Abstract
PDE6 (phosphodiesterase-6) is the effector molecule in the vertebrate phototransduction cascade. Progress in understanding the structure and function of PDE6 has been hindered by lack of an expression system of the enzyme. Here we report ectopic expression and analysis of compartmentalization and membrane dynamics of the enhanced green fluorescent protein (EGFP) fusion protein of human cone PDE6C in rods of transgenic Xenopus laevis. EGFP-PDE6C is correctly targeted to the rod outer segments in transgenic Xenopus, where it displayed a characteristic striated pattern of EGFP fluorescence. Immunofluorescence labeling indicated significant and light-independent co-localization of EGFP-PDE6C with the disc rim marker peripherin-2 and endogenous frog PDE6. The diffusion of EGFP-PDE6C on disc membranes investigated with fluorescence recovery after photobleaching was markedly slower than theoretically predicted. The enzymatic characteristics of immunoprecipitated recombinant PDE6C were similar to known properties of the native bovine PDE6C. PDE6C was potently inhibited by the cone- and rod-specific PDE6 γ-subunits. Thus, transgenic Xenopus laevis is a unique expression system for PDE6 well suited for analysis of the mechanisms of visual diseases linked to PDE6 mutations.
Introduction
Phosphodiesterases of cyclic nucleotides (PDEs)2 are essential enzymes controlling cellular levels of cAMP and cGMP. Eleven families of PDEs have been identified in mammals on the grounds of sequence homology, substrate selectivity, and regulation (1). Photoreceptor-specific PDEs in rods and cones comprise the sixth PDE family (PDE6) and serve as the effector enzymes in the vertebrate phototransduction cascade (1–5). Rod PDE6 is composed of homologous catalytic α-subunit (PDE6A) and β-subunit (PDE6B) and two copies of a small inhibitory γ-subunit (Pγ) (3). Cone PDE6 is a catalytic dimer of two identical α′-subunits (PDE6C) (3). A cone-specific inhibitory Pγ-subunit is highly homologous to the rod Pγ (6). In rod photoreceptors, PDE6 is located in the specialized compartments called rod outer segments (ROS), where it associates with disc membranes. The membrane attachment of PDE6 is mediated by farnesylation of the PDE6A C terminus and geranylgeranylation of the PDE6B C terminus (7). In cones, geranylgeranylated PDE6C resides on infoldings of the cone outer segment plasma membrane (8). Following photoexcitation of rod or cone photoreceptor cells, PDE6 is activated by the GTP-bound transducin α-subunit (GαtGTP) that relieves the Pγ inhibition of the enzyme. cGMP hydrolysis by active PDE6 leads to a cellular response due to a closure of cGMP-gated channels in the photoreceptor plasma membrane (2–5).
Although PDE6 plays a prominent role in vertebrate vision, the structure-function relationships of PDE6 are poorly understood in comparison with other key phototransduction proteins. The lack of an expression system for PDE6 has become a major impediment for PDE6 research. Importantly, an expression system for PDE6 is required to elucidate the mechanisms of visual diseases linked to mutations in PDE6 catalytic subunits. Mutations in the PDE6A and PDE6B genes are responsible for 3–4% and ∼4% of cases of recessive retinitis pigmentosa, respectively (9, 10). Retinitis pigmentosa is a common hereditary disease of retinal degeneration that results in vision loss caused by the death of the rod and cone photoreceptors. Recently, mutations in the PDE6C gene have been identified in human patients with achromatopsia (11). Achromatopsia results from a loss of cone function and is characterized by low visual acuity and lack of color discrimination.
The problem of PDE6 expression was partially addressed through generation and characterization of chimeras between PDE6 and PDE5 (cGMP-binding, cGMP-specific PDE) (12–15). The two PDE enzymes share similar domain organization, a relatively high homology of the catalytic domains, specificity for cGMP relative to cAMP, and sensitivity to common catalytic site inhibitors (1, 3). The chimeric PDE5/PDE6 approach facilitated delineation of the Pγ-binding sites of PDE6, but it has severe limitations in identification of the unique catalytic determinants of the visual effector (15). The reason for the inability of PDE6 to fold correctly in various cell types, besides photoreceptor cells, is unknown. One possibility is that expression of functional PDE6 requires photoreceptor specific chaperone proteins, such as AIPL1 (aryl hydrocarbon receptor-interacting protein-like 1). AIPL1 was shown to be a specialized chaperone obligatory for expression and stability of PDE6 in rod photoreceptors (16, 17). Mice lacking or expressing reduced levels of AIPL1 show reduced expression and destabilization of PDE6 and develop retinal degeneration (16, 17). In humans, mutations in AIPL1 cause Leber congenital amaurosis, a severe early onset retinopathy (18), apparently by compromising PDE6 expression. Thus, expression of PDE6 in living photoreceptor cells may represent a sole approach to produce and mutagenize PDE6. In this study, we demonstrate the utility of transgenic frogs for expression and studies of PDE6 by ectopically expressing EGFP fusion protein of human cone PDE6C in rods of Xenopus laevis. We investigated subcellular localization, membrane diffusion, and biochemical properties of the recombinant PDE6 enzyme.
EXPERIMENTAL PROCEDURES
Generation of Transgenic X. laevis
cDNA for the full-length human PDE6C was assembled from two DNA fragments coding amino acids 1–445 and 446–858 of PDE6C, which were obtained by PCR amplification from a human retinal cDNA library. To prepare the transgene construct (Fig. 1A), the pXOP(−508/+41)EGFP vector (19) was first modified to eliminate the stop codon after the EGFP sequence and introduce a new restriction site (XmaI) downstream of EGFP. Human PDE6C was cloned into the vector using NotI and XmaI sites to obtain the pXOP(−508/+41)EGFP-PDE6C vector. The construct sequence was confirmed by automated DNA sequencing. DNA was purified using a Qiagen Miniprep kit, digested with XhoI to linearize the plasmid, and repurified with the Qiagen PCR purification kit with final elution in water.
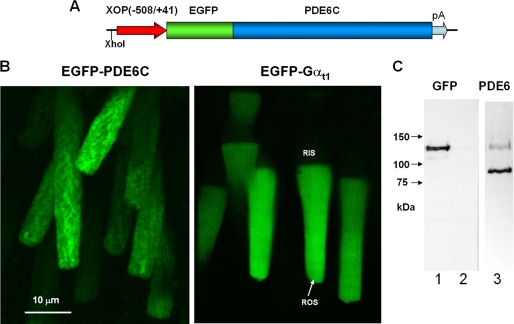
FIGURE 1.
Expression of EGFP-PDE6C in transgenic rods. A, map of the transgene. The XhoI site was used to linearize the pXOP(508/+41)EGFP-PDE6C plasmid for production of transgenic X. laevis embryos. B, EGFP fluorescence in living photoreceptor cells expressing EGFP-PDE6C and EGFP-Gαt. C, immunoblotting of retinal extracts from transgenic (lanes 1 and 3) and non-transgenic retinas (lane 2) with anti-GFP B-2 monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) (lanes 1 and 2) and anti-PDE6 MOE antibodies (CytoSignal) (lane 3).
All experimental procedures involving the use of frogs were carried out in accordance with the protocol approved by the University of Iowa Animal Care and Use Committee. Transgenic X. laevis expressing EGFP-PDE6C in rods were produced using the method of restriction enzyme-mediated integration (20). Transgenic X. laevis expressing EGFP-Gαt with EGFP insertion within the helical domain of Gαt were generated similarly as described previously (21). Transgenic animals were identified 6 days postinjection by visual examination for EGFP fluorescence using a fluorescence microscope MZ16 Leica equipped with a GFP filter. Several transgenic tadpoles were maintained through metamorphosis into adult frogs. Transgenic male adult frogs at age ∼10 months were mated with wild-type female X. laevis to produce a large number of transgenic tadpoles for biochemical characterization of PDE6C. At about stage 50, Xenopus retinas from transgenic tadpoles were dissected into small pieces in Ringer's buffer, and the EGFP fluorescence in living photoreceptor cells was imaged using a confocal fluorescence microscope LSM510 (Zeiss).
Immunofluorescence
For dark adaptation, tadpoles were kept in the dark overnight. For light adaptation, dark-adapted tadpoles were exposed to room light (∼500 lux) for at least 60 min. Detached tadpole eyeballs were fixed in 4% formaldehyde in phosphate-buffered saline for 2 h at 22 °C. After fixation, the eyeballs were submersed in a 30% sucrose solution in phosphate-buffered saline for 5 h at 4 °C and then embedded in tissue freezing medium (Tris-buffered saline) and frozen on dry ice. Radial sectioning (10 μm) of the retina was performed using a cryomicrotome Microm HM 505E. Retinal cryosections were air-dried and kept at −80 °C until use. Before staining, sections were incubated in 0.1% Triton/phosphate-buffered saline for 30 min. Labeling with anti-Xenopus peripherin-2 Xper5A11 monoclonal antibody (1:20) (22) or rabbit anti-PDE6 MOE antibody (1:1000) (CytoSignal) was performed in 0.1% Triton/phosphate-buffered saline containing 3% bovine serum albumin, 1 mm MgCl2, and 1 mm CaCl2 for 2 h at 25 °C. Following a 2-h incubation with goat anti-mouse AlexaFluor 543 or goat anti-rabbit AlexaFluor 555 secondary antibodies (Molecular Probes) (1:1000), the sections were visualized using a Zeiss LSM 510 confocal microscope.
Analysis of EGFP-PDE6C Diffusion
The lateral and longitudinal mobilities of nonactivated and Gαt-activated EGFP-PDE6C were assessed by measuring fluorescence recovery after photobleaching (FRAP) in living Xenopus rods essentially as previously described for EGFP-Gαt (21). A typical FRAP recording involved imaging of a transgenic rod cell attached to a small piece of retina on an LSM 510 confocal microscope using Plan-Neofluar ×40/1.3 numerical aperture oil lens. With the zoom setting at 8, a field of 28.4 × 28.4 μm (256 × 256 pixels, 9 pixels/μm) was scanned at maximal speed (196 ms/frame). Bleaching was induced with maximal power of an argon laser (488 nm) and a 50-ms bleach time. The bleach region was selected as a 3-pixel-wide stripe. Typically, 150 images were recorded at 0.5% laser power in each time series with 1- and 2-s intervals between the scans for lateral and longitudinal diffusion measurements, respectively. Average fluorescence intensities of bleached, nonbleached, and background regions were recorded for each time point, and the intensities of the bleached region Istripe(t) were corrected for the background and fading of fluorescence during recordings. The actual width, ω (∼1.4–1.6 μm), of a bleached stripe with a Gaussian profile was determined from the analysis of the first afterbleach image (21). Intensities integrated along the y axis for each pixel position along the x axis (perpendicular to the bleached stripe) were determined for selected regions of every first afterbleach image using ImageJ. The profiles of integrated intensities for each pixel at t = 0 corrected for the background I (x, 0) were fit to equation, I (x, 0) = I (0)·(1 − A exp(−4x2/ω2)), to estimate the depth of bleach A and the width ω. The diffusion coefficients and immobile fractions were calculated by fitting the time courses Istripe(t) to Equation 3 in Wang et al. (21) using constrained ω and the estimated A as an initial value. Diffusion of nonactivated EGFP-PDE6C was examined using Ringer's buffer supplemented with 6 mm 2-deoxyglucose and 10 mm sodium azide, whereas diffusion of the activated enzyme was monitored using Ringer's buffer containing 10 mm glucose and saturated with 95% O2, 5% CO2. The theoretical diffusion coefficient for PDE6C freely diffusing on the surface of the disc membrane was calculated according to the Saffman-Delbruck equation (23), D = (kBT/4πμh)·(ln(μh/μ′α) − γ), where kB is the Boltzmann constant (1.3807 × 10−23 J/K), T is the absolute temperature (298 K), μ is the viscosity of the disc membrane (0.7 pascal·s), μ′ is the viscosity of the cytosol (0.002 pascal·s), h is the thickness of a disk membrane spanned by the PDE6 lipid tails (4 nm), γ is Euler's constant (0.5772), and α is the radius of the two lipid tails (0.35 nm).
Extraction, Immunoprecipitation, and Isolation of PDE6C
Eyeballs were excised from tadpoles at about stage 50 and stored at −80 °C until use. Typically, 100–120 eyeballs were homogenized with a pestle in an Eppendorf tube using 20 mm Tris-HCl (pH 7.5) buffer containing 120 mm NaCl, 1 mm MgSO4, 1 mm β-mercaptoethanol, and complete protease inhibitor mixture (Roche Applied Science). The homogenate was centrifuged (20,000 × g, 20 min, 4 °C), and the resulting pellet was resuspended in hypotonic 10 mm Tris-HCl (pH 7.5) buffer containing 1 mm β-mercaptoethanol and complete protease inhibitor mixture. The supernatant containing PDE6 was obtained by centrifugation (70,000 × g, 60 min, 4 °C) and was used immediately or stored at −80 °C. For immunoprecipitation, Dynabeads with Protein G (30 mg/ml) (Invitrogen) were washed with 20 mm Tris-HCl (pH 7.5) buffer containing 500 mm NaCl, 1 mm MgSO4, and 1 mm β-mercaptoethanol (buffer A) and incubated with rabbit anti-GFP antibodies ab290 (Abcam) or sheep anti-GFP antibodies (Elmira Biologicals) for 60 min at 25 °C. The beads were washed from unbound proteins two times with buffer A, followed by the addition of PDE6 extracts from transgenic or non-transgenic retinas and incubation with rotation for 3 h at 4 °C. The beads were then washed four times with buffer A, one time with buffer A minus NaCl, and finally one time with buffer A. PDE6C was eluted by incubating 15 μl of beads with 40 μl of buffer A containing 5 μg of trypsin for 15 min at 4 °C. Typically, this treatment eluted more than 95% of PDE6C activity. The eluates were removed, placed into tubes containing 40 μg of soybean trypsin inhibitor, and analyzed immediately or stored at −20 °C in the presence of 40% glycerol.
Production of Sheep Anti-GFP Antibodies
DNA coding EGFP was PCR-amplified using pEGFP-C1 vector (Clontech) as the template and cloned into the pET15b vector using NdeI and BamHI restriction enzymes. The His-tagged EGFP was expressed in BL21 Escherichia coli cells and purified over His-bind resin (Novagen). The His tag was removed with thrombin, and EGFP was used for custom antibody production in sheep (Elmira Biologicals, Iowa City, IA).
Cloning of Human Cone Pγ
The nucleotide sequence of human cone Pγ was obtained using four overlapping synthetic oligonucleotides 1–4 (75–80 bp) covering the entire Pγ sequence. Oligonucleotides 2 (sense) and 3 (antisense) were combined in a PCR to obtain a template for PCR with oligonucleotides 1 and 4 containing flanking NdeI and BamHI restriction sites. The resulting PCR product was cloned into the pET15b vector using the NdeI/BamHI sites. The cone and rod Pγ-subunits were expressed and purified as described previously (24).
PDE Activity Assay and Data Analysis
PDE activity was measured using 5 μm [3H]cGMP and 1 pm PDE6 or 200 μm [3H]cGMP and 20 pm PDE6 as described (15, 25). To determine Km values for cGMP, PDE activity was measured using 10–1000 μm cGMP, and the data were fit to the equation, Y = Vmax·X/(Km + X). The kcat values for cGMP hydrolysis were calculated as Vmax/[PDE]. The Ki values for PDE6 inhibition by Pγ were calculated by fitting data to the equation, Y(%) = 100/(1 + 10̂(X − log Ki)), where X is the logarithm of total Pγ concentration. Fitting the experimental data to the equations was performed with nonlinear least squares criteria using GraphPad Prism version 4 software. Experimental results are shown as mean ± S.E. for three separate experiments.
RESULTS
Expression of EGFP-PDE6C in Transgenic Rods
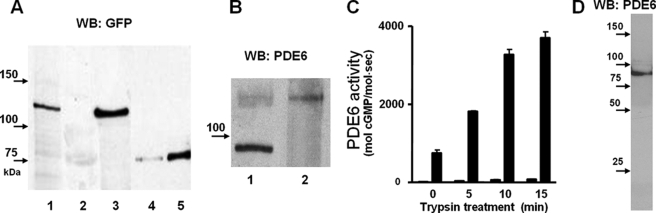
Transgenic expression of the EGFP fusion protein of human cone PDE6C in rods was directed by the well characterized Xenopus opsin promoter (19) (Fig. 1A). Cone PDE6C was chosen for the study because (a) its homodimeric nature facilitates analysis of the transgene product and (b) PDE6C is not expected to form dimers with endogenous rod PDE6AB. The C termini of PDE6 are the sites of isoprenylation and are the key determinants of subcellular localization of PDE6. The extreme PDE6 N termini do not appear to be essential to any PDE6 function and, thus, were selected for the attachment of EGFP. Examination of EGFP fluorescence in the retina of transgenic tadpoles indicated that EGFP-PDE6C is properly localized to the rod outer segments, suggesting intact transport of the protein (Fig. 1B). The expression level of EGFP-PDE6C varied between transgenic tadpoles and also between different rods within a single retina. This mosaic expression has been observed for other transgenic X. laevis models utilizing the opsin promoter (21, 26). In the retinal extract from transgenic tadpoles, but not from non-transgenic animals, anti-GFP antibodies recognized a ∼125 kDa band of the predicted size for EGFP-PDE6C (Fig. 1C). Immunoblotting with anti-PDE6 MOE antibodies showed that the average levels of EGFP-PDE6C in transgenic extract are significantly lower than the level of endogenous PDE6 (Fig. 1C).
Compartmentalization of EGFP-PDE6C in Rods
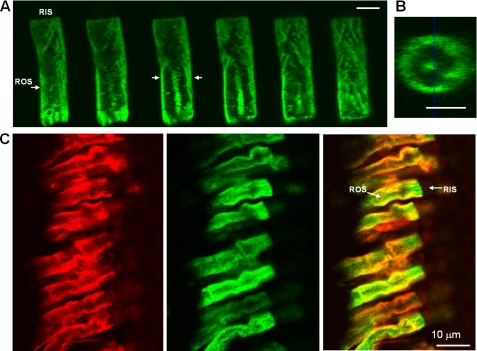
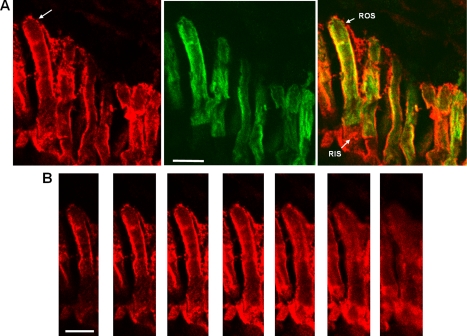
The pattern of EGFP fluorescence in transgenic EGFP-PDE6C rods was strikingly different from the known diffused distribution of EGFP-Gαt or EGFP-rhodopsin (21, 26) (Fig. 1B) The EGFP-PDE6C localization appeared similar to the striated pattern of peripherin-2-EGFP, a disc rim marker, in transgenic Xenopus rods observed previously (22). Furthermore, the Z-section EGFP fluorescence of a transgenic ROS is also consistent with the peripheral distribution of PDE6C (Fig. 2, A and B). We examined potential co-localization of EGFP-PDE6C with peripherin-2 by labeling retinal cryosections from dark-adapted or light-adapted tadpoles with anti-Xenopus peripherin-2 Xper5A11 monoclonal antibody (22). EGFP fluorescence in the retina sections was preserved following the fixation procedure. The immunofluorescence and EGFP fluorescence signals show extensive co-localization of EGFP-PDE6C and peripherin-2 in light-adapted rods, suggesting association of PDE6C with disc membranes and incisures (Fig. 2C). Similar co-localization of EGFP-PDE6 with peripherin-2 was observed in dark-adapted X. laevis (not shown). To determine EGFP-PDE6C localization relative to endogenous frog PDE6, transgenic retina cryosections were stained with anti-PDE6 MOE antibody. The MOE antibody is directed against holo-PDE6, including the Pγ-subunit. Thus, it was not surprising that, in addition to labeling ROS, the MOE antibody showed some extraneous staining in the inner segments of Xenopus rods (Fig. 3). Nonetheless, the immunofluorescence and EGFP fluorescence in the ROS portion indicate significant co-localization of EGFP-PDE6C with frog rod PDE6 (Fig. 3A). The MOE antibody immunofluorescence in the Z-sections of a transgenic ROS supports the peripheral distribution of endogenous frog PDE6 as well (Fig. 3B).
FIGURE 2.
Localization of EGFP-PDE6C in transgenic rods. A and B, EGFP fluorescence in EGFP-PDE6C rod. A, Z-scans with an interval of 0.9 μm through ROS of a light-adapted transgenic tadpole. Bar, 5 μm. B, cross-section of the ROS in A corresponding to the position identified by arrows. Bar, 5 μm. C, co-localization of EGFP-PDE6C with peripherin-2 in transgenic rods. Red, immunofluorescence of a retinal cryosection of a light-adapted transgenic tadpole stained with monoclonal antibody Xper5A11 against anti-Xenopus peripherin-2; green, EGFP fluorescence; overlay.
FIGURE 3.
Co-localization of EGFP-PDE6C with endogenous PDE6 in transgenic rods. A, red, immunofluorescence of a retinal cryosection of a light-adapted transgenic tadpole stained with anti-PDE6 MOE antibody; green, EGFP fluorescence; overlay. Bar, 10 μm. B, MOE antibody immunofluorescence. Z-Scans with an interval of 1 μm through the retina cryosection in A showing the rod cell identified by the arrow. Bar, 10 μm.
Diffusion of EGFP-PDE6C in Transgenic Rods
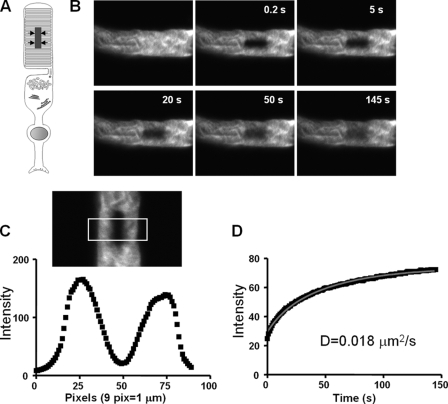
Diffusion of EGFP-PDE6C in transgenic rods was assessed using fluorescence recovery after photobleaching. The photobleaching geometry shown in Fig. 4A allows for monitoring lateral diffusion (along the plane of the disc membrane). In control experiments, we measured lateral diffusion of rhodopsin using transgenic rhodopsin-EGFP X. laevis described previously (27). The diffusion coefficient of 0.28 μm2/s for rhodopsin-EGFP was calculated after the correction for the slowing effect of incisures (28). This value is comparable with the D value for rhodopsin-EGFP obtained by Wang et al. (21), and it is in general agreement with the diffusion coefficients for rhodopsin reported previously (28, 29). Diffusion of nonactivated EGFP-PDE6C was examined under conditions of nucleotide depletion using 2-deoxyglucose and NaN3 (Fig. 4). In the absence of GTP, transducin in transgenic rods is predicted to be sequestered by photoexcited rhodopsin, thus preventing formation of the activated Gαt-PDE6 complex (21). The apparent D coefficient of 0.018 ± 0.0015 μm2/s (mean ± S.E.) for nonactivated EGFP-PDE6C was determined from FRAP recordings of 16 rods from four transgenic tadpoles. Using the correction coefficient of 2.7 for the effect of incisures, the D value of 0.049 μm2/s is still well below the predicted theoretical value for freely diffusing PDE6 (see “Discussion”). Diffusion of the Gαt-activated EGFP-PDE6C was measured in oxygenated Ringer's buffer supplemented with 10 mm glucose. The measured D value of 0.015 ± 0.0014 μm2/s (mean ± S.E., n = 12) for the activated EGFP-PDE6 was slightly lower than that for nonactivated EGFP-PDE6C. The FRAP measurements under both conditions yielded sizable immobile fractions (15–20%) of EGFP-PDE6C.
FIGURE 4.
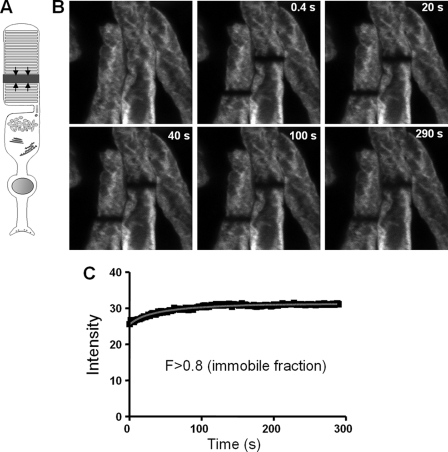
Analysis of lateral diffusion of EGFP-PDE6C by FRAP. A, photobleaching geometry to monitor lateral diffusion. The arrows indicate the direction of diffusion into the bleached stripe region. B, pre- and postbleach images of a transgenic ROS at different time points. The retina was incubated in Ringer's buffer containing 6 mm 2-deoxyglucose and 10 mm NaN3. 150 images (256 × 256 pixels) were collected with a 1-s interval between the scans. C, intensity integrated along the y axis for each x axis pixel of the selected box region from the first postbleach image. The plot is used to determine the width of a bleach region with a Gaussian profile and the depth of bleach (21). D, intensities of the bleached region during the experimental time course corrected for background and fading and the fitting curve to a one-dimensional diffusion equation described previously (21).
In addition to the lateral diffusion, transfer of PDE6 could potentially take place between the membrane discs in the longitudinal direction. The longitudinal diffusion of EGFP-PDE6C was probed by introducing a bleaching stripe perpendicular to the long axis of the transgenic ROS (Fig. 5, A and B). The FRAP analysis indicated very limited, if any, diffusion of nonactivated or activated EGFP-PDE6C in the longitudinal direction, with the immobile fraction typically exceeding 80% (Fig. 5, B and C).
FIGURE 5.
Analysis of longitudinal diffusion of EGFP-PDE6C by FRAP. A, photobleaching geometry to monitor longitudinal diffusion. The arrows indicate the direction of diffusion into the bleached stripe region. B, bleaches were introduced into two neighboring rods. Pre- and postbleach images of transgenic ROS at different time points. The retina was incubated in Ringer's buffer containing 6 mm 2-deoxyglucose and 10 mm NaN3. 150 images (256 × 256 pixels) were collected with a 2-s interval between the scans. C, intensities of the bleached region from the left rod during the experimental time course corrected for background and fading and the fitting curve to a one-dimensional diffusion equation described previously (21).
Isolation and Characterization of PDE6C
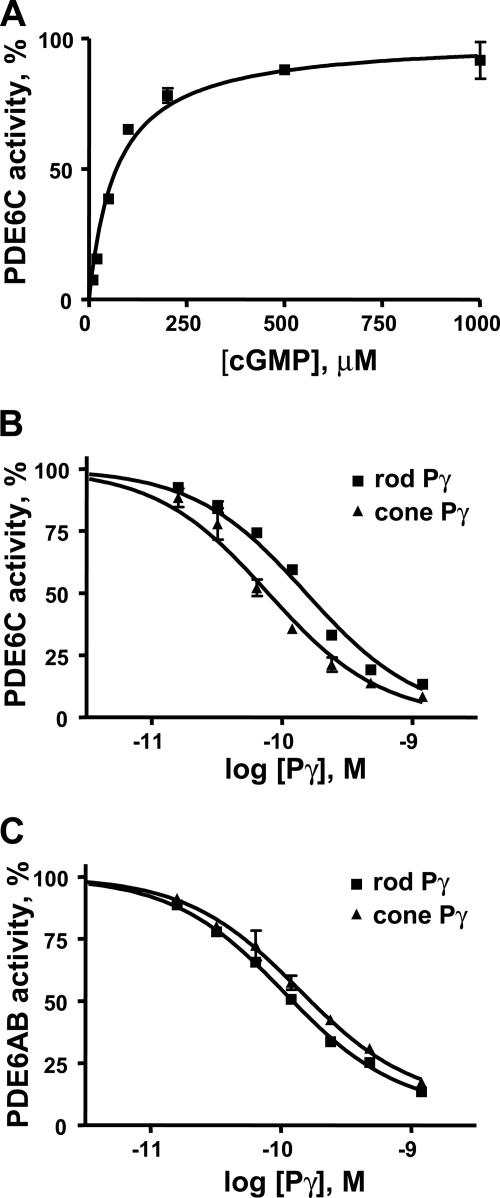
To characterize EGFP-PDE6, the enzyme was immunoprecipitated from the retinal extracts using anti-GFP antibodies and protein G beads. The anti-GFP antibody, but not an unrelated anti-GST antibody, selectively precipitated EGFP-PDE6 from the retinal extract of transgenic tadpoles (Fig. 6A). Immunoblotting with anti-PDE6 MOE antibodies (CytoSignal) showed that the Xenopus rod PDE6, which is abundantly present in the retinal extract of transgenic tadpoles, is not detectable in the sample of immunoprecipitated EGFP-PDE6C (Fig. 6B). This suggests that EGFP-PDE6C does not form heterodimers with the frog rod PDE6A and PDE6B subunits. PDE6 activities were assayed in the immunoprecipitated samples of EGFP-PDE6C prior to or after limited treatment with trypsin to cleave the Pγ-subunit and activate PDE6. The trypsin treatment resulted in a ∼5-fold activation of EGFP-PDE6C, suggesting that the enzyme precipitates in complex with the endogenous frog Pγ-subunit (Fig. 6C). Control immunoprecipitations were performed using retinal extracts of non-transgenic tadpoles. The basal and trypsin-treated PDE6 activities in immunoprecipitates from non-transgenic animals were negligible (Fig. 6C). The trypsin treatment of the immunoprecipitated EGFP-PDE6C released more than 95% of PDE6C activity from the complex with anti-GFP antibody by cleaving off EGFP (Fig. 6D). Thus, the amount of soluble PDE6C was estimated by quantifying the amount of EGFP-PDE6C in the immunoprecipitates prior to trypsin treatment using Western blotting with anti-GFP antibody and recombinant EGFP-PDE6C-(446–819) as a standard (Fig. 6A). PDE6C hydrolyzed cGMP with a Km of 65 ± 6 μm and kcat of 4400 ± 300 s−1 (Fig. 7A). For comparison, we determined the Km of 46 ± 5 μm cGMP and kcat of 4800 ± 400 s−1 for purified trypsin-activated bovine rod PDE6 (not shown). PDE6C was potently inhibited by the recombinant cone and rod Pγ-subunits with slight selectivity for the cone Pγ. The Ki values for cone and rod Pγ were 78 ± 12 pm and 155 ± 18 pm, respectively (Fig. 7B). Rod PDE6AB was similarly sensitive to both types of Pγ-subunits (Fig. 7C).
FIGURE 6.
Immunoprecipitation and solubilization of PDE6C. A, immunoblotting (WB) with anti-GFP B-2 monoclonal antibody (Santa Cruz Biotechnology, Inc.). Lane 1, retinal extract (0.5 transgenic retina equivalent); lane 2, control immunoprecipitation with unrelated rabbit anti-GST antibody; lane 3, proteins immunoprecipitated from retinal extract (four transgenic retinas) with anti-GFP antibody (ab290, Abcam). Lanes 4 and 5, EGFP standards: 5 and 15 ng of purified E. coli expressed EGFP-PDE6C (446–819). B, immunoblot with anti-PDE6 MOE antibody. Lane 1, transgenic retina extract; lane 2, proteins immunoprecipitated from retinal extract with anti-GFP antibody. C, PDE6 activity of the immunoprecipitated proteins from non-transgenic (left bars) and transgenic (right bars) extracts prior to and following trypsin treatment was measured using 200 μm [3H]cGMP. D, after immunoprecipitation of EGFP-PDE6C with anti-GFP-antibody, the beads were treated with trypsin for 15 min. Proteins released into the soluble fraction were separated from the beads, reconstituted with soybean trypsin inhibitor, and analyzed by immunoblotting with anti-PDE6 MOE antibody.
FIGURE 7.
Characterization of PDE6C. A, the rates of cGMP hydrolysis by immunoprecipitated and trypsin-solubilized PDE6C are plotted as a function of cGMP concentration. PDE6C activity is expressed as a percentage of maximal activity (4400 ± 300 mol of cGMP/mol of PDE6C/s). Results from one of three similar experiments are shown. Km = 65 ± 6 μm (mean ± S.E.). B and C, inhibition of PDE6C (B) and bovine rod PDE6AB (C) by the cone and rod Pγ-subunits. PDE6 activity was measured using 5 μm [3H]cGMP and 1 pm PDE6. Results from one of three similar experiments are shown. For PDE6C, Ki = 78 ± 12 pm with cone Pγ, and Ki = 155 ± 18 pm with rod Pγ; for PDE6AB, Ki = 140 ± 14 pm with cone Pγ, and Ki = 105 ± 20 pm with rod Pγ (mean ± S.E.).
DISCUSSION
Precise compartmentalization and membrane dynamics of PDE6 in photoreceptor cells are among many important characteristics of the visual effector that are still controversial or poorly defined. Immunoelectron microscopy of bovine retina sections first indicated uniform labeling of PDE6 on ROS membrane discs (30). However, a recent immunoelectron microscopy study suggested that a substantial portion of total PDE6 is localized near the edges of the disc membranes in dark-adapted rat ROS (31). Light exposure induced translocation of PDE6 away from the disc rims and led to a more even distribution of the enzyme (31). A generally assumed uniform distribution of PDE6 on the disc membrane clearly contrasts with the known interaction of PDE6 with GARP2 (glutamic acid-rich protein-2) (30, 32). GARP2 is an abundant soluble protein variant of the GARP region of the cGMP-gated channel β-subunit (30, 33). In ROS, GARP2 is confined to the rim region and incisures of discs, apparently due to its interaction with peripherin-2 oligomers (30, 34). Biochemical evidence suggests that GARP2 binds to PDE6 nearly stoichiometrically, and the fraction of PDE6 free of GARP2 is small (32).
Although the resolution of fluorescence microscopy is low compared with immunoelectron microscopy, the analysis of EGFP-PDE6C localization in transgenic X. laevis rods has important advantages. The distribution of EGFP-PDE6C can be examined in living rods in addition to the use of fixed retina sections. Most importantly, the analysis is not limited by the specificity of PDE6 antibodies or availability of PDE6 epitopes. Also, the large diameter of frog rods (5–7 μm) makes fluorescence microscopy adequate to the task. Peripherin-2 is a well characterized marker of the disc rims and incisures (22, 34). The pattern of EGFP-PDE6C fluorescence and co-localization of EGFP-PDE6C with frog peripherin-2 indicate that the enzyme is concentrated at the disc rim region and incisures in dark- or light-adapted rods. The quality of the available PDE6C MOE antibody limited the accuracy of localization of endogenous frog rod PDE6 and its co-localization with EGFP-PDE6C. Nonetheless, frog PDE6 also appears to have peripheral distribution within the ROS and was at least partially co-localized with EGFP-PDE6C. The FRAP analysis revealed markedly slower than predicted lateral diffusion of EGFP-PDE6C. The theoretical diffusion coefficient of 0.76 μm2/s was calculated for freely diffusing EGFP-PDE6 according to the Suffman-Delbruck equation (23), assuming the two geranylgeranyl tails of PDE6C submerged into the lipid bilayer. The observed D value is 0.018 ± 0.0015 μm2/s (or 0.049 μm2/s when corrected by a factor of 2.7 to account for the effect of incisures of frog membrane discs (28)) (i.e. ∼15-fold lower than the theoretical value). The slow lateral diffusion of EGFP-PDE6C is consistent with its association with oligomeric rim protein complexes involving peripherin-2. The soluble GARP proteins may anchor PDE6 to peripherin-2.
Our findings suggest that PDE6 is localized to the disc rim regions in rods. Retinal guanylate cyclase was also shown to concentrate at the marginal region of the disc membrane (35). Thus, the two enzymes controlling cGMP levels, PDE6 and guanylate cyclase, and the plasma membrane cGMP-gated channel might be localized in close proximity. Such compartmentalization of the phototransduction components may ensure rapid activation and recovery of photoresponses.
In contrast to holotransducin (21), EGFP-PDE6C does not appreciably diffuse in the longitudinal direction. Holotransducin and PDE6 each have two lipid tails. Thus, interactions with the rim protein complexes possibly contribute to the lack of interdisc transfer of the enzyme. The slow lateral diffusion and the absence of any significant interdisc transfer of EGFP-PDE6C both indicate that it is predominantly membrane-bound in Xenopus rods. This notion differs from the hypothesized existence of soluble PDE6 in photoreceptor cells (36, 37). The hypothesis is based on the observation that upon extraction from bovine retina, a significant fraction of PDE6 is found in a soluble complex with the 17-kDa prenyl-binding protein PrBP/δ, also known as the δ-subunit of PDE6 (36). PrBP/δ was shown to interact with the methylated prenylated C termini of PDE6A and PDE6B and solubilize the enzyme from the membrane in vitro (37). However, immunohistochemical analysis has shown that PrBP/δ localizes to the IS or near the RIS/ROS junction and does not co-localize with PDE6 (38). Therefore, solubilization of PDE6 may represent an artifact of the isolation procedure.
One of the central implications of this study is the demonstration that transgenic X. laevis is an effective system for expression of recombinant PDE6. To date, there are no unequivocal reports of functional expression of PDE6 or its mutants using conventional approaches, such as heterologous cell cultures. Even the human retinoblastoma cell line Y79, which exhibits some biochemical characteristics of photoreceptor cells and contains natural transcripts of PDE6, fails to express functional protein (39). Here, we show that functional EGFP-PDE6C is expressed in Xenopus rods and can be readily isolated without trace contaminations by frog PDE6. The isolation procedure involves selective immunoprecipitation of EGFP-PDE6C with anti-GFP antibodies and limited treatment with trypsin to release soluble PDE6C. The enzymatic characteristics of the recombinant PDE6C (kcat = 4400 s−1, Km = 65 μm) are consistent with known properties of the native bovine PDE6C (40, 41). PDE6C was potently inhibited by the cone and rod-specific Pγ-subunits. The cone and rod PDE6 enzymes appear to be similar in terms of their sensitivity to the Pγ-subunits. Overall, the expression system of human cone PDE6C in Xenopus rods provides an excellent tool for mutational analysis of PDE6 structure and function, including examination of molecular mechanisms of PDE6C mutations leading to achromatopsia.
Acknowledgments
We thank Dr. D. Weeks (University of Iowa) for generous advice, Dr. R. S. Molday (University of British Columbia) for the gift of anti-peripherin-2 antibody, and Dr. D. Oprian for providing the vector for rhodopsin-EGFP expression in X. laevis rods.
This work was supported, in whole or in part, by National Institutes of Health Grant EY-10843 (to N. O. A.).
- PDE
- cyclic nucleotide phosphodiesterase
- Pγ
- γ-subunit of PDE6
- Gαt
- transducin α-subunit
- FRAP
- fluorescence recovery after photobleaching
- ROS
- rod outer segment(s)
- RIS
- rod inner segment(s).
REFERENCES
- 1.Conti M., Beavo J. (2007) Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 2.Burns M. E., Arshavsky V. Y. (2005) Neuron 48, 387–401 [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Cote R. H. (2005) Front. Biosci. 10, 1191–1204 [DOI] [PubMed] [Google Scholar]
- 4.Lamb T. D., Pugh E. N., Jr. (2006) Invest. Ophthalmol. Vis. Sci. 47, 5137–5152 [DOI] [PubMed] [Google Scholar]
- 5.Fu Y., Yau K. W. (2007) Pflugers Arch. 454, 805–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton S. E., Hurley J. B. (1990) J. Biol. Chem. 265, 11259–11264 [PubMed] [Google Scholar]
- 7.Anant J. S., Ong O. C., Xie H. Y., Clarke S., O'Brien P. J., Fung B. K. (1992) J. Biol. Chem. 267, 687–690 [PubMed] [Google Scholar]
- 8.Karan S., Zhang H., Li S., Frederick J. M., Baehr W. (2008) Vision Res. 48, 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dryja T. P., Rucinski D. E., Chen S. H., Berson E. L. (1999) Invest. Ophthalmol. Vis. Sci. 40, 1859–1865 [PubMed] [Google Scholar]
- 10.McLaughlin M. E., Ehrhart T. L., Berson E. L., Dryja T. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3249–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang B., Grau T., Dangel S., Hurd R., Jurklies B., Sener E. C., Andreasson S., Dollfus H., Baumann B., Bolz S., Artemyev N., Kohl S., Heckenlively J., Wissinger B. (2009) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granovsky A. E., Natochin M., McEntaffer R. L., Haik T. L., Francis S. H., Corbin J. D., Artemyev N. O. ( 1998) J. Biol. Chem. 273, 24485– 24490 [DOI] [PubMed] [Google Scholar]
- 13.Granovsky A. E., Artemyev N. O. (2000) J. Biol. Chem. 275, 41258–41262 [DOI] [PubMed] [Google Scholar]
- 14.Granovsky A. E., Artemyev N. O. (2001) J. Biol. Chem. 276, 21698–21703 [DOI] [PubMed] [Google Scholar]
- 15.Muradov H., Boyd K. K., Artemyev N. O. (2006) Vision Res. 46, 860–868 [DOI] [PubMed] [Google Scholar]
- 16.Liu X., Bulgakov O. V., Wen X. H., Woodruff M. L., Pawlyk B., Yang J., Fain G. L., Sandberg M. A., Makino C. L., Li T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13903–13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramamurthy V., Niemi G. A., Reh T. A., Hurley J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13897–13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohocki M. M., Perrault I., Leroy B. P., Payne A. M., Dharmaraj S., Bhattacharya S. S., Kaplan J., Maumenee I. H., Koenekoop R., Meire F. M., Birch D. G., Heckenlively J. R., Daiger S. P. (2000) Mol. Genet Metab. 70, 142–150 [DOI] [PubMed] [Google Scholar]
- 19.Mani S. S., Batni S., Whitaker L., Chen S., Engbretson G., Knox B. E. (2001) J. Biol. Chem. 276, 36557–36565 [DOI] [PubMed] [Google Scholar]
- 20.Kroll K. L., Amaya E. (1996) Development 122, 3173–3183 [DOI] [PubMed] [Google Scholar]
- 21.Wang Q., Zhang X., Zhang L., He F., Zhang G., Jamrich M., Wensel T. G. (2008) J. Biol. Chem. 283, 30015–30024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewen C. J., Moritz O. L., Tam B. M., Papermaster D. S., Molday R. S. (2003) Mol. Biol. Cell 14, 3400–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffman P. G., Delbrück M. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 3111–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muradov H., Boyd K. K., Kerov V., Artemyev N. O. (2007) Biochemistry 46, 9992–10000 [DOI] [PubMed] [Google Scholar]
- 25.Hamilton S. E., Prusti R. K., Bentley J. K., Beavo J. A., Hurley J. B. (1993) FEBS Lett. 318, 157–161 [DOI] [PubMed] [Google Scholar]
- 26.Moritz O. L., Tam B. M., Papermaster D. S., Nakayama T. (2001) J. Biol. Chem. 276, 28242–28251 [DOI] [PubMed] [Google Scholar]
- 27.Jin S., McKee T. D., Oprian D. D. (2003) FEBS Lett. 542, 142–146 [DOI] [PubMed] [Google Scholar]
- 28.Poo M., Cone R. A. (1974) Nature 247, 438–441 [DOI] [PubMed] [Google Scholar]
- 29.Wey C. L., Cone R. A., Edidin M. A. (1981) Biophys. J. 33, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Körschen H. G., Beyermann M., Müller F., Heck M., Vantler M., Koch K. W., Kellner R., Wolfrum U., Bode C., Hofmann K. P., Kaupp U. B. (1999) Nature 400, 761–766 [DOI] [PubMed] [Google Scholar]
- 31.Chen J., Yoshida T., Bitensky M. W. (2008) Mol. Vis. 14, 2509–2517 [PMC free article] [PubMed] [Google Scholar]
- 32.Pentia D. C., Hosier S., Cote R. H. (2006) J. Biol. Chem. 281, 5500–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Körschen H. G., Illing M., Seifert R., Sesti F., Williams A., Gotzes S., Colville C., Müller F., Dosé A., Godde M. (1995) Neuron 15, 627–636 [DOI] [PubMed] [Google Scholar]
- 34.Poetsch A., Molday L. L., Molday R. S. (2001) J. Biol. Chem. 276, 48009–48016 [DOI] [PubMed] [Google Scholar]
- 35.Liu X., Seno K., Nishizawa Y., Hayashi F., Yamazaki A., Matsumoto H., Wakabayashi T., Usukura J. (1994) Exp. Eye Res. 59, 761–768 [DOI] [PubMed] [Google Scholar]
- 36.Gillespie P. G., Prusti R. K., Apel E. D., Beavo J. A. (1989) J. Biol. Chem. 264, 12187–12193 [PubMed] [Google Scholar]
- 37.Cook T. A., Ghomashchi F., Gelb M. H., Florio S. K., Beavo J. A. (2000) Biochemistry 39, 13516–13523 [DOI] [PubMed] [Google Scholar]
- 38.Norton A. W., Hosier S., Terew J. M., Li N., Dhingra A., Vardi N., Baehr W., Cote R. H. (2005) J. Biol. Chem. 280, 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White J. B., Thompson W. J., Pittler S. J. (2004) Mol. Vis. 10, 738–749 [PubMed] [Google Scholar]
- 40.Gillespie P. G., Beavo J. A. (1988) J. Biol. Chem. 263, 8133–8141 [PubMed] [Google Scholar]
- 41.Gillespie P. G., Beavo J. A. (1989) Mol. Pharmacol. 36, 773–781 [PubMed] [Google Scholar]