Abstract
Phospholipid hydroperoxide glutathione peroxidase (GPx4) is an intracellular antioxidant enzyme that directly reduces peroxidized phospholipids. GPx4 is strongly expressed in the mitochondria of testis and spermatozoa. We previously found a significant decrease in the expression of GPx4 in spermatozoa from 30% of infertile human males diagnosed with oligoasthenozoospermia (Imai, H., Suzuki, K., Ishizaka, K., Ichinose, S., Oshima, H., Okayasu, I., Emoto, K., Umeda, M., and Nakagawa, Y. (2001) Biol. Reprod. 64, 674–683). To clarify whether defective GPx4 in spermatocytes causes male infertility, we established spermatocyte-specific GPx4 knock-out mice using a Cre-loxP system. All the spermatocyte-specific GPx4 knock-out male mice were found to be infertile despite normal plug formation after mating and displayed a significant decrease in the number of spermatozoa. Isolated epididymal GPx4-null spermatozoa could not fertilize oocytes in vitro. These spermatozoa showed significant reductions of forward motility and the mitochondrial membrane potential. These impairments were accompanied by the structural abnormality, such as a hairpin-like flagella bend at the midpiece and swelling of mitochondria in the spermatozoa. These results demonstrate that the depletion of GPx4 in spermatocytes causes severe abnormalities in spermatozoa. This may be one of the causes of male infertility in mice and humans.
Introduction
A frequent cause of male infertility is defective sperm function, which is the main problem for close to a quarter of couples who attend infertility clinics (1–4). Considerable efforts are now focused on the identifying ultrastructural and/or molecular defects in the spermatozoa or seminal plasma to develop solutions to various types of male infertility.
Phospholipid hydroperoxide glutathione peroxidase (GPx4)2 is an intracellular selenoprotein that directly reduces peroxidized phospholipids produced in cell membranes (5). The GPx4 gene has a complex intron/exon structure (6, 7). Three different transcripts of GPx4 exist, differing in their 5′ extension and coding for a cytosolic protein (non-mitochondrial GPx4), a mitochondrial protein (mitochondrial GPx4), and a nuclear protein (nucleolar GPx4), respectively (6, 7). After cleavage of the N-terminal mitochondrial import sequence of mitochondrial GPx4, the mature protein becomes identical to the 20-kDa non-mitochondrial GPx4 (8, 9). Nuclear GPx4 was recently identified as a sperm nucleus-specific 34-kDa selenoprotein (called snGPx, for sperm nucleus-specific glutathione peroxidase) (10). It is formed by use of an alternative promoter and start codon localized in the first intron of the GPx4 gene (7, 10, 11). We previously reported that 34-kDa GPx4 localized in nucleoli in several cell lines by using an N-terminal nucleolar import signal (11). We call hereafter nuclear GPx4 nucleolar GPx4, because non-mitochondrial 20-kDa GPx4 exists both in cytosol and in the nucleus (12).
Expression of three types of GPx4 is induced significantly in testis during spermatogenesis, especially in late spermatocytes, spermatids, and spermatozoa in both humans and mice (13–15). The most abundant GPx4 of testis is associated with mitochondria (16). Overexpression of mitochondrial GPx4 in RBL2h3 cells suppresses mitochondrial-derived reactive oxygen species produced by the respiratory chain inhibitors (17) and suppresses apoptosis induced by 2-deoxyglucose, staurosporine, etoposide, and UV (18, 19). In spermatozoa, GPx4 is also mainly localized in the mitochondria (13, 16). One possible role of mitochondrial GPx4 in spermatozoa is to maintain motility via antioxidant activity against mitochondrial reactive oxygen species (13). Another possible role is to maintain the structure of the mitochondrial capsule by cross-linking with itself or other proteins (16, 20). The later hypothesis proposes that an active GPx4 is converted into an enzymatically inactive protein aggregate. The nucleolar GPx4 was found to be required for chromatin condensation and thus for sperm maturation (10).
We and other groups have demonstrated a dramatic decrease in the expression of GPx4 in spermatozoa in 30% of human infertile males diagnosed with oligoasthenozoospermia (13, 21, 22). In mice, severe selenium deficiency resulted in male infertility (23), and selenoprotein P-null male mice are infertile due to depletion of selenocysteine transport into the testis via apoE receptor 2 (apoER2) (23, 24). ApoER2-null mice also exhibited male infertility and a decrease in GPx4 expression in sperm (25).
Previously, GPx4 knock-out mice have been established, and disruption of the GPx4 gene was found to be embryonic lethal at 7.5 days post coitum (26, 27). In the present study, we established spermatocyte-specific GPx4 knock-out mice by using the Cre-loxP system to address whether the depletion of GPx4 in spermatocytes causes male infertility. Spermatocyte-specific GPx4 knock-out mice exhibited oligoasthenozoospermia, resulting in male infertility, directly demonstrating that a decrease in GPx4 in spermatozoa results in male infertility in mice.
EXPERIMENTAL PROCEDURES
Construction of GPx4 Transgenic loxP Vector
To construct the loxP-GPx4 transgenic gene (Tg(loxP-GPx4), supplemental Figs. S1A and S1B), the approximate 5-kbp upstream regulatory region, EIa and E1b regions of the mouse GPx4 gene, (EcoRI/BamHI in supplemental Fig. S1A), E1b and EII regions containing a loxP site at the BglII site (BamHI/EcoRI in supplemental Fig. S1A), and EII–VII and 1-kbp downstream region (EcoRI/ApaI in supplemental Fig. S1A) were separately prepared from our previously cloned mouse GPx4 gene (7, 26) and inserted into pBluescript SK+ (Stratagene, La Jolla, CA). To identify mRNA for GPx4 transcribed from the transgene, we replaced the NheI site located after the stop codon in the GPx4 gene with a BamHI site (as shown in supplemental Fig. S1A). A transgenic screening vector, pTG1, containing a 5′-end marker from part of the hygromycin cDNA (250- to 660-bp site; 5′-CCTCGCTCCAGTCAATGACC-3′/5′-AGCGAGAGCCTGACCATATTGC-3′), a mouse genomic GPx4 insert site (XhoI), a second loxP site, and a marker sequence (M) for Tg(loxP-GPx4) transgenic gene screening sites (from the cGPx cDNA (27- to 272-bp site; 5′-GGCACAGTCCACCGTGTATG-3′/5′-AGAATCTCTTCATTCTTGCC-3′) and β-galactosidase cDNA (52- to 380-bp site, 5′-GGCATTGGTCTGGACACCA-3′/5′-ACGTTGGTGTAGATGGGCG-3′)) was constructed between two SalI sites in pBluescript SK+. Mouse genomic DNA containing all GPx4-encoding exons digested with SalI was inserted into the XhoI site of the pTG1 vector. Finally, the loxP-GPx4 transgenic gene, Tg(loxP-GPx4), was excised with SalI from the pTG1 vector and purified.
Generation of Tg(loxP-GPx4) Transgenic Rescued GPx4 Knock-out Mice (Tg(loxP-GPx4):GPx4−/− Mice)
The loxP-GPx4 transgenic gene was injected into fertilized eggs derived from BDF1 parents. Transgenic loxP-GPx4 mice (Tg(loxP-GPx4):GPx4+/+) were generated by standard methods. To facilitate genotyping of Tg(loxP-GPx4):GPx4+/+ mice, the following primer pair specific for Tg(loxP-GPx4) was used: PHGP BAMS/LOXSCR-AS, 5′-CTCTAGGGATCCTAGCCCTACAAGTGTGTCCC-3′/5′-CTTGCCATTCTCCTGATGTCCGAAC-3′. We established eight transgenic loxP-GPx4 mice lines (Tg(loxP-GPx4):GPx4+/+). Tg(loxP-GPx4):GPx4+/+ mice were then mated with GPx4 heterozygous (GPx4+/−) mice that we had established previously (26). Tg(loxP-GPx4) transgenic GPx4 heterozygous mice (Tg(loxP-GPx4):GPx4+/−) were mated with GPx4 heterozygous mice (GPx4+/−) to obtain Tg(loxP-GPx4) transgene-rescued GPx4 knock-out mice (Tg(loxP-GPx4):GPx4−/−). All Tg(loxP-GPx4):GPx4−/− mice examined in this study were from a mixed genetic background, with contributions from TT2, ICR, and BDF1 strains. To facilitate genotyping of Tg(loxP-GPx4):GPx4−/− mice, three primer pairs were used; one specific for Tg(loxP-GPx4) as descried above, one specific for the wild type (103/G1R, 5′-CTGCGTGGTGAAGCGCTAT-3′/5′-AGCGTCATCCACTTCAGCC-3′), and one specific for the knock-out (NEOF3/G1R, 5′-CGATGCCTGCTTGCCGAAT-3′/5′-AGCGTCATCCACTTCAGCC-3′). Three transgenic Tg(loxP-GPx4) mice lines (Fκ, Fα, and Fγ) rescued GPx4 knock-out mice from embryonic lethality. More than three independent mice from each line were used for examining body weights, and expression of GPx4 mRNA and protein in several tissues compared with wild-type mice.
Generation of Spermatocyte-specific GPx4 Knock-out Mice (pgk2-Cre:Tg(loxP-GPx4):GPx4−/− Mice; Tg(loxP):GPx4−/−)
Transgenic male mice (C57BL) expressing Cre recombinase under the control of the mouse pgk-2 promoter (28) were first mated with GPx4 heterozygous mice (GPx4+/−). Pgk2-Cre:GPx4+/− mice were next mated with Tg(loxP-GPx4):GPx4−/− mice to obtain spermatocyte-specific GPx4 knock-out mice (pgk2-Cre:Tg(loxP-GPx4):GPx4−/−; Tg(loxP):GPx4−/−). To facilitate genotyping of Tg(loxP):GPx4−/−mice, four primer pairs were used; three each specific for Tg(loxP-GPx4), wild type, and knock-out as descried above, and one specific for Cre (Cre5F/Cre3R, 5′-GCAGAACCTGAAGATGTTCGCGAT-3′/5′-AGGTATCTCTGACCAGAGTCATC-3′). To clarify the deletion of Tg(loxP-GPx4) gene in genomic DNA of several tissues by Cre recombinase, the following primer pair was used: (Gen80/LoxSCR-AS, 5′-ATAGGATCCGGCCGCCGCCGAGATGAGCTGG-3′/5′-CTTGCCATTCTCCTGATGTCCGAAC-3′). The non-recombinant fragment size and the recombinant fragment size detected by LA-PCR (Long and Accurate PCR, TAKARA BIO Inc.) were ∼4000 and 1000 bp, respectively, in the Tg(loxP-GPx4) gene, as shown in supplemental Fig. S1A.
RNA Isolation, Semiquantitative Reverse Transcription-PCR, and Quantitative Real-time PCR
Isolation of RNA from several tissues and reverse transcription were carried out as described previously (7, 29). To identify mRNA derived from the Tg(loxP-GPx4) and endogenous GPx4 genes, primer pairs specific for the endogenous GPx4 gene (F-N2/NHETG-AS, 5′-AATGCGGCCGCTAGCTGGTCTGGCAGGCACCATG-3′/5′-CACACACTTGTAGGGCTAGC-3′) and the Tg(loxP-GPx4) gene (F-N2/BamTG-AS, 5′-AATGCGGCCGCTAGCTGGTCTGGCAGGCACCATG-3′/5′-CACACACTTGTAGGGCTAGGATCC-3′) were used. Primer pairs specific for the Cre gene (Cre5F/Cre3R) were used as described above. Quantitative TaqMan real-time PCR for GPx4 (total GPx4) mRNA was carried out as described previously (7).
Western Blotting
5 × 104 sperm cells were pelleted and dissolved in radioimmune precipitation assay buffer (0.1% Nonidet P-40, 50 μg/ml deoxycholate, 10 μg/ml SDS, 100 μg/ml phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 mm Na3VO4). Several tissues (including testis) were homogenized in homogenizing buffer 1 (250 mm sucrose, 1 mm EDTA, 3 mm imidazole) supplemented with protease inhibitors. GPx4 protein levels in the homogenates (5 μg) and spermatozoa (5 × 104 cells) were determined by Western blotting using anti-GPx4 monoclonal antibody (6F10) as described previously (13). The levels of antioxidant enzyme for CuZn-superoxide dismutase (CuZn-SOD), Mn-superoxide dismutase (Mn-SOD), and voltage-dependent anion channel were determined by Western blotting using anti-CuZn-SOD pAb (StressGen), Mn-SOD pAb (Stressgen), and anti-Porin31 HL monoclonal antibody (Calbiochem).
Fertility Assessment
We investigated the reproductive capacities of Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice by mating one male with two ICR female mice for 2 weeks. Female mice were checked for vaginal plugs each morning, and litter sizes were recorded on delivery, after three successive matings.
Evaluation of Epididymal Spermatozoa
The cauda and caput regions of the epididymis obtained from ∼12-week-old mice were placed in TYH medium (Mitsubishi Chemical Medience Corp.) and gently minced with a surgical blade and incubated at 37 °C for 2 h. Spermatozoa were examined for structure, motility, measurement of mitochondrial membrane potential, and oxygraphic studies. To measure the mitochondrial membrane potential, spermatozoa were examined by monitoring the fluorescence of DiOC6 (Molecular Probes). 1 × 105 spermatozoa were incubated for 2 h and then stained with 10 μg/ml DiOC6, 1 μg/ml Hoechst 33258 for 20 min and washed with PBS. The stained sperm cells were dropped onto glass slides. Fluorescence due to DiOC6 and Hoechst in spermatozoa was monitored and photographed with a fluorescence microscope (BIOZERO, BZ-8000, Keyence).
Oxygraphic Studies
Spermatozoa were subjected to hypotonic treatment essentially as described previously (30). Briefly, spermatozoa were kept in ice-chilled hypotonic medium (10 mm potassium phosphate, pH 7.4, 2 g/liter bovine serum albumin) for 1.5 h. Spermatozoa were then washed three times using isotonic salt medium (2 g/liter bovine serum albumin, 113 mm KCl, 12.5 mm KH2PO4, 2.5 mm K2HPO4, 3 mm MgCl2, 0.4 mm EDTA, and 20 mm Tris-HCl, pH 7.4). Oxygen uptake by spermatozoa was measured using a Clark-type oxygen probe (YSI Model 5300A, YSI Japan) immersed in a magnetically stirred, 1-ml sample chamber in a water bath. Samples were stirred vigorously in the reaction chamber (1 ml) at 36 °C in an isotonic salt medium without EDTA. In each experiment, cells were temperature-equilibrated at 36 °C for 15 min prior to substrates or ADP addition. The rate of oxygen uptake by spermatozoa (V) was expressed as nanomoles of O2.ml/min/108 cells. The respiratory control ratio was calculated by dividing V3 (rate of oxygen uptake measured in the presence of substrates plus ADP, i.e. respiration state 3) by V4 (rate of oxygen uptake measured with substrates alone, i.e. respiration state 4).
Transmission Electron Microscopy
The suspension of epididymal spermatozoa was placed on silane-coated glass and fixed in a solution of 2.5% glutaraldehyde in 0.1 m phosphate buffer. The sample was postfixed in a solution of 1% OsO4 in 0.1 m phosphate buffer, dehydrated through a graded series of ethanol solutions, and embedded in Epon 812. Ultrathin sections were double-stained with uranyl acetate and lead citrate and then examined by transmission electron microscopy (Hitachi H-7100, Hitachinaka, Japan) operated at 75 kV. We examined 10 longitudinal profiles of sperm in 10 longitudinal sections of the midpiece.
In Vitro Fertilization Assay
C57BL/B6 mice were induced to superovulate by consecutive injections of pregnant mare serum gonadotropin (Sigma) and human chorionic gonadotropin (Teikoku Zouki, Japan), with an interval of 48 h between injections. Unfertilized oocytes were collected from the oviducts 15 h after the human chorionic gonadotropin injection. In vitro fertilization was carried out in an incubator using sperm collected from Tg(loxP):GPx4−/−, Tg(loxP):GPx4+/−, and Tg(loxP-GPx4):GPx4−/− mice in TYH medium. After incubation overnight for 1 or 2 days, the developmental frequency of two-cell stage or four-cell stage embryos was used as a measure of the rate of successful fertilization.
Histological Procedures
Testis from Tg(loxP):GPx4−/−, Tg(loxP):GPx4+/−, and Tg(loxP-GPx4):GPx4−/− mice was fixed in Bouin's solution and embedded in paraffin, and sections were stained with hematoxylin and eosin. Sections were also stained by indirect immunostaining with anti-GPx4 monoclonal antibody (8B8 (14)), EE2 pAb (4), BC7 pAb (4), and anti-calmegin pAb (31), followed by LSAB2 Kit/HRP (DAKO, Japan). Frozen sections (10 μm thick) of testes mounted on glass slides were used as samples for the analysis. Sections were immunostained with Cy3-conjugated anti-cleaved caspase 3 (Asp175) pAb (Daiichi Kagaku, Japan). Photomicrographs were obtained using a BIOZERO (BZ-8000).
Flow Cytometry Analysis of Testicular Cells for DNA Content and Intracellular Reactive Oxygen Species
A monocellular suspension of testicular cells was prepared as described previously (32). Briefly, the tunica albuginea was removed, and the seminiferous tubules were minced in PBS to release the testicular cells. The minced tissue was incubated with 0.5 mg/ml collagenase IV in PBS for 15 min at 32 °C and 1.0 μg/ml trypsin for 15 min at 32 °C, followed by addition of 1.0 μg/ml trypsin inhibitor for 2 min. The buffer was then gently aspirated, and the cells were washed in PBS and spun down at 800 × g for 10 min. Cells were resuspended in PBS, filtered through 80-μm nylon mesh, fixed in cold 70% ethanol, and kept at 4 °C until further analysis. For the DNA content assay, 1 × 106 cells were washed twice with PBS and incubated in 500 μl of 0.2% pepsin for 10 min at 37 °C. After centrifugation, the cells were stained with a solution containing 25 μg/ml propidium iodide, 40 μg/ml RNase, and 0.3% Tween 20 in PBS at room temperature for 20 min. The stained cells were analyzed with a FACScan flow cytometer (EPICS@Elite Flow cytometer, Coulter, Hialeah, FL). We used an oxidation-sensitive fluorescent probe, dihydrorhodamine (DHR), to assess levels of intracellular peroxides as follows. 1 × 106 cells were washed with PBS and incubated with 1 μg/ml DHR in PBS at 32 °C for 15 min. DHR-loaded cells were then analyzed with a flow cytometer (EPICS@Elite Flow cytometer).
Assay for Depletion of GPx4 in Murine Embryonic Fibroblasts
Timed matings of male Tg(loxP-GPx4):GPx4−/− and female GPx4+/− mice were carried out to obtain murine embryonic fibroblasts (MEFs) from Tg(loxP-GPx4):GPx4−/− mice. MEFs derived from 13.5-day post coitum embryos were transformed by SV40 T antigen and cultured in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. The immortalized MEF cells were infected with a retrovirus with or without Cre cDNA using the pMXs-Puro vector (33) and incubated for 4 days with 5 μg/ml puromycin with or without 400 μm Trolox. The MEF cells were simultaneously infected by retrovirus with or without non-mitochondrial, mitochondrial, and nucleolar GPx4 (Sec) cDNA, non-mitochondrial GPx4(Ser), an inactive form, cDNA, SOD1, SOD2, and GPx1 cDNA using pMXs-IG vector (33). The transfection efficiency by each retrovirus was >85% as assessed using Tg wild (Tg(loxP-GPx4):GPx4+/+) MEF cells. The expression efficiencies by retrovirus transfection of three types of GPx4 and GFP protein were examined by immunoblot analysis using Trolox-rescued Cre expressing Tg(loxP-GPx4):GPx4−/− MEF cells (GPx4-depleted MEF cells). We monitored cell growth by light and fluorescence microscopy BIOZERO (BZ-8000). The percentage of rescued GFP-positive cells by retroviral infections with Cre-SV40-Puro gene and several antioxidant enzyme-IRES-GFP gene was calculated relative to total GFP-positive cells by retroviral infections with only SV40-Puro gene and several antioxidant enzyme-IRES-GFP gene. Data are shown as mean ± S.D. (n = 4).
RESULTS
Generation of Spermatocyte-specific GPx4 Knock-out Mice
We had previously established GPx4 heterozygous mice by homologous recombination (26). In the present study, we introduced the transgene carrying the mouse GPx4 gene and two loxP sites into the GPx4 heterozygous mice to establish conditional GPx4 knock-out mice. We first introduced the loxP sequence into the transgene expressing GPx4 under the regulation of a 5.0-kbp upstream sequence and 1.0-kbp downstream sequence of the GPx4 gene (Tg(loxP-GPx4), supplemental Fig. S1A) and then generated eight independent loxP-GPx4 transgenic mouse lines (Tg(loxP-GPx4):GPx4+/+ mice). These mice were crossed with GPx4+/− mice to obtain Tg(loxP-GPx4):GPx4+/− mice, which were further crossed with GPx4+/− mice to generate Tg(loxP-GPx4) transgene-rescued GPx4−/− mice (Tg(loxP-GPx4):GPx4−/−). Genotyping analysis using the probes as indicated (supplemental Fig. S1B) in adult mouse tail revealed that we successfully obtained Tg(loxP-GPx4):GPx4−/− mice (supplemental Fig. S1C). We established three lines of Tg(loxP-GPx4):GPx4−/− mice, all of which were viable and fertile, with body weights indistinguishable from those of wild-type mice. Tg(loxP-GPx4):GPx4−/− mice grew normally and lived for at least more than 1 year.
We examined the expression of GPx4 mRNAs from the transgene and endogenous GPx4 genes in wild-type mice and Tg(loxP-GPx4):GPx4−/− mice by reverse transcription-PCR. To distinguish the endogenous GPx4 gene-derived GPx4 mRNA from the loxP-GPx4 transgene-derived mRNA, the NheI site after the stop codon was replaced with a BamHI site in the loxP-GPx4 transgene (supplemental Fig. S1, A and B). By reverse transcription-PCR using endogenous (NheI site)- and transgene (BamHI site)-specific antisense primers and a common forward primer P1, we detected the loxP-GPx4 transgene-derived and the endogenous GPx4 gene-derived GPx4 mRNA only from Tg(loxP-GPx4):GPx4−/− and wild-type mice, respectively (supplemental Fig. S1, D and E). As in the wild-type mice, GPx4 mRNA and protein were expressed at almost the same level in most of the murine tissues examined, with the highest expression in the testis and epididymal spermatozoa (supplemental Fig. S1, D–F). In the seminiferous tubules of the testes, GPx4 protein was expressed most strongly in the late spermatocytes and spermatids of both the wild-type and Tg(loxP-GPx4):GPx4−/− mice (supplemental Fig. S1, G and H).
These results indicated that the loxP-GPx4 transgene could transcribe GPx4 mRNA in the Tg(loxP-GPx4):GPx4−/− mice at nearly the same level as in the wild-type mice and rescued GPx4 knock-out mice from embryonic lethality. We used one of the Tg(loxP-GPx4):GPx4−/− line (Fκ) to produce spermatocyte-specific GPx4 knock-out mice in the following study.
Using a Cre-loxP conditional knock-out strategy, we next mated Tg(loxP-GPx4):GPx4−/− mice with spermatocyte-specific pgk-2 promoter-driven Cre:GPx4+/− mice to generate spermatocytes-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−) (supplemental Fig. S2A). Phosphoglycerate kinase-2 (pgk-2) is expressed specifically in testicular germ cells (4). Previous study demonstrated that Cre recombinase activity in a transgenic line possessing pgk-2-driven expression of the Cre recombinase was present in spermatocytes and spermatogenic cells at later differentiation stages (28). By genotype PCR analysis, we detected the bands for Cre fragment (600 bp), Tg(loxP-GPx4) transgene fragment (1.7 kbp, supplemental Fig. S1B), endogenous GPx4 knock-out (Neo) fragments (1.8 kb, supplemental Fig. S1B), and endogenous wild-type allele (1.4 kbp, supplemental Fig. S1B) in tail genomic DNA of 21-day-old mice to find Tg(loxP-GPx4):GPx4−/− mice, Tg(loxP):GPx4−/− mice, and Tg(loxP):GPx4+/− mice (supplemental Fig. S2A). Thus we successfully obtained Tg(loxP-GPx4):GPx4−/− mice, Tg(loxP):GPx4−/− mice, and Tg(loxP):GPx4+/− mice.
Using genomic DNA harvested from various tissues of 14-week-old Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice, we examined whether the loxP-GPx4 transgenic gene was specifically deleted in testis by spermatocyte-specific Cre recombinase expression (supplemental Figs. S1A and S2B). Only testes from Tg(loxP):GPx4−/− mice and Tg(loxP):GPx4+/− mice showed deletion of GPx4 exons II–VII as revealed by the appearance of the PCR product (1000 bp, supplemental Fig. S1A) with a sense primer specific for exon Ia and a transgene-specific antisense primer (supplemental Fig. S2B). These findings suggest that selective disruption of the loxP-GPx4 transgene occurred in the testes of Tg(loxP):GPx4−/− and Tg(loxP):GPx4+/− mice.
In the following experiments, we compared three types of mice: Tg(loxP):GPx4−/− (spermatocyte-specific GPx4 knock-out mice), Tg(loxP-GPx4):GPx4−/− (control mice), and Tg(loxP):GPx4+/− mice (endogenous GPx4-restored Tg(loxP):GPx4−/− mice) (supplemental Fig. S2A). Tg(loxP):GPx4+/− mice, in which the loxP-GPx4 transgene was disrupted by spermatocyte-specific Cre recombinase (supplemental Fig. S2A), have one copy of the endogenous GPx4 gene in testis and other tissues.
Characterization of Spermatocyte-specific GPx4 Knock-out (Tg(loxP):GPx4−/−) Mice
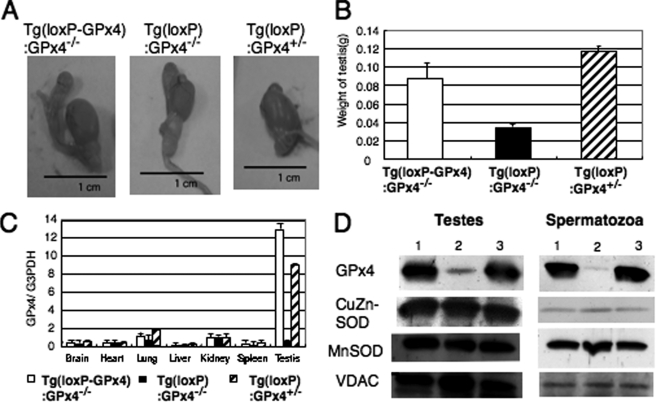
Phenotype analysis showed that a significant decrease in total body weight was observed in 9-week-old Tg(loxP):GPx4−/− mice (24.0 ± 0.4 g), but not Tg(loxP-GPx4):GPx4−/− mice (28.0 ± 0.5 g) and Tg(loxP):GPx4+/− mice (28.0 ± 0.3 g, n = 10). A significant decrease in the weight and size of testes was observed only in Tg(loxP):GPx4−/− mice and not Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice (Fig. 1, A and B).
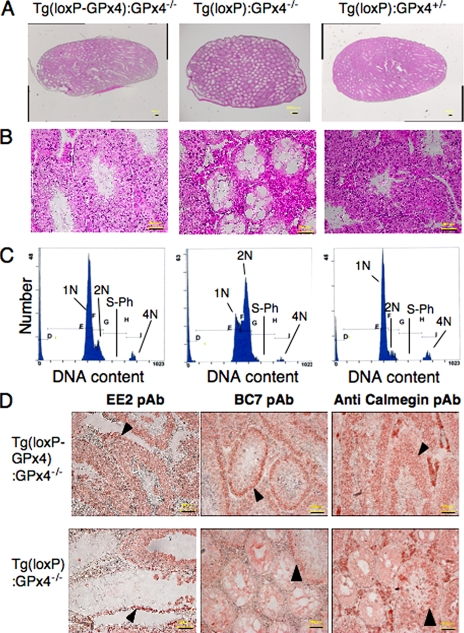
FIGURE 1.
Characterization of testes from 14-week-old spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−). A and B, decreased testis size (A) and weight (B) in Tg(loxP):GPx4−/− mice. C, quantification of expression of GPx4 mRNA in several tissues of Tg(loxP-GPx4):GPx4−/− (white), Tg(loxP):GPx4−/− (black), and Tg(loxP):GPx4+/− (hatch) mice by quantitative real-time PCR. D, expression of GPx4, CuZn-SOD, Mn-SOD, and voltage-dependent anion channel protein in testes and epididymal spermatozoa of Tg(loxP-GPx4):GPx4−/− (1), Tg(loxP):GPx4−/− (2), and Tg(loxP):GPx4+/− (3) mice by immunoblot analysis with antibodies specific to each protein.
In Tg(loxP):GPx4−/− mice, the expression of GPx4 mRNA in the testis was only one-tenth of that in Tg(loxP-GPx4):GPx4−/− mice (Fig. 1C), but it was not significantly decreased in other tissues. The expression of GPx4 protein in Tg(loxP):GPx4−/− mice was also decreased in testis and epididymal spermatozoa (Fig. 1D), but not in liver, brain, and kidney (data not shown). Expressions of other antioxidant enzymes, including mitochondrial Mn-superoxide dismutase (Mn-SOD) and CuZn-superoxide dismutase (CuZn-SOD), were unchanged in the testis or spermatozoa of Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice. These results demonstrated that GPx4 expression was lost selectively in spermatocytes and spermatozoa of Tg(loxP):GPx4−/− mice. Even when the GPx4 transgene was disrupted by Cre recombinase in Tg(loxP):GPx4+/− mice, introduction of endogenous GPx4 gene rescued the expression of GPx4, indicating that Cre recombinase expression itself does not affect the GPx4 mRNA and protein expressions from endogenous GPx4 gene (Fig. 1, C and D).
Impaired Fertility and Abnormal Epididymal Spermatozoa of Spermatocyte-specific GPx4 Knock-out (Tg(loxP):GPx4−/−) Mice
The results of the mating assay showed that male Tg(loxP):GPx4−/− mice were infertile, whereas male Tg(loxP):GPx4+/− mice and Tg(loxP-GPx4):GPx4−/− mice were fully fertile (Table 1). To clarify the mechanisms by which the deletion of GPx4 in spermatocytes induced male infertility, we examined spermatozoa in the epididymis of male Tg(loxP):GPx4−/− mice. Spermatozoa number in epididymal semen was significantly lower in Tg(loxP):GPx4−/− mice than in Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice (n = 10, p < 0.05, Table 1).
TABLE 1.
Fertility assessment and epididymal sperm analysis
| Mice | Mate number |
Sperm count (106)/epididymis | Forward motility | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| % | |||||
| Tg(loxP-GPx4):GPx4−/− | 13.6 ± 0.5 | 12.3 ± 0.7 | 12.3 ± 0.9 | 13.8 ± 0.9 | 74.3 ± 3.2 |
| Tg(loxP):GPx4−/− | 0 | 0 | 0 | 1.88 ± 2.1 | 17.8 ± 4.1 |
| Tg(loxP):GPx4+/− | 13.6 ± 0.5 | 13.3 ± 0.2 | 15.3 ± 0.5 | 14.8 ± 2.1 | 72.0 ± 2.6 |
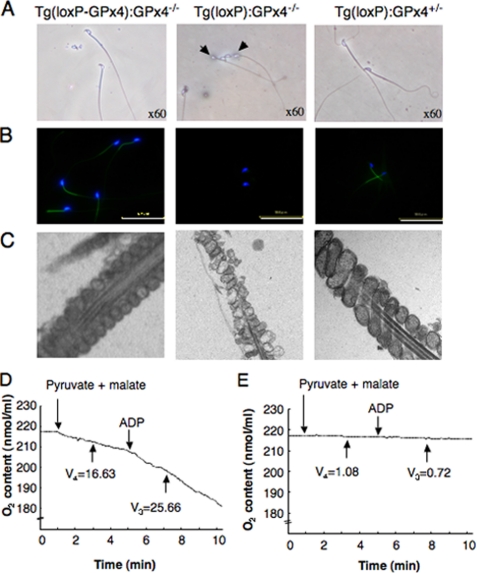
Light and electron microscopic analyses of cauda epididymal spermatozoa revealed a severe flagellar defects in spermatozoa from Tg(loxP):GPx4−/− mice (Fig. 2A). In the entire sperm population, the flagellum was folded at the midpiece-principal junction into a sharp hairpin configuration (Fig. 2A).
FIGURE 2.
Morphological and functional abnormalities of cauda spermatozoa in spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−). A–C, spermatozoa were flushed from the cauda epididymis of Tg(loxP-GPx4):GPx4−/− (left panel), Tg(loxP):GPx4−/− (center panel), and Tg(loxP):GPx4+/− mice (right panel) and analyzed by light and fluorescence microscopy. A, light microscopy showed that most epididymal spermatozoa of Tg(loxP):GPx4−/− mice exhibit a hairpin flagellar configuration (arrow). B, the mitochondrial membrane potential in epididymal spermatozoa was measured by incorporation of DiOC6. Fluorescence due to DiOC6 (green) and Hoechst 33258 (blue) was photographed under a fluorescence microscope. Scale bars, 50 μm. C, ultrastructure of mitochondria in the midpiece of spermatozoa by electron microscopy. Magnification, ×13,000. D and E, respiration capacity of cauda epididymal spermatozoa collected from Tg(loxP-GPx4):GPx4−/− (D) and Tg(loxP):GPx4−/− mice (E). Oxygen uptake by spermatozoa was measured polarographically in the presence of 10 mm malate and 10 mm pyruvate and 0.76 μm ADP. The rate of oxygen uptake in respiration states 4 (V4) and 3 (V3) is expressed as nanomoles of O2·ml/min/108 cells.
To examine the sperm motility, isolated cauda spermatozoa were incubated in TYH medium for 2 h at 37 °C as previously described (34). The forward motility of spermatozoa from Tg(loxP):GPx4−/− mice was dramatically reduced to 24% compared with spermatozoa from Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice (Table 1). Indeed, hairpin configuration spermatozoa frequently displayed weak flagellar beating, particularly at the principal segment.
GPx4 is known to be expressed predominantly in the mitochondria of spermatozoa (13). We next examined mitochondrial function in the incubated spermatozoa by fluorescence microscopy using DiOC6, which is selectively taken up by mitochondria at a rate that depends on the mitochondrial membrane potential. Fluorescence was clearly observed in the midpiece of spermatozoa from Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice (Fig. 2B). In contrast, spermatozoa from Tg(loxP):GPx4−/− mice failed to incorporate DiOC6 (Fig. 2B). The ultrastructure of mitochondria was assessed by transmission electron microscopy (Fig. 2C). Although the outer mitochondrial membranes were appropriately retained, the internal membrane structure of the mitochondria was irregular, with disappearance of cristae-like folds and formation of electron-dense central clumps in Tg(loxP):GPx4−/− spermatozoa (Fig. 2C). The respiration capacity of mitochondria of cauda spermatozoa was tested polarographically. Addition of 10 mm of each of the two substrates, pyruvate and malate, to 2 × 107 spermatozoa from Tg(loxP-GPx4):GPx4−/− mice promoted a significant oxygen uptake by mitochondria (respiration state 4, Fig. 2D). This was further stimulated by 0.76 mm ADP (respiratory state 3). In Tg(loxP):GPx4−/− spermatozoa, a significant reduction in the active respiration state was observed (Fig. 2E). The V4 and V3 values (V4 = 1.08 ± 1.3 and V3 = 0.72 ± 1.0 nmol of O2/ml/min/108 cells, respectively) of Tg(loxP):GPx4−/− mice were significantly lower than those (V4 = 16.33 ± 2.5 and V3 = 25.66 ± 1.3 O2/ml/min/108 cells, respectively) of Tg(loxP-GPx4):GPx4−/− mice. The respiratory control ratio (1.57) indicated a good coupling between respiration and phosphorylation in Tg(loxP-GPx4):GPx4−/− spermatozoa. However, a significant decrease was found in the respiratory efficiency (0.66) of Tg(loxP):GPx4−/− spermatozoa. These functional and structural defects in the mitochondria of the spermatozoa may result in asthenozoospermia of Tg(loxP):GPx4−/− mice.
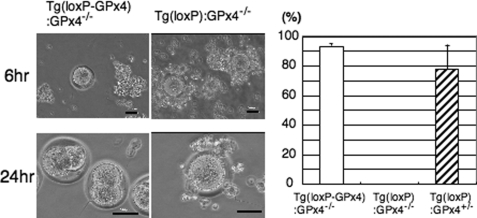
We also performed an in vitro fertilization assay using the same number of sperms prepared from Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice. Fertilization rate, measured as the developmental frequency of two-cell-stage or four-cell-stage embryos, was almost negligible in Tg(loxP):GPx4−/− mice (Fig. 3).
FIGURE 3.
In vitro fertilization assay. The same number of spermatozoa collected from Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice (n = 3) were used in the assay. Phase-contrast microscopy of in vitro fertilization by spermatozoa from Tg(loxP-GPx4):GPx4−/− mice (left) and Tg(loxP):GPx4−/− mice (right) after 6- and 24-h incubation. Spermatozoa from Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice were clearly able to inseminate eggs as shown by fertilized eggs at the two-cell stage (left bottom panel). Eggs incubated with spermatozoa from Tg(loxP):GPx4−/− mice showed no evidence of fertilization (bottom right panel). Scale bars, 50 μm. The development frequency of two-cell-stage embryos after 24 h was used as the measure of successful fertilization rate. No fertilization of spermatozoa from Tg(loxP):GPx4−/− mice was observed. All values are the means ± S.D.
Developmental Progression of Defects in Flagellar Structure and Mitochondrial Membrane Potential in the Spermatozoa of Spermatocyte-specific GPx4 Knock-out (Tg(loxP):GPx4−/−) Mice
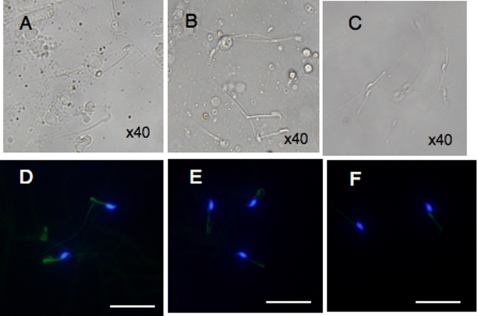
We next examined when the flagellar structure and the mitochondrial membrane potential were disrupted during spermatogenesis in GPx4-depleted spermatozoa. GPx4-depleted testicular spermatozoa were of full form and had extended flagellum (Fig. 4A). In contrast, GPx4-depleted caput spermatozoa displayed abnormal flagellar bending in the posterior midpiece of the flagellum (Fig. 4B), and GPx4-depleted cauda spermatozoa were sharply bent at the midpiece-principal piece junction and exhibited a hairpin flagellar configuration (Fig. 4C). As mentioned above, the mitochondria membrane potential of GPx4-depleted cauda spermatozoa was significantly decreased after 2 h in vitro incubation (Figs. 2B and 4F). However, we found that the mitochondrial membrane potential of GPx4-depleted cauda spermatozoa were normally maintained when in vitro incubation was omitted (Fig. 4, D and E). These results suggest that loss of the mitochondrial membrane potential of GPx4-depleted spermatozoa occurs only after in vitro incubation in TYH medium and that loss of mitochondrial membrane potential is not a direct cause of abnormal flagella bending.
FIGURE 4.
The developmental progression of defects in the flagellar structure and dysfunction of the mitochondrial membrane potential in spermatozoa of spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−). A–C, phase-contrast photomicrographs show the developmental progression of sperm defects in spermatocyte-specific GPx4 knock-out mice. Spermatozoa were flushed from seminiferous tubules (A), the caput epididymis (B), and cauda epididymis (C) from Tg(loxP):GPx4−/− mice observed by light microscopy. d–F, time course of the decrease of the mitochondrial membrane potential of GPx4-depleted cauda epididymal spermatozoa. Spermatozoa collected from cauda epididymis were cultivated for the indicated times, 0 h (D), 1 h (E), and 2 h (F), and stained with 10 μg/ml DiOC6 (green) and 1 μg/ml Hoechst 33258 (blue) for 20 min. Fluorescence from DiOC6 and Hoechst 33258 was photographed under a fluorescence microscope. Scale bars, 50 μm.
Defects of Spermatogenesis in Testes of Spermatocyte-specific GPx4 Knock-out (Tg(loxP):GPx4−/−) Mice
The occurrence of oligospermia suggests abnormal spermatogenesis in infertile Tg(loxP):GPx4−/− mice. To investigate how oligospermia was induced in Tg(loxP):GPx4−/− mice, we histopathologically analyzed the testes of Tg(loxP):GPx4−/− mice. In 70% of seminiferous tubules, hematoxylin and eosin staining detected little or no spermatocytes, spermatids, or sperm (Fig. 5, A and B).
FIGURE 5.
Histopathological observation of testes from spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−). A and B, histological observation of testes. Testis sections from Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice were stained with hematoxylin and eosin. A, scale bars, 200 μm. B, scale bars, 50 μm. C, analysis of germ cell DNA content in testes from Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice by flow cytometry. 1N represents haploid cells, 2N represents diploid cells, and 4N represents tetraploid cells. S-phase (S-Ph) represents spermatogonial cells, and preleptotene spermatocytes synthesizing DNA. D, immunohistochemical analysis of the distribution of stage-specific germ cell. Testis sections from Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4−/− mice were stained with EE2 pAb, BC7 pAb, and anti-calmegin pAb to visualize spermatogonia (EE2), early spermatocytes (BC7), and late spermatocytes and spermatids (Calmegin). Arrows indicate the location of spermatogonia, early spermatocytes, and late spermatocytes in each panel. Scale bars, 50 μm.
We analyzed spermatogenesis by determining the relative distribution of germ cell populations in the testes of Tg(loxP):GPx4−/− mice. Using flow cytometric scanning of propidium iodide-labeled Tg(loxP-GPx4):GPx4−/− germ cells (32), we detected four main histogram peaks of DNA content that corresponded to haploid (1N, round and elongated spermatids and spermatozoa), diploid (2N, spermatogonia, preleptotene and diplotene primary spermatocytes, secondary spermatocytes, and somatic cells), tetraploid (4N, G2 spermatogonia, leptotene, zygotene, and pachytene primary spermatocytes), and S-phase cells between peaks 2N and 4N (S-Ph, spermatogonial cells and preleptotene spermatocytes synthesizing DNA) (Fig. 5C). Somatic cells comprise <3% of the total testicular cells in the mouse testis (33). Tg(loxP-GPx4):GPx4−/− and Tg(loxP):GPx4+/− mice showed normal germ cell distribution ratios that were consistent with the hematoxylin and eosin staining results (Fig. 5, A– C). In contrast, Tg(loxP):GPx4−/− mice showed a significant increase in diploid cells, whereas cells with haploid DNA content were greatly reduced.
To investigate the distribution of the germ cell population in the testis of Tg(loxP):GPx4−/− mice in more detail, testis sections were immunostained with the anti-testis stage-specific antibodies, EE2 pAb (4), BC7 pAb (4), and anti-calmegin pAb (31). EE2 pAb, BC7 pAb, and anti-calmegin pAb recognize antigens specific to mouse stage-specific cells, mainly spermatogonia, spermatocytes and late spermatocytes, and spermatids, respectively (4, 27). In damaged seminiferous tubules of Tg(loxP):GPx4−/− mice, we detected only EE2 pAb staining in the external layer of the most damaged seminiferous tubules in Tg(loxP):GPx4−/− mice (Fig. 5D) but no BC7 pAb or anti-calmegin pAb staining. We detected normal stage-specific staining by EE2 pAb, BC7 pAb, and anti-calmegin pAb in undamaged seminiferous tubules of Tg(loxP):GPx4−/− mice (Fig. 5D). Thus, immunohistochemical analysis demonstrated that only spermatogonia were observed among germinal cells in damaged seminiferous tubules of GPx4-depleted testis, indicating that germinal cells died before postnatal mitosis but not before meiosis.
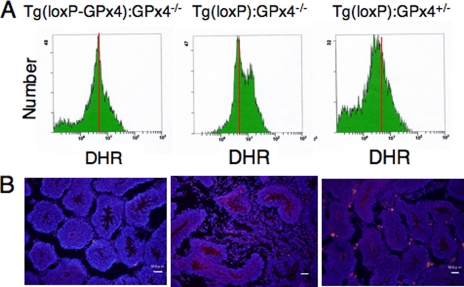
Generation of Hydroperoxide and Induction of Apoptotic Cell Death in Germ Cells of Spermatocyte-specific GPx4 Knock-out (Tg(loxP):GPx4−/−) Mice
We next examined the generation of hydroperoxide in germ cells of Tg(loxP):GPx4−/− mice by using the oxidative stress-sensitive fluorescent probe, DHR. Flow cytometric analysis revealed significant elevation of intracellular hydroperoxide in germ cells of Tg(loxP):GPx4−/− mice due to depletion of GPx4 (Fig. 6A). We also examined whether apoptosis was enhanced in Tg(loxP):GPx4−/− mice by antibody staining for activated caspase 3, a positive regulator of the apoptosis pathway. We observed more seminiferous tubules with activated caspase 3-positive spermatocytes in Tg(loxP):GPx4−/− mice than in Tg(loxP-GPx4):GPx4−/− mice and Tg(loxP):GPx4+/− mice (Fig. 6B). These results demonstrated that severe depletion of GPx4 in germ cells induced the elevation of intracellular hydroperoxide and subsequent germ cell death.
FIGURE 6.
Detection of hydroperoxide and caspase 3 activation in germ cells of spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/−). A, flow cytometric analysis of intracellular hydroperoxides in germ cells of Tg(loxP-GPx4):GPx4−/− (left panel), Tg(loxP):GPx4−/− (center panel), and Tg(loxP):GPx4+/− mice (right panel). To assess the levels of intracellular peroxides, flow cytometric analysis was performed with the fluorescent probe, dihydrorhodamine (DHR). The intensity of fluorescence from DHR of cells was quantified by flow cytometry and is plotted on a logarithmic scale, in arbitrary units, against the number of cells. B, detection of activated caspase 3, an apoptotic cell death regulator, in testis of Tg(loxP-GPx4):GPx4−/−, Tg(loxP):GPx4−/−, and Tg(loxP):GPx4+/− mice. Testis sections from each mouse were stained with Cy3-conjugated anti-activated caspase 3 pAb. Scale bars, 50 μm.
DISCUSSION
Previously, low GPx4 contents in spermatozoa were observed in human infertile males (12, 20, 21). The diagnosis for infertile patients with low GPx4 contents in their spermatozoa was oligoasthenozoospermia due to a significantly lower number of spermatozoa in semen and significantly lower motility of spermatozoa than in fertile males. However, it was not clear whether low GPx4 content in sperm was a cause of infertility or whether other impairments of testicular function could lead to a decrease in the GPx4 content of sperm. To clarify the relationship between low sperm GPx4 content and male infertility, we used the Cre-loxP system to establish mice with a spermatocyte-specific deletion of the GPx4 gene. In this study, we demonstrate that mice with spermatocyte-specific deletion of the GPx4 gene showed male infertility with two significant conspicuous phenotypes: first, a decreased number of spermatozoa in the epididymis caused by the depletion of spermatogenic cells in seminiferous tubules, and second, loss of forward motility of spermatozoa due to mitochondrial dysfunction and/or hairpin-like bending of the tail in the distal midpiece region. The finding of similar phenotypes in spermatozoa of infertile patients and GPx4-deficient mice suggests that our mouse model will be useful for understanding of oligoasthenozoospermia in human GPx4-deficient infertile patients.
GPx4 is a major selenoprotein in spermatozoa and is highly expressed in their mitochondria at the midpiece. Recent knock-out studies demonstrated that plasma selenoprotein P delivers selenocysteine to testis (22) through the uptake of apoER2 in Sertoli cells (23). Spermatozoa with hairpin-like bending of the tail were observed in selenoprotein P knock-out mice, apoER2 knock-out mice and mice fed a selenium-deficient diet for 6 months (22, 23). Reduced GPx4 expression in spermatozoa was also observed in apoER2 knock-out mice (24). Our data demonstrate that hairpin-like tail bending and the loss of motility in spermatozoa are due to loss of GPx4 function in spermatozoa. The mitochondrial ultrastructure of spermatozoa in the GPx4-deficient mice was more severely damaged than in selenoprotein P knock-out mice (22) and apoER2 knock-out mice (24). Our data also demonstrate that GPx4-deficient spermatozoa lacked mitochondrial membrane potential and showed significantly reduced mitochondrial respiration capacity. These results suggest that mitochondrial dysfunction in spermatozoa is a major functional defect brought on by GPx4 deficiency. During submission of the manuscript for this report, Conrad and his colleagues demonstrated that depletion of mitochondrial GPx4 causes impaired sperm quality, severe structural abnormalities, and male infertility (35), which also confirms our ideas.
In the present study, we showed that two severe defects, including abnormal flagellar structure and reduced mitochondrial membrane potential, occur independently in GPx4-depleted spermatozoa. In human infertile GPx4-deficient spermatozoa, the ultrastructure of mitochondria are damaged in the midpiece of spermatozoa and the motility sharply decreases during in vitro incubation, but the flagellum is of full form and normally extended (13). These results also support the idea that mitochondrial dysfunction is not a direct cause of flagellar bending. One possible role of mitochondrial GPx4 in spermatozoa is to maintain motility via antioxidant activity against mitochondrial reactive oxygen species (13). Another possible role is to maintain the structure of the mitochondrial capsule by cross-linking with itself or other proteins (16, 20). In our preliminary experiments, cauda spermatozoa isolated from Tg(loxP):GPx4−/− mice fed with excess vitamin E did not lose membrane potential for at least 1 h during in vitro incubation, but showed abnormal flagellar structure. These results may indicate that GPx4 plays two important roles in maintaining mitochondrial membrane potential via antioxidant activity and in maintaining the structure of flagella.
Three isoforms of GPx4 mRNA, non-mitochondrial GPx4, mitochondrial GPx4 and nucleolar GPx4, are transcribed from one gene (6, 7). All three GPx4 mRNAs were expressed in late spermatocytes in testis (13–15). Nucleolar GPx4 was first found as a sperm nucleus-specific 34-kDa selenoprotein (10). However, specific disruption of the nucleolar GPx4 gene in mice did not result in male infertility (36). According to the study by Conrad et al. (35), depletion of mitochondrial GPx4 causes abnormalities in spermatozoa but has no effect on proliferation or apoptosis of germinal or somatic tissues. We showed that deletion of all GPx4 isoforms induces dramatic loss of spermatocytes and spermatozoa. These results suggest that the depletion of non-mitochondrial GPx4, but not mitochondrial GPx4 and nucleolar GPx4, significantly lowers the number of spermatozoa as a result of the loss of germ cells, and that non-mitochondrial GPx4 has an important role in normal germ cell growth in testis. We obtained MEF from Tg(loxP-GPx4):GPx4−/− mice and depleted the GPx4 transgene by retroviral expression of Cre recombinase (supplemental Fig. S3A). Complete loss of the GPx4 gene resulted in severe cell death 3 days after infection (supplemental Fig. S3B). We also found that overexpression of non-mitochondrial GPx4 but not other isoforms such as the mitochondrial and nucleolar forms could completely rescue the cell death (supplemental Fig. S3, E–G). Expression of an enzymatically inactive form of GPx4 did not rescue the viability (supplemental Fig. S3G). Trolox, a vitamin E derivative, also efficiently rescued the viability GPx4-deleted MEF (supplemental Fig. S3, C and D). Overexpression of other antioxidant enzymes such as cytosolic glutathione peroxidase (GPx1), CuZn-SOD, and mitochondrial Mn-SOD could not rescue the viability (supplemental Fig. S3G). These results suggest that non-mitochondrial GPx4 plays an essential role in cell growth of somatic and germinal cells most probably by scavenging lipid peroxide. Consistent with this hypothesis, we observed significant elevation of intracellular hydroperoxide in the GPx4-depleted germ cells (Fig. 6A).
We established floxed GPx4 mice (Tg(loxP-GPx4):GPx4−/− mice) using the transgenic complementation rescue method (37). Transgenically rescued floxed mice exhibited normal growth and fertility. These results reveal that the ∼5-kbp promoter regions in the loxP-GPx4 transgene were sufficient for normal regulation of GPx4 expression in embryogenesis and spermatogenesis. The loxP-GPx4 transgene was effectively deleted in testis by Cre expression under the control of the pgk-2 promoter in this study. The sperm abnormalities and disorders of seminiferous tubules in spermatocyte-specific GPx4 knock-out mice (Tg(loxP):GPx4−/− mice) were restored by the gain of one copy of the endogenous GPx4 gene in Tg(loxP):GPx4+/− mice. In this study, we first generated Tg(loxP-GPx4):GPx4+/+ mice (GPx4-overexpressing transgenic mice). Thus, Tg(loxP-GPx4):GPx4−/− mice and Tg(loxP-GPx4):GPx4+/+ mice established in this study can be used for functional analysis of GPx4 in other murine tissues using the tissue-specific Cre-loxP system. Our results also demonstrate that the loxP-GPx4 transgene is useful for functional analysis of the three isoforms of GPx4 in mice by mutation of their start codons. These spermatocyte-specific GPx4 knock-out mice might be useful for drug screening and for the development of treatment methods for human male infertility.
Supplementary Material
Acknowledgments
We are most grateful to N. Okawa, R. Suzuki, S. Kuwabara, N. Matsui, and M. Shibata for excellent technical assistance; T. Kitamura for the kind gifts of retrovirus vector and PlatE cells; H. Tanaka and Y. Nishimune for the kind gifts of anti-EE2 pAb and BC7 pAb; and N. Inoue and M. Okabe for the kind gift of anti-calmegin pAb. We greatly thank H. Arai for preparation of the manuscript.
This work was supported by Grant-in-aid 20590067 from the Ministry of Education, Science and Culture of Japan, by PRESTO from the Japan Science and Technology Agency, and by a SHISEIDO grant for science research.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-Bank™/EBI Data Bank with accession number(s).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GPx4
- phospholipid hydroperoxide glutathione peroxidase
- apoER2
- apoE receptor 2
- Tg(loxP-GPx4)
- loxP-GPx4 transgenic gene
- SOD
- superoxide dismutase
- pAb
- polyclonal antibody
- PBS
- phosphate-buffered saline
- DHR
- dihydrorhodamine
- MEF
- mouse embryonic fibroblast
- GFP
- green fluorescent protein
- pgk-2
- phosphoglycerate kinase-2.
REFERENCES
- 1.Sharma R. K., Agarwal A. (1996) Urology 48, 835–850 [DOI] [PubMed] [Google Scholar]
- 2.Lipschultz L. I., Howards S. S. (1983) in Infertility in the Male (Lipschultz L. I., Howards S. S. eds) pp. 187– 206, Churchill Livingstone, New York [Google Scholar]
- 3.Hull M. G., Glazener C. M., Kelly N. J., Conway D. J., Foster P. A., Hinton R. A., Coulson C., Lambert P. A., Watt E. M., Desai K. M. (1985) Br. Med. J. 291, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimune Y., Tanaka H. (2006) J. Androl. 27, 326–334 [DOI] [PubMed] [Google Scholar]
- 5.Imai H., Nakagawa Y. (2003) Free Radic. Biol. Med. 34, 145–169 [DOI] [PubMed] [Google Scholar]
- 6.Boschan C., Borchert A., Ufer C., Thiele B. J., Kuhn H. (2002) Genomics 79, 387–394 [DOI] [PubMed] [Google Scholar]
- 7.Imai H., Saito M., Kirai N., Hasegawa J., Konishi K., Hattori H., Nishimura M., Naito S., Nakagawa Y. (2006) J. Biochem. 140, 573–590 [DOI] [PubMed] [Google Scholar]
- 8.Pushpa-Rekha T. R., Burdsall A. L., Oleksa L. M., Chisolm G. M., Driscoll D. M. (1995) J. Biol. Chem. 270, 26993–26999 [DOI] [PubMed] [Google Scholar]
- 9.Arai M., Imai H., Sumi D., Imanaka T., Takano T., Chiba N., Nakagawa Y. (1996) Biochem. Biophys. Res. Commun. 227, 433–439 [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer H., Conrad M., Roethlein D., Kyriakopoulos A., Brielmeier M., Bornkamm G. W., Behne D. (2001) FASEB J. 15, 1236–1238 [PubMed] [Google Scholar]
- 11.Nakamura T., Imai H., Tsunashima N., Nakagawa Y. (2003) Biochem. Biophys. Res. Commun. 311, 139–148 [DOI] [PubMed] [Google Scholar]
- 12.Imai H., Narashima K., Arai M., Sakamoto H., Chiba N., Nakagawa Y. (1998) J. Biol. Chem. 273, 1990–1997 [DOI] [PubMed] [Google Scholar]
- 13.Imai H., Suzuki K., Ishizaka K., Ichinose S., Oshima H., Okayasu I., Emoto K., Umeda M., Nakagawa Y. (2001) Biol. Reprod. 64, 674–683 [DOI] [PubMed] [Google Scholar]
- 14.Conrad M., Schneider M., Seiler A., Bornkamm G. W. (2007) Biol. Chem. 388, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 15.Tramer F., Vetere A., Martinelli M., Paroni F., Marsich E., Boitani C., Sandri G., Panfili E. (2004) Biochem. J. 383, 179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohé L. (1999) Science 285, 1393–1396 [DOI] [PubMed] [Google Scholar]
- 17.Arai M., Imai H., Koumura T., Yoshida M., Emoto K., Umeda M., Chiba N., Nakagawa Y. (1999) J. Biol. Chem. 274, 4924–4933 [DOI] [PubMed] [Google Scholar]
- 18.Nomura K., Imai H., Koumura T., Arai M., Nakagawa Y. (1999) J. Biol. Chem. 274, 29294–29302 [DOI] [PubMed] [Google Scholar]
- 19.Imai H., Koumura T., Nakajima R., Nomura K., Nakagawa Y. (2003) Biochem. J. 371, 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flohé L. (2007) Biol. Chem. 388, 987–995 [DOI] [PubMed] [Google Scholar]
- 21.Foresta C., Flohé L., Garolla A., Roveri A., Ursini F., Maiorino M. (2002) Biol. Reprod. 67, 967–971 [DOI] [PubMed] [Google Scholar]
- 22.Diaconu M., Tangat Y., Böhm D., Kühn H., Michelmann H. W., Schreiber G., Haidl G., Glander H. J., Engel W., Nayernia K. (2006) Andrologia 38, 152–157 [DOI] [PubMed] [Google Scholar]
- 23.Olson G. E., Winfrey V. P., Nagdas S. K., Hill K. E., Burk R. F. (2005) Biol. Reprod. 73, 201–211 [DOI] [PubMed] [Google Scholar]
- 24.Olson G. E., Winfrey V. P., Nagdas S. K., Hill K. E., Burk R. F. (2007) J. Biol. Chem. 282, 12290–12297 [DOI] [PubMed] [Google Scholar]
- 25.Andersen O. M., Yeung C. H., Vorum H., Wellner M., Andreassen T. K., Erdmann B., Mueller E. C., Herz J., Otto A., Cooper T. G., Willnow T. E. (2003) J. Biol. Chem. 278, 23989–23995 [DOI] [PubMed] [Google Scholar]
- 26.Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., Kitamoto T., Hayasaka M., Hanaoka K., Nakagawa Y. (2003) Biochem. Biophys. Res. Commun. 305, 278–286 [DOI] [PubMed] [Google Scholar]
- 27.Yant L. J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J. G., Motta L., Richardson A., Prolla T. A. (2003) Free Radic. Biol. Med. 34, 496–502 [DOI] [PubMed] [Google Scholar]
- 28.Kido T., Arata S., Suzuki R., Hosono T., Nakanishi Y., Miyazaki J., Saito I., Kuroki T., Shioda S. (2005) Dev. Growth Differ. 47, 15–24 [DOI] [PubMed] [Google Scholar]
- 29.Hattori H., Imai H., Kirai N., Furuhama K., Sato O., Konishi K., Nakagawa Y. (2007) Biochem. J. 408, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferramosca A., Focarelli R., Piomboni P., Coppola L., Zara V. (2008) Int. J. Androl. 31, 337–345 [DOI] [PubMed] [Google Scholar]
- 31.Ikawa M., Wada I., Kominami K., Watanabe D., Toshimori K., Nishimune Y., Okabe M. (1997) Nature 387, 607–611 [DOI] [PubMed] [Google Scholar]
- 32.Zhang C., Yeh S., Chen Y. T., Wu C. C., Chuang K. H., Lin H. Y., Wang R. S., Chang Y. J., Mendis-Handagama C., Hu L., lardy H., Chang C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17718–17723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeyaraj D. A., Grossman G., Petrusz P. (2003) Reprod. Biol. Endocrinol. 1, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L., Nutter L. M., Law N., McKerlie C. ( 2009) J. Am. Assoc. Lab. Anim. Sci. 48, 39– 43 [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider M., Forster H., Boersma A., Seiler A., Wehnes H., Sinowatz F., Neumüller C., Deutsch M. J., Walch A., Hrabé de Angelis M., Wurst W., Ursini F., Roveri A., Maleszewski M., Maiorino M., Conrad M. (2009) FASEB J. 23, 3233–3242 [DOI] [PubMed] [Google Scholar]
- 36.Conrad M., Moreno S. G., Sinowatz F., Ursini F., Kölle S., Roveri A., Brielmeier M., Wurst W., Maiorino M., Bronkamm G. W. (2005) Mol. Cell. Biol. 25, 7637–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. (2008) Mol. Cell. Biol. 28, 2758–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.