-type ERdj3 (Fig. 5C and data not shown); however, the IVLFa mutant stimulated the ATPase activity of Ssa1 only about half as efficiently as wild-type ERdj3. We also noted that the mutant proteins were expressed at similar levels as CaaX-ERdj3 as determined by indirect immunofluorescence microscopy (supplemental Fig. S4 and data not shown).
FIGURE 5.
ERdj3 substrate-binding mutants fail to enhance the growth of hlj1Δydj1-151 yeast. Cytosolic forms (CaaX) of wild-type or mutant ERdj3 were expressed in wild-type (HLJ1YDJ1) (A) or hlj1Δydj1-151 mutant (B) yeast strains. Cells were plated and incubated as described in the legend to Fig. 3A. C, the ATPase activity of Ssa1 was measured in the absence (−) or presence of the indicated wild-type and mutant ERdj3 proteins as described in the legend to Fig. 2D except that the molar ratio of Ssa1 to the indicated JDPs was 1:2. Data represent the means of a minimum of three independent experiments ± S.E.
In contrast to the inability of the substrate-binding mutants to restore viability, the J domain mutants rescued the growth of the hlj1Δydj1-151 strain as efficiently as wild-type CaaX-ERdj3 (Fig. 5B). As anticipated, the D55N mutant protein was unable to stimulate Ssa1 ATPase activity in vitro (Fig. 5C). Together the data argue that the substrate-binding properties of CaaX-ERdj3 are required to complement the growth defect of the hlj1Δydj1-151 strain and that this does not simply depend on (or even require) the functional interaction of JDPs with a cognate Hsp70.
Cytosolically Expressed ERdj3 Compensates for Cell Wall Defects in the hlj1Δydj1-151 Strain
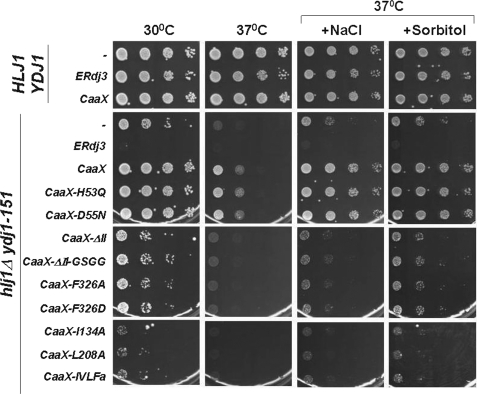
Yeast cells are fortified by a cell wall that is composed of two layers: an inner layer composed of β-1,3-glucans, β-1,6-glucans, and small amounts of chitin and a protective outer layer composed of glycosylphosphatidylinositol and Pir glycoproteins. Cell wall integrity, which is essential during growth, cell division, and stress, is maintained by the coordinated action of several signaling pathways (52). Recently, we demonstrated that Ydj1 also plays a role in the maintenance of cell wall integrity (29). For example, the slow growth phenotype of ydj1Δ mutant yeast was rescued by osmostabilizing agents such as sodium chloride and sorbitol. However, the role of Ydj1 in cell wall integrity appeared to be independent of Ssa1 function. This was because yeast cells carrying a temperature-sensitive mutant allele in SSA1, ssa1–45, were not rescued by the overexpression of genes that improve cell wall integrity. Based on these data, we next asked if hlj1Δydj1-151 yeast demonstrated cell wall defects, and if so, whether CaaX-ERdj3 expression could rescue cell wall-associated phenotypes.
First, HLJ1YDJ1 and hlj1Δydj1-151 cells were transformed with a vector control and serially diluted on growth medium supplemented with NaCl or sorbitol. We found that the growth of hlj1Δydj1-151 was partially restored at 37 °C under these conditions (Fig. 6, row labeled with “-”), suggesting that the mutant strain has a cell wall phenotype. Next, hlj1Δydj1-151 yeast were transformed with plasmids expressing either wild-type or mutant versions of CaaX-ERdj3 and were tested for growth on NaCl- or sorbitol-supplemented medium. We observed that the expression of wild-type and J domain mutants of CaaX-ERdj3, but not the substrate-binding mutants, significantly rescued the cell wall defects of hlj1Δydj1-151 yeast (Fig. 6). These data further support our hypothesis that the role of Ydj1 in the maintenance of cell wall integrity is not reliant upon its interaction with Ssa1 (29).
FIGURE 6.
CaaX-ERdj3 complements the cell wall phenotype of the hlj1Δydj1-151 strain. Either an empty vector (−), a vector containing an ER-targeted form (ERdj3), or a vector engineered to produce a cytosolically localized form (CaaX) of ERdj3 was transformed into wild-type (HLJ1YDJ1) and mutant (hlj1Δydj1-151) yeast strains. In addition, cytosolic forms (CaaX) of mutant ERdj3 were expressed in hlj1Δydj1-151 yeast. Cells were serially diluted onto selective medium containing or lacking 0.4 m NaCl or 1 m sorbitol and were incubated as described in the legend to Fig. 3A.
CaaX-ERdj3 Expression Restores ERAD in hlj1Δydj1-151 Yeast
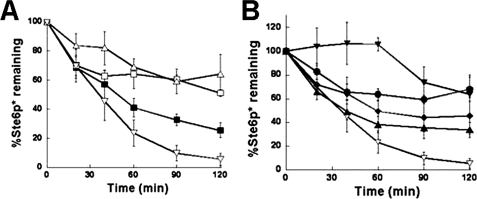
In the previous section, we reported that ERdj3 rescues a JDP-dependent cellular process that is independent of Ssa1. Next, we wished to examine the effects of ERdj3 on an Hsp70-dependent event, namely ERAD. ERAD is a quality control process during which misfolded ER-localized proteins are retrotranslocated to the cytosol and targeted to the 26 S proteasome for degradation (53). We previously showed that Hlj1 and Ydj1 act as Ssa1 cofactors during ERAD recognition and the subsequent targeting of integral membrane proteins such as Ste6p* and cystic fibrosis transmembrane conductance regulator. In keeping with this, the hlj1Δydj1-151 strain is defective for the ERAD of these substrates (26, 34). To determine whether CaaX-ERdj3 could substitute for Hlj1 and Ydj1 during ERAD, we co-expressed wild-type or mutant CaaX-ERdj3 proteins with Ste6p* in the hlj1Δydj1-151 strain and performed a cycloheximide chase analysis to measure the degradation efficiency of Ste6p*. Although the hlj1Δydj1-151 strain degraded Ste6p* poorly, we found that CaaX-ERdj3 overexpression significantly accelerated the Ste6p* degradation (Fig. 7A). In contrast, none of the ERdj3 mutants were as efficient as wild-type CaaX-ERdj3 in compensating for the ERAD defect in the mutant strain (Fig. 7B). Intriguingly, the strongest defect was observed when the ability of the D55N J domain mutant form of CaaX-ERdj3 was tested, which is unable to stimulate Ssa1 ATPase activity (Fig. 5C). These data indicate that the ability of CaaX-ERdj3 to improve ERAD is primarily dependent on its interaction with the cytosolic Hsp70, Ssa1. However, peptide binding by the JDP appears to be important for ERAD substrate recognition and/or targeting.
FIGURE 7.
CaaX-ERdj3 substitutes for Hlj1 and Ydj1 during ERAD. Cycloheximide chase assays were performed to measure the degradation of Ste6p* in wild-type (HLJ1YDJ1) yeast transformed with an empty vector (A, ■) and hlj1Δydj1-151 yeast transformed with either an empty vector (□), a vector containing an ER-lumenal form of ERdj3 (▵), or a vector engineered to produce a cytosolically localized form of ERdj3 (▿) or in hlj1Δydj1-151 yeast transformed with vectors engineered to produce cytosolically localized forms of wild-type (B, ▿) and the following mutant ERdj3 proteins: D55N (▾), ΔII-GSGG (●), F326D (♦), and IVLFa (▾). Data represent the means of a minimum of three independent experiments ± S.E.
DISCUSSION
Ydj1 and ERdj3 are Type I JDPs and share a similar domain organization (Fig. 1). However, the chemical environments within the resident organelles of these JDPs are significantly different. Ydj1 resides in the yeast cytosol, an environment that is relatively less oxidizing than the native milieu of ERdj3, the mammalian ER. Consequently, the cysteines of ERdj3 are involved in intramolecular disulfide bridges (41), whereas the cysteine-rich region of Ydj1 coordinates Zn2+, similar to other type I DnaJ homologs (54). Given these differences and the low overall sequence identity shared by Ydj1 and ERdj3 (i.e. 37%), one might expect that these JDPs would be unable to substitute for one another in vivo. To our surprise, we observed that Ydj1 could function in the mammalian ER and ERdj3 could function in the yeast cytosol, indicating that these JDPs attain their native or near-native conformations in diverse environments. In contrast, ERdj3 was unable to compensate for two ER-localized yeast JDPs, Scj1 and Jem1, most likely due to its failure to efficiently stimulate the ATPase activity of the yeast BiP homolog, Kar2 (Fig. 3B). Together, these results demonstrate that Ydj1 and ERdj3 associate productively with Hsp70s in unique subcellular compartments and can exhibit a relaxed Hsp70 specificity. In addition, given our previous observation that Ydj1 cannot interact productively with Kar2 (24), these data also indicate that Kar2 is more fastidious than Ssa1 or BiP with regards to JDP association.
We also discovered that, although cytosolically expressed ERdj3 could complement the temperature-sensitive growth of the hlj1Δydj1-151 strain, it could not rescue the slow growth phenotype of ydj1Δ yeast. This was somewhat surprising given that the overexpression of isolated J domains is sufficient to compensate for the loss of Ydj1 (13). This phenomenon probably arises due to the activation of the ATPase and folding activities of cytosolic Hsp70s by the examined J domains. Accordingly, we show that ERdj3 robustly stimulates the ATPase activity of a cytosolic Hsp70, Ssa1 (Fig. 4A). However, we observed that ERdj3 poorly stimulates the Ssa1-mediated refolding of denatured firefly luciferase (data not shown). Taken together with results in Figs. 6 and 7, these data suggest that ydj1Δ yeast, but not hlj1Δydj1-151 yeast, are deficient for protein folding, and that CaaX-ERdj3 supports several other essential functions that are defective in the hlj1Δydj1-151 strain. Alternately, because the level of J domain expression also influences the efficiency of ydj1Δ growth rescue (13), it is possible that CaaX-ERdj3 was not expressed to sufficiently high levels in vivo to either interact with Ssa1 or localize correctly; hence, CaaX-ERdj3 was unable to compensate for the loss of Ydj1. Another possibility is that the interaction of ERdj3 with a second cytosolic Hsp70 such as Ssb1 (55) may be required to complement ydj1Δ growth, and ERdj3 is unable to stimulate the activity of this Hsp70. Finally, it is formally possible that ERdj3 is more suited to complement the functions of Hlj1 than those of Ydj1. Because Hlj1-specific functions and phenotypes have not been reported, there is no ready way to test this possibility.
We determined that ERdj3 mutants that fail to associate with Ssa1 are competent to rescue the temperature-sensitive and cell wall phenotypes associated with hlj1Δydj1-151 mutant yeast. This suggests that these phenotypes result from defects in processes that are independent of Ssa1 interaction. Based on the observation that substrate-binding mutants of ERdj3 were unable to rescue hlj1Δydj1-151 phenotypes (Figs. 5 and 6), and that hlj1Δ yeast do not exhibit slow growth and cell wall phenotypes (data not shown) but ydj1Δ do (29, 44), we surmise that cell wall function and robust growth at elevated temperature are linked to the ability of Ydj1 to bind to specific substrates. Formally, the cell wall phenotypes may also arise from the ability of Ydj1 to partner with Hsp82, the cytosolic Hsp90 that plays a role in high osmotic stress response in yeast (29, 56, 57) and acts in conjunction with Ydj1 in the folding and maturation of certain client proteins (54, 58, 59). The cell wall phenotype may additionally arise from altered association with Sse1, the cytosolic Hsp110 (56), which has previously been shown to genetically interact with YDJ1 (60).
The substrate binding-dependent effects of ERdj3 expression on hlj1Δydj1-151 growth, but not on ydj1Δ growth, are reminiscent of prior studies. It was observed that, although Ydj1 and Sis1, an essential yeast cytosolic JDP (61), perform unique essential cellular activities, they also exhibit substrate binding promiscuity: (i) Ydj1 overexpression is unable to support the viability of sis1Δ yeast, although Sis1 overexpression rescues the growth defect of the ydj1Δ strain (13, 61), and (ii) the substrate-binding domains of either Ydj1 or Sis1, but not both, are required for optimal yeast growth and survival (15). Taken together with our results, we infer that although certain substrate proteins can interact with multiple JDPs for delivery to cognate Hsp70s for folding/assembly, others may require specific JDPs for cellular targeting or protein folding.
To our knowledge, this study is the first demonstration that a full-length mammalian JDP can function in a non-native organelle in a divergent organism. Several other reports have analyzed the effects of overexpression of the J domains of cognate or non-cognate JDPs in related/divergent model systems (13, 62, 63), but rarely has the specific impact of the JDP substrate-binding domain been investigated. Our data, together with those of others, reveal novel insights into the biology of DnaJ family proteins. For example, because isolated J domains can complement a subset of JDP-related defects (13), it appears that for many cellular processes, the JDP-stimulated high affinity binding of partner Hsp70s to substrates is sufficient. In other cases (i.e. ERAD), the JDP must interact both with the substrate and the Hsp70, presumably to aid in the recruitment of the Hsp70 to its substrate. Finally, for other functions (i.e. cell wall integrity), the JDPs do not have to interact with an Hsp70, but merely need to recognize their substrates. In keeping with these diverse models/pathways, ERdj3 was able to function both in ERAD and in maintaining cell wall integrity, arguing that this JDP is able to perform both Hsp70-dependent and Hsp70-independent functions through its substrate-binding activity. Very recently, a novel class of Type IV JDPs has been identified, constituting proteins that lack the canonical HPD motif that is essential for interacting with Hsp70s but possess putative substrate-binding domains (64). Thus, this group of JDPs may have evolved to simply bind to substrates, presumably in an Hsp70 independent fashion.
Finally, our study opens up a new avenue to determine the functions of mammalian JDPs in ERAD. In particular, we are now positioned to examine the ability of any JDP to substitute for cytosolic homologs during this quality control process.
Supplementary Material
Acknowledgments
We thank D. Cyr for the kind provision of purified Ydj1 protein and T. Weaver for the pIRES2-EGFP-ERdj4 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants GM75061 (to J. L. B.) and GM54068 (to L. M. H.). This work was supported by Cancer Center CORE Grant CA21765 and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital (to L. M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and additional references.
- Hsp70
- heat shock protein of 70 kDa
- JDP
- J domain-containing protein
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- ss
- signal sequence
- HA
- hemagglutinin
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Munro S., Pelham H. R. (1986) Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 2.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 3.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer M. P., Bukau B. (2005) Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gässler C. S., Buchberger A., Laufen T., Mayer M. P., Schröder H., Valencia A., Bukau B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh W. C., Burkholder W. F., Lu C. Z., Zhao X., Gottesman M. E., Gross C. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rüdiger S., Schneider-Mergener J., Bukau B. (2001) EMBO J. 20, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig E. A., Huang P., Aron R., Andrew A. (2006) Rev. Physiol. Biochem. Pharmacol. 156, 1–21 [DOI] [PubMed] [Google Scholar]
- 9.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 10.Gall W. E., Higginbotham M. A., Chen C., Ingram M. F., Cyr D. M., Graham T. R. (2000) Curr. Biol. 10, 1349–1358 [DOI] [PubMed] [Google Scholar]
- 11.Hennessy F., Nicoll W. S., Zimmermann R., Cheetham M. E., Blatch G. L. (2005) Protein Sci. 14, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheetham M. E., Caplan A. J. (1998) Cell Stress Chaperones 3, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahi C., Craig E. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caplan A. J., Cyr D. M., Douglas M. G. (1992) Cell 71, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 15.Johnson J. L., Craig E. A. (2001) J. Cell Biol. 152, 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z., Cyr D. M. (1998) J. Biol. Chem. 273, 27824–27830 [DOI] [PubMed] [Google Scholar]
- 17.Lu Z., Cyr D. M. (1998) J. Biol. Chem. 273, 5970–5978 [DOI] [PubMed] [Google Scholar]
- 18.Caplan A. J., Tsai J., Casey P. J., Douglas M. G. (1992) J. Biol. Chem. 267, 18890–18895 [PubMed] [Google Scholar]
- 19.Mumberg D., Müller R., Funk M. (1995) Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 20.Jin Y., Zhuang M., Hendershot L. M. (2009) Biochemistry 48, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendershot L., Wei J., Gaut J., Melnick J., Aviel S., Argon Y. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5269–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung K. T., Shen Y., Hendershot L. M. (2002) J. Biol. Chem. 277, 47557–47563 [DOI] [PubMed] [Google Scholar]
- 23.Wei J., Hendershot L. M. (1995) J. Biol. Chem. 270, 26670–26676 [DOI] [PubMed] [Google Scholar]
- 24.McClellan A. J., Endres J. B., Vogel J. P., Palazzi D., Rose M. D., Brodsky J. L. (1998) Mol. Biol. Cell 9, 3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cyr D. M., Lu X., Douglas M. G. (1992) J. Biol. Chem. 267, 20927–20931 [PubMed] [Google Scholar]
- 26.Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. (2004) Mol. Biol. Cell 15, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corsi A. K., Schekman R. (1997) J. Cell Biol. 137, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa S., Endo T. (1997) J. Biol. Chem. 272, 12889–12892 [DOI] [PubMed] [Google Scholar]
- 29.Wright C. M., Fewell S. W., Sullivan M. L., Pipas J. M., Watkins S. C., Brodsky J. L. (2007) Genetics 175, 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y., Hendershot L. M. (2005) Mol. Biol. Cell 16, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y., Meunier L., Hendershot L. M. (2002) J. Biol. Chem. 277, 15947–15956 [DOI] [PubMed] [Google Scholar]
- 34.Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 35.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 36.Lorenz M. C., Muir R. S., Lim E., McElver J., Weber S. C., Heitman J. (1995) Gene 158, 113–117 [DOI] [PubMed] [Google Scholar]
- 37.Bhangoo M. K., Tzankov S., Fan A. C., Dejgaard K., Thomas D. Y., Young J. C. (2007) Mol. Biol. Cell 18, 3414–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlenstedt G., Harris S., Risse B., Lill R., Silver P. A. (1995) J. Cell Biol. 129, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silberstein S., Schlenstedt G., Silver P. A., Gilmore R. (1998) J. Cell Biol. 143, 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus N. Y., Marcus R. A., Schmidt B. Z., Haslam D. B. (2007) Arch. Biochem. Biophys. 468, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodsky J. L., Schekman R. (1993) J. Cell Biol. 123, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodsky J. L., Goeckeler J., Schekman R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caplan A. J., Douglas M. G. (1991) J. Cell Biol. 114, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker J., Walter W., Yan W., Craig E. A. (1996) Mol. Cell. Biol. 16, 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oka M., Nakai M., Endo T., Lim C. R., Kimata Y., Kohno K. (1998) J. Biol. Chem. 273, 29727–29737 [DOI] [PubMed] [Google Scholar]
- 47.Kushnirov V. V., Kryndushkin D. S., Boguta M., Smirnov V. N., Ter-Avanesyan M. D. (2000) Curr. Biol. 10, 1443–1446 [DOI] [PubMed] [Google Scholar]
- 48.Park S. H., Bolender N., Eisele F., Kostova Z., Takeuchi J., Coffino P., Wolf D. H. (2007) Mol. Biol. Cell 18, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S. (2008) J. Biol. Chem. 283, 32302–32316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y., Awad W., Petrova K., Hendershot L. M. (2008) EMBO J. 27, 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai J., Douglas M. G. (1996) J. Biol. Chem. 271, 9347–9354 [DOI] [PubMed] [Google Scholar]
- 52.Lesage G., Bussey H. (2006) Microbiol. Mol. Biol. Rev. 70, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fliss A. E., Rao J., Melville M. W., Cheetham M. E., Caplan A. J. (1999) J. Biol. Chem. 274, 34045–34052 [DOI] [PubMed] [Google Scholar]
- 55.Nelson R. J., Ziegelhoffer T., Nicolet C., Werner-Washburne M., Craig E. A. (1992) Cell 71, 97–105 [DOI] [PubMed] [Google Scholar]
- 56.Shaner L., Gibney P. A., Morano K. A. (2008) Curr. Genet. 54, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X. X., Maurer K. C., Molanus M., Mager W. H., Siderius M., van der Vies S. M. (2006) FEMS Yeast Res. 6, 195–204 [DOI] [PubMed] [Google Scholar]
- 58.Cintron N. S., Toft D. (2006) J. Biol. Chem. 281, 26235–26244 [DOI] [PubMed] [Google Scholar]
- 59.Caplan A. J., Langley E., Wilson E. M., Vidal J. (1995) J. Biol. Chem. 270, 5251–5257 [DOI] [PubMed] [Google Scholar]
- 60.Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. (2002) Mol. Biol. Cell 13, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luke M. M., Sutton A., Arndt K. T. (1991) J. Cell Biol. 114, 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pesce E. R., Acharya P., Tatu U., Nicoll W. S., Shonhai A., Hoppe H. C., Blatch G. L. (2008) Int. J. Biochem. Cell Biol. 40, 2914–2926 [DOI] [PubMed] [Google Scholar]
- 63.Nicoll W. S., Botha M., McNamara C., Schlange M., Pesce E. R., Boshoff A., Ludewig M. H., Zimmermann R., Cheetham M. E., Chapple J. P., Blatch G. L. (2007) Int. J. Biochem. Cell Biol. 39, 736–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Botha M., Pesce E. R., Blatch G. L. (2007) Int. J. Biochem. Cell Biol. 39, 1781–1803 [DOI] [PubMed] [Google Scholar]
- 65.Li J., Qian X., Sha B. (2003) Structure 11, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 66.Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. (1992) Mol. Biol. Cell 3, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.