Abstract
Glycogen synthase kinase 3β (GSK3β) has been identified to play important roles in neuronal death. Evidence from both in vitro and in vivo studies indicates that increased GSK3β activity contributes to neurodegeneration and to the pathogenesis of Alzheimer disease. But the molecular mechanisms that underlie GSK3β-mediated neurotoxicity remain poorly understood. We reported here that myocyte enhancer factor 2D (MEF2D), a nuclear transcription factor known to promote neuronal survival, is directly phosphorylated by GSK3β. Our data showed that phosphorylation of MEF2D by GSK3β at three specific residues in its transactivation domain inhibits MEF2D transcriptional activity. Withdrawal of neuronal activity in cerebellar granule neurons activated GSK3β in the nucleus, leading to GSK3β-dependent inhibition of MEF2 function. This inhibition contributed to GSK3β-mediated neuronal toxicity. Overexpression of MEF2D mutant that is resistant to GSK3β inhibition protected cerebellar granule neurons from either GSK3β activation- or neuronal activity deprivation-induced toxicity. These results identify survival factor MEF2D as a novel downstream effector targeted by GSK3β and define a molecular link between activation of GSK3β and neuronal survival machinery which may underlie in part GSK3β-mediated neurotoxicity.
Introduction
Neuronal survival plays an important role in both immature neurons during development and mature neurons under stress. Although the molecular mechanisms that underlie neuronal death are complex, some key regulators have been identified. One such regulator is glycogen synthase kinase 3β (GSK3β).3 Indeed, in vitro and in vivo evidence suggests that the GSK3β signaling pathway plays a prominent role in neurodegeneration and in the formation of plaque and neurofibrillary tangle in Alzheimer disease (1, 2). In vitro, withdrawal of trophic support from cultured neurons increases GSK3β activity, which is accompanied with neuronal death (3), whereas overexpression of it is sufficient to induce apoptosis (4). In vivo, overexpression of GSK3β also leads to activation of caspase, neuronal loss, and gliosis (5). Consistent with its role in neuronal death, GSK3β activity is inhibited by the insulin signaling pathway. But how GSK3β acts to induce neuronal death and promote neurodegeneration is not entirely clear and remains an area of active investigation.
As a kinase, one of the most likely modes of action by which GSK3β modulates neuronal viability is to phosphorylate downstream effectors. Indeed, although identified first to regulate glycogen synthesis (6), GSK3β is now known to target many protein substrates to modulate many processes including apoptosis (7, 8). An interesting feature of GSK3β is how it recognizes its substrates. Some GSK3β substrates require prior phosphorylation by another kinase to prime the substrates for GSK3β phosphorylation. A second category of GSK3β substrates can be phosphorylated directly by GSK3β without the priming event. Although GSK3β has been shown to regulate several substrates implicated in the pathogenesis of Alzheimer disease, including microtubule-associated protein Tau (9), much remains to be learned on whether GSK3β directly regulates factors required for neuronal survival.
Members of myocyte enhancer factor 2 (MEF2A-D) belong to the MADS (MCM1, agamous, deficiens, serum-response factor) family of transcription factor. They have been shown to play critical roles in neuronal development and survival. Studies have shown that MEF2s are required for the survival of various types of neurons in several model systems. For example, recent studies have shown that various isoforms of MEF2s are required for neuronal activity-dependent survival of cerebellar granule neurons (10–12) and for trophic factor-induced survival of developing cortical neurons (13). MEF2s are the targets for several key intracellular signaling pathways that regulate cell survival and apoptosis. For example, the phosphorylation of MEF2 by p38 mitogen-activated protein kinase (10), extracellular-regulated kinase 5 (ERK5), or protein kinase A (14, 15) promotes MEF2 function, enhancing neuronal survival. In contrast, the phosphorylation of MEF2 by cyclin-dependent kinase 5 (Cdk5) in response to overt neurotoxic insults inhibits MEF2 function (11). However, the kinases that mediate inhibitory effects on MEF2 upon neuronal activity withdrawal have not been identified.
In this study we examined the regulating of MEF2D in cerebellar granule neuron death induced by potassium withdrawal. We found that potassium withdrawal triggered activation of GSK3β. GSK3β directly phosphorylated MEF2D at multiple sites, which inhibits MEF2 transactivation activity. MEF2 mutants that are resistant to GSK3β phosphorylation rescued neurons from potassium withdrawal-induced apoptosis.
EXPERIMENTAL PROCEDURES
Plasmids, Chemicals, and Antibodies
Wild type and mutated MEF2-dependent luciferase reporter constructs were described previously (11). Anti-GSK3β, anti-MEF2D, and anti-Tyr(P)-216-GSK3β antibodies were purchased from BD Transduction Laboratories; anti-Ser(P)-9-GSK3β, anti-poly(ADP-ribose) polymerase, anti-Raf-1, and anti-MAP2 antibodies were from Cell Signaling; anti-NeuN was from Millipore; anti-actin antibody was from Sigma. Propidium iodide (PI), poly-l-lysine, LiCl, kenpaullone, insulin, valproate, and SB215763 were purchased from Sigma, mammalian transfection system calcium phosphate was from Promega, purified GSK3β was from New England Biolabs, catenin was from Upstate, and restore Western blot stripping buffer was from Pierce.
Culture of Rat Primary Cerebellar Granule Neurons (CGNs)
Cultures of primary cerebellar granule neurons were performed as described previously (12). Briefly, cerebellum from postnatal day 6 rat pups was dissected and subjected to enzymatic dissociation. Dissociated cells were cultured in basal medium Eagle's medium in poly-l-lysine-coated plates. Neurons were treated with serum/potassium withdrawal on DIV 7 unless indicated otherwise.
Immunofluorescence Staining
Cells were plated on glass covers precoated with poly-l-lysine. After treatment, cells were washed twice with cold PBS solution, fixed with 4% paraformaldehyde in PBS solution at 4 °C for 10 min, and then treated with 0.5% Triton-X100 in PBS solution for 30 min. After blocking with 5% goat serum in PBS solution for 1 h, cells were incubated with anti-GSK3β antibodies (1:500) at 4 °C overnight. After washing 3 times with PBS solution, cells were incubated with fluorescence fluorescein isothiocyanate-conjugated secondary antibodies (1:200) and 5 ng/ml of Hoechst for 5 min in a dark room. The immunofluorescence signals were visualized with an Olympus BX51 fluorescence microscope.
Electrophoretic Mobility Shift Assays
DNA binding activity of MEF2 was studied by electrophoretic mobility shift assay with radiolabeled double-stranded oligonucleotides. Oligonucleotides corresponding to MEF2 binding sites (MEF2 probe: wile type, 5′-AGCTTCGCTCTAAAAATAACCCTGATC-3′; for the mutant probe, the three nucleotides in italics were mutated to GGC) were annealed and labeled with 32P using T4 polynucleotide kinase. Cell lysates were incubated at 4 °C for 10 min in binding buffer. Radiolabeled wild or mutant probe (1pmol) was added to the reactions (total volume 20 μl) and incubated for an additional 20 min. Reaction mixtures were analyzed by electrophoresis at 4 °C on pre-run 5% native polyacrylamide gels.
Transfection of Cells
Primary neurons in Dulbecco's modified Eagle's medium were transfected by calcium phosphate method at DIV 4–6 as described by Mao et al. (10). In general, neurons were returned to conditioned full media after transfection and then treated if needed. A 3:1 DNA ratio of effector versus green fluorescent protein (GFP) vector was used for survival assays. Vector DNA was used to balance the amount of total DNA.
Cytoplasmic and Nuclear Fractionation
Cytoplasmic and nuclear fractionation was performed using an EZ nuclei isolation kit (Sigma NUC-101) according to the manufacturer's protocol that involves three cycles of thorough cell lysis and washing.
In Vitro Kinase Assays
Catenin were incubated with GSK3β immunoprecipitated from cell lysates by anti-GSK3β monoclonal antibody in a kinase reaction buffer containing [γ-32P]ATP and cold ATP. Nuclear extracts from cultured primary CGNs were prepared as described above. For immunoprecipitation, 100 μg of nuclear lysates were incubated with anti-GSK3β antibody for 1 h at 4 °C and then incubated with protein G plus agarose beads for an additional 2 h. The beads were washed with kinase buffer five times. The kinase reaction was carried out for 30 min at 30 °C after the manufacturer's protocol (New England Biolabs) and terminated by the addition of Laemmli sample buffer. Reaction products were resolved by SDS-PAGE, and 32P-labeled proteins were visualized by autoradiography. For the in vitro phosphorylation of MEF2D by GSK3ββ, purified GSK3β was incubated with different MEF2D fragments.
Luciferase Reporter Gene Assays
Primary neurons were transiently transfected with various constructs using calcium phosphate transfection procedure as described by Mao et al. (10). A β-galactosidase expression plasmid was used to determine the efficiency in each transfection. The total amount of DNA for each transfection was kept constant by using control vectors. Cell lysates were analyzed for luciferase (Roche Applied Science) following the manufacturer's instruction.
RNA Interference
For GSK3β small interfering RNA (siRNA), the following siRNA duplexes were purchased from Qiagen: GSK3β sense (CGAUUACACGUCUAGUAUAdTdT) and antisense (UAUACUAGACGUGUAAUCGdGdT); control nonsilencing siRNA, sense (UUCUCCGAACGUGUCACGUdTdT) and antisense (ACGUGACACGUUCGGAGAAdTdT). The transfection procedures have been described by Mussmann et al. (16) previously. After 48 h of interference, cells were treated as indicated.
Survival Assays
The survival assays were carried out as described previously (11, 17). Neurons were stained with propidium iodide (PI) without permeation. GFP-positive cells with or without PI were counted using an Olympus IX51 fluorescence microscope in a blind manner. Three hundred or more transfected cells were counted for each treatment. Apoptotic rates were calculated as the apoptotic cells in the total number of GFP-positive cells counted.
Statistical Methods
The results were analyzed using one-way analysis of variance with Bonferroni multiple comparison to test significance between experimental groups. p < 0.05 was considered significant.
RESULTS
Potassium Withdrawal Induced Increased GSK3β in the Nucleus
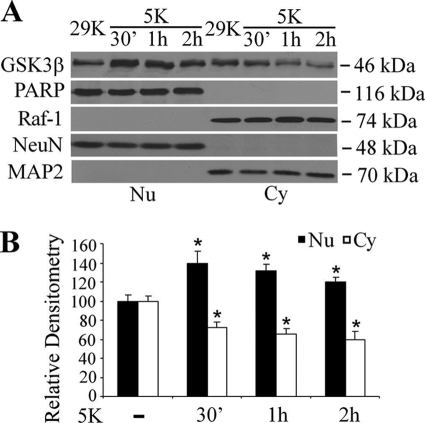
It is reported that under serum withdrawal in SH-SY5Y cells, GSK3β is accumulated in nuclei (18). To test if nuclear GSK3β is regulated in cerebellar granule neurons under low concentration of potassium chloride, CGNs cultured in medium containing serum and depolarizing concentrations of KCl (29 mm) were switched to medium containing no serum with either depolarizing concentrations or low concentrations of KCl (5 mm). The nuclear and cytosolic fractions were made, and GSK3β expression levels were determined by Western blot using anti-GSK3β antibody. Consistent with other reports, GSK3β appeared as a double band by immunoblot analysis (19). Neuronal activity withdrawal reduced the total levels of GSK3β in the cytoplasm but caused a consistent and significant increase in its levels in the nuclei after adjusting for neuronal markers (Fig. 1A). Consistent with the immunoblot data, immunofluorescence staining showed increased nucleus staining of GSK3β 1 h after neuronal activity withdrawal (data not shown). These data indicated that a decrease in neuronal activity in CGNs results in an increase of nuclear GSK3β.
FIGURE 1.
Increase of GSK3β in the nucleus of cerebellar granule neurons after neuronal activity withdrawal. A, expression of GSK3β in the cytoplasm (Cy) and nuclei (Nu) of CGNs is shown. CGNs at DIV 7 were exposed to 5 mm KCl medium without serum. At the time points indicated the levels of GSK3β were determined by Western blot analysis (Raf-1 and MAP2, cytoplasm marker; poly(ADP-ribose) polymerase (PARP) and NeuN, nuclear marker; MAP2 and NeuN also served as neuronal marker). No Raf-1 and MAP2 were detected in the nuclear lysates, and no poly(ADP-ribose) polymerase and NeuN was detectable in cytoplasm. B, Densitometry analysis indicates the relative fold of GSK3β (*, p < 0.05; n = 3).
Low Concentrations of Potassium-induced Activation of GSK3β Both in the Cytoplasm and Nuclei
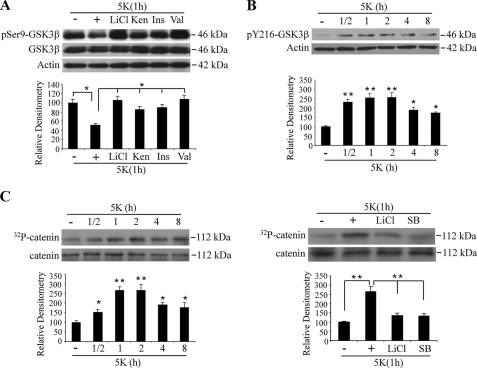
GSK3β activity increases under cellular stress and plays a critical role in regulation of neuronal apoptosis (4). Previous studies have confirmed that phosphorylation of GSK3β at serine 9 and tyrosine 216 inhibits or enhances its activity, respectively (20, 21). We determined GSK3β activity by measuring the levels of phosphorylation at Ser-9 or Tyr-216 using phospho-Ser-9 or phosphor-Tyr-216 antibodies in whole cell lysates and cytosolic fractions, respectively. Potassium chloride withdrawal caused a decrease in Ser-9 phosphorylation in whole cell lysates, whereas the total level of GSK3β didn't change (Fig. 2A). In cytosolic fractions, even though the level of GSK3β decreased upon KCl withdrawal as shown in Fig. 1A, the level of phosphorylation at Tyr-216 increased (Fig. 2B). We next directly measured the nuclear GSK3β kinase activity by in vitro kinase assay after immunoprecipitation. KCl withdrawal led to a robust increase in GSK3β activity in the nucleus over time (Fig. 2C). Two widely used GSK3β inhibitors, LiCl and SB216763, greatly attenuated the kinase activity measured after KCl withdrawal, indicating that our assay specifically measures GSK3β.
FIGURE 2.
Changes in GSK3β phosphorylation and kinase activity after KCl withdrawal. A, shown is low concentration of potassium-induced decrease in inhibitory phosphorylation at Ser-9 of GSK3β. CGNs pretreated with LiCl (10 mm), kenpaullone (Ken, 10 μm), insulin (Ins, 10 μg/ml), valproic acid (Val, 1 mm), or vehicle cultured in medium containing serum and 29 mm KCl were switched to 5 mm KCl with the re-addition of the indicated GSK3β inhibitors for 1 h. Whole cell lysates were analyzed for phospho-Ser-9 (*, p < 0.05; n = 3). B, shown is low concentration of potassium-induced increase in phosphor-Tyr-216 (pY216) signal in the cytoplasmic fraction of CGNs. Cytoplasmic lysates prepared from CGNs were analyzed using phospho-Tyr216 antibody (*, p < 0.05; **, p < 0.01; n = 3). C, shown is activation of nuclear GSK3β activity by KCl withdrawal. CGNs cultured at DIV 7 were treated as described in B. GSK3β immunoprecipitated from nuclear lysates were assayed for kinase activity using catenin as a substrate (left panel). For the right panel, CGNs were co-treated with inhibitors, 10 mm LiCl, or 10 μm SB216763 (SB) when exposed to 5 mm KCl. Data shown are representative of three independent experiments (*, p < 0.05; **, p < 0.01).
Regulation of MEF2 Levels and Activity by GSK3β in CGNs
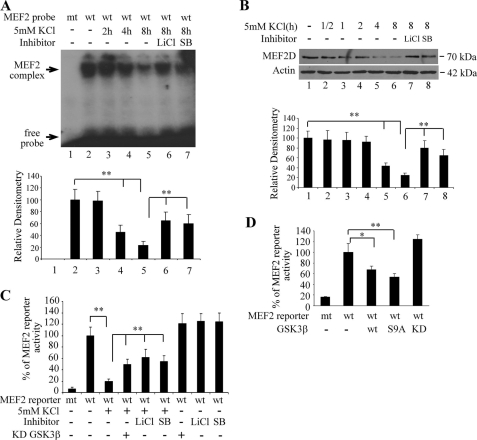
Our previous studies show that MEF2 is required for neuronal survival mediated by membrane depolarization (10, 15, 22). To determine whether GSK3β inhibited MEF2 function, we tested MEF2 DNA binding by electrophoretic mobility shift assay upon activation of GSK3β. In association with the increased GSK3β activity in nuclei of CGNs after potassium withdrawal, MEF2 DNA binding activity decreased markedly over time compared with that in CGNs cultured in media containing 29 mm KCl (Fig. 3A). This decline of MEF2 DNA binding activity correlated well with a gradual reduction in the levels of MEF2D protein (Fig. 3B). Inhibition of GSK3β kinase activity with small molecule inhibitors largely blocked KCl-induced decline of levels of MEF2D and partially restored the level of MEF2 DNA binding.
FIGURE 3.
GSK3β-dependent inhibition of MEF2 DNA binding and transactivation activity after KCl withdrawal. A, analysis of MEF2 DNA binding after KCl withdrawal is shown. Cell lysates (5 μg) from CGNs treated as indicated were examined in an electrophoretic mobility shift assay. Data shown are representative of three experiments (**, p < 0.01). mt, mutant; wt, wild type. B, KCl withdrawal induces degradation of MEF2D. CGNs treated as described in A with or without GSK3β inhibitors were analyzed for MEF2D by Western blot. The experiments were repeated three times (**, p < 0.01). C, attenuation of KCl withdrawal-induced inhibition of MEF2D activity by GSK3β inhibitors is shown. CGNs were transfected for 12 h, switched to the indicated medium with or without the addition of LiCl (10 mm) or SB216763 (SB, 10 μm), and determined for luciferase activity 8 h later. Luciferase activity is shown as the percentage of wild type MEF2 reporter activity under 29 mm KCl conditions (**, p < 0.01, n = 3). D, shown is inhibition of MEF2 function by increased GSK3β activity. CGNs were treated as described under C (*, p < 0.05; **, p < 0.01; n = 3). KD, kinase dead.
To further prove that MEF2 activity is regulated by GSK3β, we employed MEF2-dependent reporter gene assay to correlate GSK3β activity and MEF2 activity. CGNs were transiently transfected with a luciferase reporter gene whose expression is under the control of two MEF2 DNA binding elements. MEF2 activity was measured by luciferase assay. A significant decline in MEF2-luciferase activity was observed when CGNs were exposed to 5 mm KCl for 8 h (Fig. 3C). Co-treatment of CGNs with GSK3β inhibitor LiCl or SB216763 significantly restored MEF2-dependent gene expression. Furthermore, overexpression of a K85A dominant negative mutant GSK3β (supplemental Fig. S1) (23) also attenuated KCl withdrawal-induced loss of MEF2-dependent luciferase activity. Conversely, increasing the levels of GSK3β activity by overexpression of either the wild type or constitutive active form (S9A, Ser-9 to Ala mutant, which cannot be phosphorylated and, therefore, inhibited by Akt) (supplemental Fig. S1) of GSK3β resulted in great inhibition of the endogenous MEF2 function even when CGNs were maintained in the presence of depolarizing level of KCl (Fig. 3D). Taken together, these results suggest that potassium chloride withdrawal-triggered inhibition of MEF2 activity in CGNs is mediated at least in part through a GSK3β-dependent mechanism.
Direct Phosphorylation of MEF2D by GSK3β
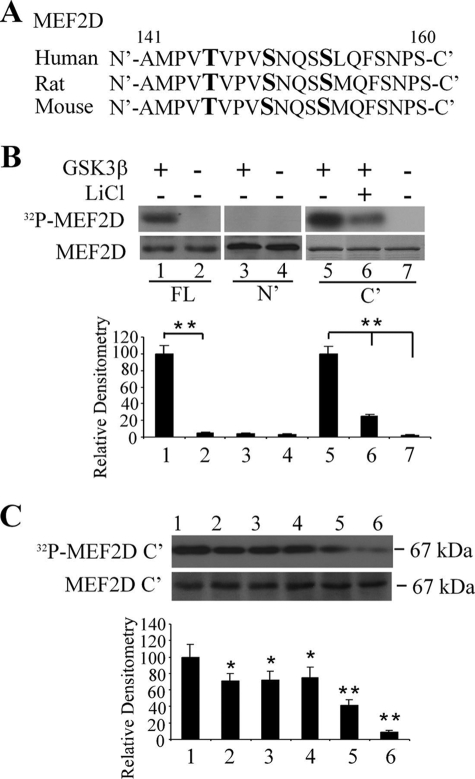
Given that the GSK3β pathway affects MEF2 activity as shown in Fig. 3, we investigated the mechanisms by which GSK3β may regulate MEF2D. Examining the protein sequence of MEF2D revealed the presence of consecutive putative GSK3β phosphorylation sites in a region shortly after the DNA binding motif but within the transactivation domain (Fig. 4A), suggesting that MEF2D may serve as a substrate recognized directly by GSK3β. We tested whether GSK3β phosphorylates MEF2D directly by in vitro kinase assay. The purified GSK3β and immunoprecipitated full-length MEF2D or purified recombinant fragments encompassing various regions of MEF2D were co-incubated in a kinase reaction buffer containing [γ-32P]ATP. Incubation of the full-length MEF2D or C-terminal fragment (MEF2D C′) but not the N′ fragment (MEF2D N′) with GSK3β resulted in significant phosphorylation of the MEF2D fragment. This phosphorylation was greatly inhibited by the inclusion of GSK3β inhibitor lithium chloride in the reaction buffer (Fig. 4B). To localize the GSK3β phosphorylation sites within MEF2D, we selectively mutated the putative serine or threonine residues identified in Fig. 3A to alanine and tested their effects on phosphorylation by GSK3β. Mutation of Thr-145, Ser-149, or Ser-153 individually to Ala had little effect on the overall extent of MEF2D phosphorylation by GSK3β in vitro (Fig. 4C). However, combined mutation of Thr-145/Ser-149, Ser-149/Ser-153, or Thr-145/Ser-149/Ser-153 substantially reduced MEF2D phosphorylation, suggesting that all three amino acid residues were phosphorylated by GSK3β and contributed to the signal in vitro.
FIGURE 4.
Direct phosphorylation of MEF2D by GSK3β. A, shown is the regional sequence alignment of MEF2D from the indicated species. The numbers indicate the starting and ending positions of the amino acid residues of MEF2D protein shown. Bold letters mark the putative phosphorylatable threonine and serine residues that match the GSK3β consensus site. B, shown is direct phosphorylation of MEF2D by GSK3β. The top panel shows representative results of an in vitro kinase assay after co-incubation of purified GSK3β with full-length (FL) MEF2D (FLAG-MEF2D), N′-terminal MEF2D (GST-MEF2D1–86), or C′-terminal MEF2D (GST-MEF2D87–507). The bottom panel shows the loading level of MEF2D. The experiments were repeated three times (**, p < 0.01). C, the effects of mutation at Thr-145, Ser-149, and Ser-153 on phosphorylation of MEF2D by GSK3β are shown. Kinase assay was carried out as described in B (lane 1, wild type (WT) MEF2D C′; lane 2, T145A; lane 3, S149A; lane 4, S153A; lane 5, T145AS149A; lane 6, T145AS149AS153A). The bottom panel shows the loading of C′-MEF2D. Data shown are representative of three experiments (*, p < 0.05; **, p < 0.01).
Effects of GSK3β-mediated Phosphorylation on MEF2 Activity
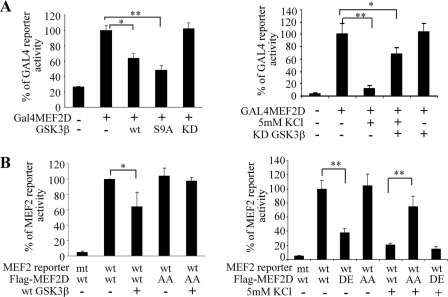
To investigate if potassium withdrawal inhibited MEF2D activity via GSK3β phosphorylation, we first determined whether an increase in GSK3β activity affects the function of Gal4-MEF2D in a Gal4-dependent reporter gene assay. Gal4-MEF2D is a fusion protein with a Gal4 DNA binding domain and MEF2D transactivation domain, which contains the GSK3β phosphorylation sites identified in Fig. 4. Co-transfection of CGNs with constructs encoding Gal4-MEF2D and Gal4 reporter led to increased reporter activity under depolarizing conditions (Fig. 5A, left panel). Under this condition, overexpression of either wild type GSK3β or S9A mutant inhibited Gal4-MEF2D reporter activity. Similarly, Gal4-MEF2D activity was also reduced when CGNs were exposed to 5 mm KCl (Fig. 5A, right panel). Under the latter condition, inhibition of endogenous GSK3β activity by overexpression of the kinase dead form of GSK3β significantly attenuated KCl withdrawal-induced inhibition of Gal4-MEF2D, indicating that GSK3β regulates Gal4-MEF2D likely by modulating its transactivation domain.
FIGURE 5.
Phosphorylation by GSK3β in inhibition of MEF2 transactivation activity. A, inhibition of Gal4-dependent reporter gene expression is shown. Gal4 reporter and Gal4-MEF2D were transfected into cerebellar granule neurons with or without GSK3β as indicated. Luciferase activity was measured 12 h later (left panel). In the right panel, CGNs after transfection were switched to 29 or 5 mm KCl media for 8 h before luciferase activity was determined (*, p < 0.05; **, p < 0.01). KD, kinase dead. wt, wild type. B, the requirement of phosphorylation sites in GSK3β-induced inhibition of MEF2 transactivation activity is shown. Experiments were carried out as described in A (AA, MEF2D T145A/S153A; DE, MEF2D T145D/S149D/S153E). Data shown are representative of three experiments (*, p < 0.05; **, p < 0.01).
To demonstrate directly that GSK3β-mediated inhibition of MEF2 transactivation requires the phosphorylation sites at Thr-145, Ser-149, and Ser-153, we examined the response of GSK3β phosphorylation mutants of MEF2D to either GSK3β or KCl withdrawal-induced inhibition of MEF2 reporter gene expression. Overexpression of FLAG-MEF2D stimulated, whereas co-expression of wild type GSK3β reduced, MEF2- dependent reporter activity in cerebellar granule neurons cultured in the presence of 29 mm KCl (Fig. 5B, left panel). Similarly, overexpression of FLAG-MEF2D mutant with both Thr-145 and Ser-153 mutated to Ala also enhanced MEF2 reporter activity. However, this enhancement was resistant to wild type GSK3β-induced inhibition. Moreover, overexpression of this T145A/S153A mutant allowed CGNs to largely escape 5 mm KCl-induced loss of MEF2 activity (Fig. 5B, right panel). Consistently, the phosphorylation mimic mutant of MEF2D, in which the three GSK3β sites were mutated to charged residue Asp or Glu, failed to stimulate the MEF2 reporter in the presence of 29 mm KCl as compared with the wild type MEF2D. Together, these results indicate that phosphorylation of MEF2D at Thr-145/Ser-149/Ser-153 by GSK3β is inhibitory to its transcriptional potential, which at least partly mediates KCl withdrawal-induced inhibition of MEF2D transactivation function in CGNs.
Correlation of the Phosphorylation Status of MEF2D by GSK3β at Thr-145, Ser-149, and Ser-153 with CGN Survival
Previous studies showed that membrane depolarization by KCl promotes the survival of CGNs, and this process is dependent on MEF2 transactivation activity (10, 15, 22). In contrast, activation of GSK3β by KCl withdrawal induces neuronal apoptosis (24). Thus, phosphorylation of MEF2D by GSK3β in response to KCl withdrawal suggests that it may suppress the pro-survival effect of MEF2. We showed that unlike MEF2D (Fig. 5), MEF2CVP16, a constitutively active MEF2D mutant that is resistant to phosphorylation by GSK3β due to the replacement of MEF2D transactivation domain by viral protein VP16 transactivation domain, is resistant to GSK3β-induced inhibition (Fig. 6A, left panel). We then tested whether MEF2DVP16 may attenuate GSK3β-induced neuronal apoptosis by survival assays. CGNs were transfected with constructs encoding GFP and the indicated constructs. After transfection, neurons were stained with nuclear dye propidium iodide without permeation, and GFP-positive neurons were examined. Overexpression of GSK3β or S9A mutant induced apoptosis of CGNs cultured in medium containing 29 mm KCl (Fig. 6A, left panel). Co-overexpression of MEF2CVP16 attenuated GSK3β S9A-induced neuronal apoptosis.
FIGURE 6.
Protection of CGNs from apoptosis by phosphorylation resistant mutants of MEF2D. A, resistance to GSK3β-mediated inhibition by MEF2DVP16 is shown. The experiments in the left panel were carried out as described in Fig. 5A. For the right panel, CGNs transfected with various constructs indicated, including GFP, were treated as described in Fig. 5A, right panel, before being scored by propidium iodide staining for viability in a blind manner. Data shown are the percentage of propidium iodide (PI)-positive cells divided by GFP-positive cells under each condition (*, p < 0.05; **, p < 0.01; n = 3). mt, mutant; wt, wild type; KD, kinase dead. B, protection of CGNs from KCl withdrawal-induced toxicity by GSK3β or MEF2D mutant is shown. The experiments were carried out as described under A (AA, MEF2D T145A/S153A; DE, MEF2D T145D/S149D/S153E). Data shown are representative of three experiments (*, p < 0.05; **, p < 0.01). C, shown is protection of CGNs by knocking down GSK3β. CGNs with GSK3β knocked down were treated with KCl withdrawal. The levels of apoptosis were scored (**, p < 0.01; n = 3).
To determine the role of this regulatory pathway in KCl withdrawal triggered CGN apoptosis, we first showed that overexpression of the kinase dead GSK3β can reduce the number of apoptotic CGNs triggered by KCl withdrawal (Fig. 6B, right panels), indicating that the GSK3β pathway plays a critical role in KCl withdrawal-induced neuronal death. Using the same experimental paradigm, we showed that overexpression of wild type MEF2D reduced the number of apoptotic CGNs after KCl withdrawal. More importantly, GSK3β phosphorylation mutant MEF2D T145A/S153A provided CGNs significantly better protection (Fig. 6B, right panels). Furthermore, knocking down GSK3β attenuated 5 mm KCl-induced CGN apoptosis (Fig. 6C). Together, these results indicate that enhancing MEF2 activity is sufficient to block GSK3β-induced neuronal apoptosis triggered by KCl withdrawal.
DISCUSSION
Strong evidences indicate that transcription factor MEF2s play a critical role in the survival of different types of neurons (10–13, 15). MEF2 activity is tightly controlled by both survival and death signals. Multiple key pathways converge on MEF2 to regulate its activity and neuronal survival. For example, membrane depolarization promotes the survival of CGNs via p38MAPK-mediated phosphorylation and activation of MEF2s (10), whereas brain-derived neurotrophic factor seems to activate MEF2 through a ERK5-mediated mechanism (13). Protein kinase A has also been shown to modulate MEF2 activity (15). In contrast to our knowledge on the positive regulation of MEF2 by survival signals, how death signals regulate MEF2 is not well understood. It is known that Cdk5 phosphorylates the transactivation domain of MEF2s, which facilitates caspase-dependent degradation (11, 12). This mechanism appears to function predominantly in response to overt excitotoxic and oxidative stress as KCl withdrawal does not appear to induce significant activation of Cdk5 in the nucleus (25). In this study we identified GSK3β-mediated phosphorylation of MEF2D as a major mode of inhibitory regulation in response to KCl withdrawal. Using potassium withdrawal-induced apoptosis in CGNs as a model, we found that KCl withdrawal increases GSK3β activity in the nucleus. GSK3β directly phosphorylates the transactivation domain of MEF2D, which leads to inhibition of MEF2 activity. MEF2D mutants that are resistant to GSK3β phosphorylation can effectively rescue cerebellar granule neurons from potassium withdrawal-induced apoptosis. Taken together, our data define GSK3β as a novel negative regulator of MEF2D in mediating apoptotic signal in cerebellar granule neurons.
In addition to the positive regulators that directly phosphorylate MEF2 and stimulate its activity, MEF2 function is also enhanced upon the activation of phosphatidylinositol 3-kinase-Akt pathway (26, 27). However, Akt does not seem to phosphorylate MEF2D efficiently (27). Because Akt is a potent inhibitor of GSK3β, our finding that MEF2 is phosphorylated and inhibited by GSK3β provides a link between MEF2 and Akt.
GSK3β has been implicated in the induction of apoptosis in response to many forms of apoptotic signals, including trophic signal withdrawal and neurotoxicity (28, 29). Activation of GSK3β has been proposed to underlie the pathogenic process in neurodegenerative disorders including Alzheimer disease. Despite the important role for GSK3β in controlling neuronal apoptosis, the molecular mechanisms by which GSK3β regulates this process were not well illustrated. The identification of survival factor MEF2D as a direct substrate of GSK3β during neuronal apoptotic process expands the targets by which GSK3β transmits apoptotic signal. It also provides a mechanism that underlies GSK3β-mediated neurotoxicity. Because both GSK3β and MEF2D are known to play important roles in non-neuronal tissues such as mediating insulin signal and regulating glucose metabolism (30), it would be interesting to investigate whether GSK3β-dependent regulation of MEF2 also plays a role in this and other processes.
An interesting feature of GSK3β is its substrate specificity. Many GSK3β substrates require prior phosphorylation by different kinases at a downstream S/T site to generate a motif (S/T)XXX(S/T)P, where the upstream S/T can serve as the phosphorylation site for GSK3 (31). Other GSK3β substrates seem not to require prior phosphorylation for their recognition by the kinase (32). Our data suggest that GSK3β can directly phosphorylate MEF2D at 145TXXXSXXXS153. Residues Thr-145, Ser-149, and Ser-153 are required for efficient phosphorylation (Fig. 4). Because robust phosphorylation of MEF2D at these sites by GSK3β can be easily achieved in vitro without prior incubation of MEF2D with another kinase, it is consistent with the notion that MEF2D is efficiently recognized as an unprimed substrate by GSK3β. Hyperphosphorylation of microtubule associate protein Tau is the pathological hallmark of Alzheimer disease (33). GSK3β phosphorylates Tau at both primed and unprimed sites. Moreover, the GSK3β mediated phosphorylation of Tau at the primed sites, which are prephosphorylated by Cdk5/p25 (32). Given that MEF2 is phosphorylated by Cdk5 in response to neurotoxic insults and given the involvement of Cdk5 in Alzheimer disease (11), it would be important to investigate if MEF2 may be synergistically regulated by GSK3β and Cdk5 under the pathological conditions associated with the development of Alzheimer disease.
Supplementary Material
Acknowledgment
We thank Dr. James R. Woodgett for providing constructs for hemagglutinin (HA)-GSK3|gb, HA-GSK3|gb(K85A), and HA-GSK3|gb(S9A).
This work was supported, in whole or in part, by National Institutes of Health Grants NS048254, AG023695, ES015317, and ES016731-010002 (to Z. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- GSK3β
- glycogen synthase kinase 3β
- MEF2
- myocyte enhancer factor 2
- ERK5
- extracellular-regulated kinase 5
- CGN
- cerebellar granule neurons
- PI
- propidium iodide
- GFP
- green fluorescent protein
- DIV
- day(s) in vitro
- PBS
- phosphate-buffered saline
- Cdk5
- cyclin-dependent kinase 5.
REFERENCES
- 1.Kaytor M. D., Orr H. T. (2002) Curr. Opin. Neurobiol. 12, 275–278 [DOI] [PubMed] [Google Scholar]
- 2.Mora A., Sabio G., González-Polo R. A., Cuenda A., Alessi D. R., Alonso J. C., Fuentes J. M., Soler G., Centeno F. (2001) J. Neurochem. 78, 199–206 [DOI] [PubMed] [Google Scholar]
- 3.Hetman M., Cavanaugh J. E., Kimelman D., Xia Z. (2000) J. Neurosci. 20, 2567–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pap M., Cooper G. M. (1998) J. Biol. Chem. 273, 19929–19932 [DOI] [PubMed] [Google Scholar]
- 5.Lucas J. J., Hernández F., Gómez-Ramos P., Morán M. A., Hen R., Avila J. (2001) EMBO J. 20, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embi N., Rylatt D. B., Cohen P. (1980) Eur. J. Biochem. 107, 519–527 [PubMed] [Google Scholar]
- 7.Jope R. S., Johnson G. V. (2004) Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 8.Beurel E., Jope R. S. (2006) Prog. Neurobiol. 79, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashima A., Honda T., Yasutake K., Michel G., Murayama O., Murayama M., Ishiguro K., Yamaguchi H. (1998) Neurosci. Res. 31, 317–323 [DOI] [PubMed] [Google Scholar]
- 10.Mao Z., Bonni A., Xia F., Nadal-Vicens M., Greenberg M. E. (1999) Science 286, 785–790 [DOI] [PubMed] [Google Scholar]
- 11.Gong X., Tang X., Wiedmann M., Wang X., Peng J., Zheng D., Blair L. A., Marshall J., Mao Z. (2003) Neuron 38, 33–46 [DOI] [PubMed] [Google Scholar]
- 12.Tang X., Wang X., Gong X., Tong M., Park D., Xia Z., Mao Z. (2005) J. Neurosci. 25, 4823–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., Cavanaugh J. E., Wang Y., Sakagami H., Mao Z., Xia Z. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Tang X., Li M., Marshall J., Mao Z. (2005) J. Biol. Chem. 280, 16705–16713 [DOI] [PubMed] [Google Scholar]
- 16.Mussmann R., Geese M., Harder F., Kegel S., Andag U., Lomow A., Burk U., Onichtchouk D., Dohrmann C., Austen M. (2007) J. Biol. Chem. 282, 12030–12037 [DOI] [PubMed] [Google Scholar]
- 17.Blair L. A., Bence-Hanulec K. K., Mehta S., Franke T., Kaplan D., Marshall J. (1999) J. Neurosci. 19, 1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bijur G. N., Jope R. S. (2001) J. Biol. Chem. 276, 37436–37442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Y., Planel E., Herman M., Figueroa H. Y., Wang L., Liu L., Lau L. F., Yu W. H., Duff K. E. (2008) J. Neurosci. 28, 2624–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 21.Hughes K., Nikolakaki E., Plyte S. E., Totty N. F., Woodgett J. R. (1993) EMBO J. 12, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Z., Wiedmann M. (1999) J. Biol. Chem. 274, 31102–31107 [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Garcia C. A., Rehani K., Cekic C., Alard P., Kinane D. F., Mitchell T., Martin M. (2008) J. Immunol. 181, 6797–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M., Wang X., Meintzer M. K., Laessig T., Birnbaum M. J., Heidenreich K. A. (2000) Mol. Cell. Biol. 20, 9356–9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian B., Yang Q., Mao Z. (2009) Nat Cell Biol. 11, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Q., Wu Z. (2000) J. Biol. Chem. 275, 36750–36757 [DOI] [PubMed] [Google Scholar]
- 27.Wiedmann M., Wang X., Tang X., Han M., Li M., Mao Z. (2005) J. Neurosci. Res. 81, 226–234 [DOI] [PubMed] [Google Scholar]
- 28.Nair V. D., Olanow C. W. (2008) J. Biol. Chem. 283, 15469–15478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra R., Barthwal M. K., Sondarva G., Rana B., Wong L., Chatterjee M., Woodgett J. R., Rana A. (2007) J. Biol. Chem. 282, 30393–30405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M. L., Olson A. L., Edgington N. P., Moye-Rowley W. S., Pessin J. E. (1994) J. Biol. Chem. 269, 28514–28521 [PubMed] [Google Scholar]
- 31.Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 32.Cho J. H., Johnson G. V. (2003) J. Biol. Chem. 278, 187–193 [DOI] [PubMed] [Google Scholar]
- 33.Engel T., Goñi-Oliver P., Gómez de Barreda E., Lucas J. J., Hernández F., Avila J. (2008) Neurodegener. Dis. 5, 247–249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.