Abstract
The cytosolic adaptor protein Apaf-1 is a key player in the intrinsic pathway of apoptosis. Binding of mitochondrially released cytochrome c and of dATP or ATP to Apaf-1 induces the formation of the heptameric apoptosome complex, which in turn activates procaspase-9. We have re-investigated the chain of events leading from monomeric autoinhibited Apaf-1 to the functional apoptosome in vitro. We demonstrate that Apaf-1 does not require energy from nucleotide hydrolysis to eventually form the apoptosome. Despite a low intrinsic hydrolytic activity of the autoinhibited Apaf-1 monomer, nucleotide hydrolysis does not occur at any stage of the process. Rather, mere binding of ATP in concert with the binding of cytochrome c primes Apaf-1 for assembly. Contradicting the current view, there is no strict requirement for an adenine base in the nucleotide. On the basis of our results, we present a new model for the mechanism of apoptosome assembly.
Introduction
Cells of most multicellular organisms can terminate themselves conducting the evolutionary conserved program of apoptosis. As the well regulated response to a death signal from outside or inside the cell, a cascade of cysteine proteases, so-called caspases, is activated that eventually leads to the orderly disintegration of cell organelles and chromatin (1–3). In contrast to necrotic cell death, which is accompanied by inflammation and damage of the surrounding tissue, apoptotic cells keep their degraded constituents contained in intact membrane. The cell corpses are then cleanly removed by phagocytes obviating inflammatory and autoimmune responses (4). Apoptosis is vital for development and homeostasis of metazoans (5–7), and malfunction of the human apoptosis machinery is implicated in autoimmune diseases, neurodegenerative disorders, and cancer (8–11).
The intrinsic pathway of apoptosis responds to cytotoxic stress, genomic damage, and developmental cues (12, 13). The pivotal point of this pathway is the release of cytochrome c from the mitochondria through permeabilization of the outer membrane (14, 15). In human cells, transduction of this decisive death signal is achieved via the 142-kDa cytosolic protein Apaf-1, the mammalian ortholog of Caenorhabditis elegans CED-4 (16). Apaf-1 contains an N-terminal caspase recruitment domain (CARD),2 a central nucleotide binding and oligomerization domain (NOD), and a C-terminal regulatory domain of 13 WD40 repeats. It is thought that the WD40 repeat domain blocks the CARD and NOD until cytochrome c is released into the cytosol (17–20). Binding of cytochrome c to the WD40 repeat domain in the presence of ATP or dATP appears to induce a large conformational change in Apaf-1 that eventually bares the NOD and the effector-binding CARD. Mediated by the NOD, Apaf-1 oligomerizes into a wheel-like heteroheptameric protein complex (21) termed apoptosome. The apoptosome recruits the initiator caspase-9 via homophilic CARD-CARD interaction and the holoenzyme then activates downstream executioner caspases (22–24).
So far, there is no consistent view of the course of events that lead to the formation of active apoptosome. Several studies have pointed at a peculiar need for dATP as a cofactor for apoptosome assembly (16, 18, 25–28). It has been reported that recombinantly produced Apaf-1 in its autoinhibited form contains dATP and that the nucleotide cannot be hydrolyzed in the absence of cytochrome c (26). The authors of that study propose that binding of cytochrome c to Apaf-1 prompts hydrolysis of the bound nucleotide to provide energy to open Apaf-1 for oligomerization. The produced dADP is then exchanged for exogenous dATP, which finally triggers the formation of the apoptosome. The preference for deoxynucleotides and the necessity for a hydrolytic step prior to the nucleotide exchange are contradicted by a recent in vitro study by Bao et al. (29) and a high resolution crystal structure of WD40-deleted Apaf-1 (30), which identify ADP as the nucleotide bound to full-length Apaf-1 purified from insect cells and WD40-deleted Apaf-1 purified from bacteria, respectively. We have assayed different aspects of apoptosome formation in vitro to clear the controversial views on the role of the nucleotide in the transition of Apaf-1 from inactive monomer to the large apoptosomal complex that finally activates caspase-9.
EXPERIMENTAL PROCEDURES
Materials
ATP, ADP, dATP, dADP, AppNHp, and GTP were purchased from Sigma. ADP and dADP were further purified according to the method described in John et al. (31) to remove trace amounts of the corresponding triphosphates. Ac-LEHD-AFC was purchased from Biomol GmbH (Hamburg, Germany). Horse heart cytochrome c was purchased from Sigma and further purified via gel filtration.
Protein Purification
N-terminally His6-tagged human Apaf-1-XL-(1–1248) was produced in Sf21 insect cells. A recombinant baculovirus was produced using the Bac-to-Bac system (Invitrogen), and cells were infected at a density of 2 × 106/ml. Cells were harvested 72–96 h post-infection and lysed by sonication in the presence of 1% (v/v) Nonidet P-40. Apaf-1 was isolated by nickel-nitrilotriacetic acid (Qiagen) chromatography, and the eluate was subjected to SuperQ (GE Healthcare) anion exchange chromatography. The fractions containing Apaf-1 were added with recombinant tobacco etch virus protease and incubated on ice for 6 h. The protein solution was concentrated using ultrafiltration devices (Millipore) with a cutoff of 50 kDa and loaded onto a Superdex 200 (GE Healthcare) gel filtration column equilibrated with buffer A (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm dithioerythritol, 1 mm MgCl2). The fractions containing monomeric Apaf-1 were concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C. Quantitative removal of the affinity tag was verified by Western blotting. Procaspase-9 carrying a C-terminal His6 tag was overexpressed in Escherichia coli BL21-DE3 and purified by consecutive nickel-nitrilotriacetic acid, anion exchange, and size exclusion chromatography. Upon expression, the procaspase underwent quantitative autocleavage yielding active caspase-9 consisting of a large subunit (∼35 kDa) and a small subunit (∼12 kDa). All purification steps were performed at 4 °C.
Detection of Nucleotides by HPLC
Nucleotides were analyzed using a C-18 reverse phase column (Ultrasphere ODS, 4.6 × 250 mm, Beckman Coulter) connected to a Waters HPLC system (components 626 Pump, 600S Controller, 717Plus Autosampler, and 2487 Absorption Detector) at a wavelength of 254 nm. The running buffer contained 50 mm potassium phosphate, pH 6.6, 10 mm tetra-n-butyl ammonium bromide, and 10% (v/v) acetonitrile. Reference nucleotides were used to determine the respective retention time at a flow rate of 1.5 ml/min. Protein samples containing nucleotide were denatured by immersion in boiling water for 30 s. The precipitated protein was removed by centrifugation, and the supernatant was subjected to HPLC analysis. The nature of the bound nucleotide was determined by comparison with the chromatograms of the reference nucleotides.
Hydrolytic Activity of Apaf-1
To assay the hydrolysis of nucleoside triphosphates by Apaf-1, 10 μm Apaf-1 in buffer A was added with 1 mm ATP or dATP and incubated at 30 °C. Samples of 10 μl were taken automatically every 60 min and were analyzed by HPLC at a wavelength of 254 nm. The implemented integration function of the Empower software (Waters) was used to determine the peak area increase of the diphosphate signals. Reference samples of known concentration were used to calculate the increase in nucleotide concentration. To determine the background hydrolysis rate, nucleotides were incubated in buffer A without Apaf-1, and samples were taken and analyzed accordingly.
Apoptosome Assembly
To assemble apoptosomes, we essentially employed the protocols described by Shi (32). According to this, apoptosomes for analytic gel filtration experiments were produced by overnight incubation at 4 °C, whereas for the caspase activation assay short incubation times at higher temperatures (10 min at 30 °C) were used.
Analytic Gel Filtration
Protein samples of 250 μl were analyzed on a Superose 6 10/300 GL column or an S200 10/300 GL column (GE Healthcare) equilibrated with buffer A. For apoptosome assembly, Apaf-1 (10 μm) was incubated with a 5-fold excess of cytochrome c and nucleotide concentrations as indicated in buffer A at 4 °C overnight. For isolation of apoptosomes, the fractions corresponding to the apoptosome peak were pooled and concentrated using an Amicon centrifugal filter device with a cutoff of 100 kDa.
Caspase-9 Activation Assay
The reaction mixtures at a final volume of 100 μl contained 0.4 μm Apaf-1, 2 μm cytochrome c, 0.2 μm caspase-9, and an indicated amount of nucleotide in buffer A. After incubation at 30 °C for 10 min, Ac-LEHD-AFC was added to a final concentration of 200 μm, and caspase-9 activity was fluorometrically measured as the release of AFC using a Tecan Safire2 fluorospectrometer at an excitation wavelength of 400 nm and an emission wavelength of 505 nm. Determination of the caspase-9 activity of preassembled apoptosomes was performed accordingly using concentrated apoptosome at a final concentration of 0.4 μm (referring to monomeric Apaf-1) omitting cytochrome c and nucleotide from the reaction mixture.
Preparation of Recycled Apaf-1
Apoptosomes were assembled using either ATP or AppNHp and isolated via gel filtration. The fractions corresponding to the apoptosome peak were pooled and concentrated, and NaCl was added to a final concentration of 1 m. After incubation for 30 min on ice, the sample was run on a Superdex 200 equilibrated with buffer A containing 1 m NaCl. The fractions corresponding to monomeric Apaf-1 were pooled and concentrated using an Amicon centrifugal filter device with a cutoff of 50 kDa. To decrease the salt concentration to the starting level, buffer A was added to the initial volume after removal of ∼90% of the buffer volume. After repetition of this procedure, the protein was flash-frozen in liquid nitrogen and stored at −80 °C.
RESULTS
Autoinhibited Full-length Apaf-1 Contains ADP and Possesses dATPase Activity
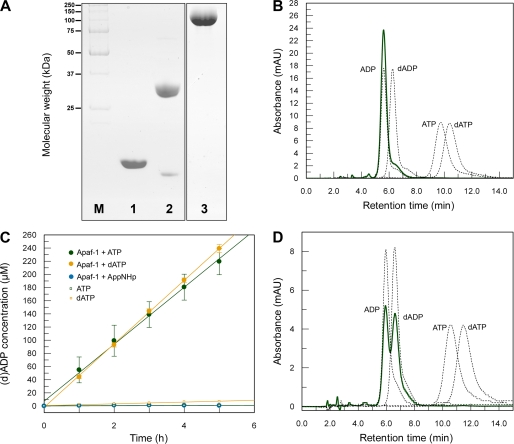
We recombinantly produced full-length human Apaf-1 (residues 1–1248) using Sf21 insect cells and purified the protein to homogeneity (Fig. 1A). We compared the retention times of the nucleotide extracted from our recombinantly produced Apaf-1 and of pure reference nucleotides (dATP, dADP, ATP, and ADP) on a high pressure liquid chromatography (HPLC) system (Fig. 1B). The nucleotide bound to Apaf-1 was unambiguously identified to be ADP. We then asked whether the presence of a nucleoside diphosphate in purified Apaf-1 reflects an intrinsic hydrolytic activity. We therefore monitored the conversion of dATP into dADP and of ATP into ADP under steady-state conditions using an HPLC system (supplemental Fig. S1). At 30 °C and in the absence of cytochrome c, ATP and dATP are hydrolyzed at a rate of 4.33 ± 0.13 and 4.83 ± 0.04 molecules per molecule of Apaf-1 per h, respectively (Fig. 1C). To verify the ability of Apaf-1 to exchange and hydrolyze dATP/ATP in the absence of cytochrome c, we analyzed by HPLC the nucleotide status of formerly ADP-bound Apaf-1 after incubation with dATP in a 100-fold excess. After 3 h at 20 °C, Apaf-1 contained approximately equal amounts of ADP and dADP but not even trace amounts of dATP (Fig. 1D).
FIGURE 1.
Nucleotide status and hydrolytic properties of Apaf-1. A, Coomassie-stained SDS-PAGE showing the purity of the proteins used in the assays. Lane M, molecular weight marker; lane 1, horse cytochrome c; lane 2, caspase-9; lane 3, Apaf-1. B, Apaf-1 purified from Sf21 insect cells contains ADP. Absorption curves corresponding to reference nucleotides are shown as black dotted lines; the absorption curve of the nucleotide isolated from purified Apaf-1 is shown as green solid line. C, Apaf-1 hydrolyzes ATP and dATP at a low rate. 10 μm Apaf-1 was incubated in buffer A at 30 °C with 1 mm ATP and dATP, respectively. Samples taken at different time points were analyzed on an HPLC system to detect the formed nucleoside diphosphate. By comparison with reference samples, the respective peak area (supplemental Fig. S1) was converted into concentration (green or yellow dots). As controls, the same was done with samples containing nucleotide but no Apaf-1 (green or yellow open squares) or Apaf-1 together with the nonhydrolyzable ATP analog AppNHp (blue dots). Mean averages of triplicate measurements are plotted against time. The resulting linear fits represent an ATPase activity of 4.33 ± 0.13 ATP molecules per molecule of Apaf-1 per h and a dATPase activity of 4.83 ± 0.04 dATP molecules per molecule of Apaf-1 per h. D, Apaf-1 is able to exchange ADP for dATP in the absence of cytochrome c. 500 μg of Apaf-1 were incubated with 1 mm dATP at 20 °C for 3 h. After separation of the protein from excess nucleotide by gel filtration, the peak fractions were concentrated and analyzed by HPLC. Absorption curves corresponding to reference nucleotides are shown as black dotted lines, and the absorption curve of the nucleotide isolated from Apaf-1 is shown as green solid line. mAU, milliabsorbance units.
Apoptosome Assembly Depends on Exogenous Supply of a Nucleoside Triphosphate but Does Not Rely on the Nature of the Base or the Hydrolyzability of the Nucleotide
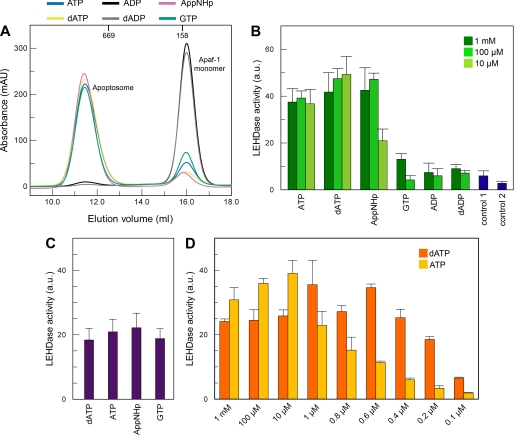
To analyze which properties of the nucleotide are required for apoptosome assembly to proceed, we incubated 10 μm Apaf-1 overnight at 4 °C with cytochrome c in a 5-fold excess and various nucleotides (ATP, ADP, dATP, dADP, and GTP or the nonhydrolyzable ATP analog AppNHp) at a concentration of 1 mm. We assayed the reaction mixtures by size exclusion chromatography. In the presence of any of the tested nucleoside triphosphates, a high molecular weight species appeared, which represents the formed apoptosome complex, and the elution peak corresponding to Apaf-1 monomer markedly decreased (Fig. 2A). In the presence of nucleoside diphosphate or without exogenous nucleotide, no apoptosome was formed. If only cytochrome c was added, approximately half of the monomeric Apaf-1 precipitated (data not shown). This behavior has already been observed by others, and it has been suggested that the protein here adopts a less stable conformation that is prone to aggregation (26).
FIGURE 2.
Cofactor requirements for apoptosome formation and caspase-9 activation. A, nucleotide dependence of apoptosome formation. Apaf-1 at a concentration of 10 μm was incubated at 4 °C overnight with a 5-fold excess of cytochrome c and 1 mm nucleotide. The reaction mixtures were analyzed by analytic gel filtration. Apoptosome formation is indicated by strong decrease of the elution volume. Addition of a nucleoside triphosphate (blue, ATP; yellow, dATP; pink, AppNHp; green, GTP) leads to formation of apoptosomes. Incubation with ADP (black curve) or dADP (gray curve) has no effect. The elution volumes of globular marker proteins are indicated. B, nucleotide dependence of caspase-9 activation. Apaf-1 at a concentration of 0.4 μm was incubated at 30 °C for 10 min with 2 μm cytochrome c, 0.2 μm caspase-9, and nucleotide as indicated. Ac-LEHD-AFC was added to a final concentration of 200 μm, and the reaction was fluorometrically monitored. Measurements were done in triplicate; error bars are standard deviations. Control 1 contained no nucleotide; control 2 contained only caspase-9. C, stimulation of caspase-9 activity by the apoptosome is independent of the nucleotide used for assembly. Apoptosomes were assembled using ATP, dATP, AppNHp, and GTP, respectively, and isolated via gel filtration. Isolated apoptosomes equivalent to 0.4 μm Apaf-1 monomer were incubated with 0.2 μm caspase-9 at 30 °C for 10 min. Ac-LEHD-AFC was added to a final concentration of 200 μm, and the reaction was fluorometrically monitored. Measurements were repeated five times; error bars are standard deviations. D, only at very low nucleotide concentrations is dATP more efficient for caspase-9 activation than ATP. Assay conditions are as in B. mAU, milliabsorbance units.
We extracted the nucleotide from the formed apoptosomes. Analysis via HPLC showed that the apoptosome exclusively contains the triphosphate used for assembly. Apparently, oligomerized Apaf-1 is no longer competent for hydrolysis (supplemental Fig. S2). When equal concentrations of 1 mm ATP and GTP were supplied for assembly, the resulting apoptosomes predominantly contained ATP (supplemental Fig. S3). To have at least half of the formed apoptosomes occupied by GTP, a 100-fold excess of GTP over ATP had to be supplied (supplemental Fig. S3).
Once Formed, Apoptosomes Are Equally Active Regardless of the Nature of the Nucleotide Bound
To investigate the functionality of apoptosomes assembled with different nucleotides, we assayed the stimulation of caspase-9 activity in the presence of Apaf-1, cytochrome c, and nucleotide. Functional apoptosomes are known to increase the basal activity of caspase-9 by up to 3 orders of magnitude (22, 33, 34). We fluorimetrically monitored the cleavage of the artificial peptide substrate Ac-LEHD-AFC by caspase-9. The increase of the fluorescence intensity over time due to the accumulation of released AFC is proportional to the caspase-9 activity. We find that the exogenous supply of ATP as well as of the nonhydrolyzable ATP analog AppNHp at concentrations of 100 μm or higher leads to caspase-9 stimulation in the range of that achieved with dATP (Fig. 2B). Even GTP shows a stimulatory effect, which is approximately one-fifth of the stimulation seen for the adenine-containing triphosphates. However, this effect is only seen for highest concentrations of 1 mm GTP.
To test whether apoptosomes assembled using different nucleotides differ in their potential to stimulate caspase-9, we isolated apoptosomes assembled using ATP, dATP, AppNHp, or GTP via size exclusion chromatography and performed the caspase activation assay with equal amounts of the isolated apoptosomes. All apoptosomes proved to be equally competent for activation of caspase-9 (Fig. 2C), indicating that different activation levels depicted in Fig. 2B are due to different amounts of apoptosome formed.
To carefully examine the possible differences in the potentials of dATP and ATP to stimulate caspase-9, we assayed caspase activation over a broad concentration range, including substoichiometric nucleotide levels. The measured activities are very similar for both nucleotides, and only at concentrations below 10 μm does a decrease in ATP-induced activation become visible (Fig. 2D).
Recovered from the Apoptosome, Apaf-1 Re-adopts Its Primary Conformation after Hydrolysis of the Bound Nucleotide in the Monomer
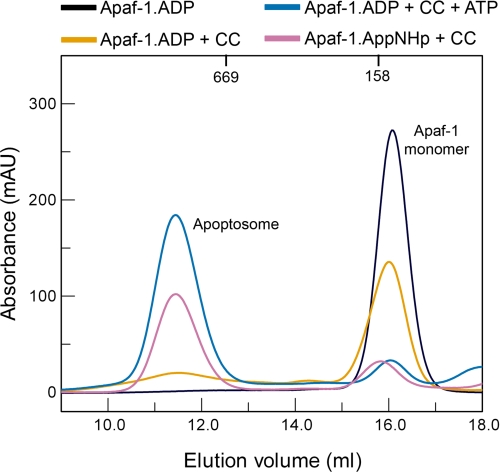
The question, whether the conformational changes upon assembly of Apaf-1 are reversible, prompted us to recycle Apaf-1 from apoptosomes. If the process was invertible, recycled Apaf-1 would need the same conditions for re-assembly as naive Apaf-1.
We disassembled isolated apoptosomes in high salt buffer. Using size exclusion chromatography under high salt conditions, we separated monomeric Apaf-1 from cytochrome c. Monomeric Apaf-1 (supplemental Fig. S4) was then transferred to low salt buffer and submitted to another round of apoptosome assembly. Prior to re-assembly, the nucleotide state of recycled Apaf-1 was determined using HPLC. When exogenous ATP was provided to assemble the initial apoptosomes, recycled monomeric Apaf-1 contained ADP. Apparently, Apaf-1 regains its hydrolytic activity as it leaves the apoptosomal complex. Using the nonhydrolyzable ATP analog AppNHp for the initial apoptosome assembly led to AppNHp-bound recycled Apaf-1. If recycled Apaf-1 was ADP-loaded, it required cytochrome c and nucleoside triphosphate for complex formation (Fig. 3), like freshly purified Apaf-1. If either cytochrome c or nucleoside triphosphate was omitted from the reaction mixture, recycled Apaf-1 remained in the monomeric state. As for naive Apaf-1, approximately half of the protein precipitated if only cytochrome c was added. In contrast to ADP-bound Apaf-1, AppNHp-bound Apaf-1 only needed cytochrome c but no exogenous nucleoside triphosphate to assemble into apoptosomes.
FIGURE 3.
Apoptosome formation using recycled Apaf-1. Recycled ADP-bound Apaf-1 remains in the monomeric state if no cofactors are supplied (black curve) or if only cytochrome c is added (yellow curve). As naive Apaf-1, recycled ADP-bound Apaf-1 requires both exogenous ATP and cytochrome c for apoptosome formation (blue curve). On the other hand, recycled AppNHp-bound Apaf-1 only requires cytochrome c for apoptosome formation (pink curve). mAU, milliabsorbance units.
DISCUSSION
We have shown that recombinantly produced monomeric Apaf-1 purified from Sf21 insect cells contains ADP (Fig. 1B). Previous studies using Hi-5 insect cells (29) or E. coli (30) for heterologous protein production share our observation. However, a third study had identified dATP as the nucleotide bound to monomeric Apaf-1 purified from Sf21 insect cells (26). This discrepancy had been attributed to the use of different expression systems, expression conditions, and/or purification procedures or unidentified cellular factors stimulating an ATPase activity of Apaf-1 (29). Having the nucleotide-binding site of Apaf-1 occupied by a nucleoside triphosphate, as proposed by Kim et al. (26), would rely on complete lack of hydrolytic activity in the absence of cytochrome c. A low dATPase activity has been reported (20, 35, 36) but was interpreted as background (20). We have shown that autoinhibited Apaf-1 indeed possesses an intrinsic hydrolytic activity (Fig. 1, C and D), which is low but nevertheless certainly fast enough to hydrolyze all potentially bound triphosphate within the time span of a typical protein purification procedure. Therefore, the presence of a nucleoside diphosphate in monomeric Apaf-1 (Refs. 29, 30, 36 and this study) is not surprising.
It has been claimed that hydrolysis of a triphosphate by Apaf-1 is necessary to render Apaf-1 competent for apoptosome assembly (26, 35, 37). Our findings contradict a requirement for hydrolysis-derived energy to prime Apaf-1 for oligomerization. We demonstrate that ADP-bound Apaf-1 does not need a hydrolyzable nucleoside triphosphate to oligomerize but that a nonhydrolyzable ATP analog like AppNHp is sufficient to trigger apoptosome formation in the presence of cytochrome c (Fig. 2A).
HPLC analysis of apoptosomes assembled with different hydrolyzable nucleoside triphosphates showed that Apaf-1 lacks any hydrolytic activity when integrated in the apoptosomal complex (supplemental Fig. S2). This finding is supported by Kim et al. (26), who arrived at the same result using radiolabeled dATP. Together with the fact that nonhydrolyzable ATP analogs support apoptosome formation and subsequent caspase-9 activation (Refs. 20, 21 and our data), these data emphasize that hydrolysis does also not occur at a later stage of apoptosomal caspase-9 activation.
A necessary and sufficient condition to prime Apaf-1 for apoptosome assembly appears to be the presence of a γ-phosphate moiety in the nucleotide-binding site of monomeric Apaf-1. If monomeric Apaf-1 is loaded with AppNHp, it only requires cytochrome c to oligomerize, bypassing the need for nucleotide exchange required in the case of ADP-bound Apaf-1 (Fig. 3). It is commonly accepted that Apaf-1 has to undergo a large conformational change to proceed from the autoinhibited monomeric structure into the apoptosomal assembly. From cryo-EM studies, it is conceivable that these conformational changes include gross structural alterations in the NOD itself, because the crystallographic model of the N-terminal half of Apaf-1 (30) could not be fitted into the cryo-EM map of the apoptosome without major modifications in the subdomain arrangement (21). We propose that these conformational changes are brought about by the γ-phosphate exerting mechanical pressure on a structural element within the NOD.
As long as the supplied nucleotide provides a γ-phosphate, Apaf-1 appears to tolerate non-adenine purine bases in the process of apoptosome formation and caspase activation at least to some extent. This contradicts earlier reports stating that only adenine-containing triphosphates support caspase-9 activation and that GTP or dGTP, for instance, have no activation potential at all (25, 26, 28). We find that at high nucleotide concentrations and in the absence of a more preferred nucleotide even GTP can be bound by Apaf-1 and induce apoptosome assembly (Fig. 2A). Moreover, in our caspase activation assay, GTP displays a slight stimulatory action (Fig. 2B). The low level of caspase-9 stimulation induced by GTP in this assay reflects in our opinion a lower efficiency of GTP for apoptosome formation. The extent of caspase-9 stimulation seems to be correlated with the amount of apoptosome formed within the limited incubation time (10 min) in this experiment. Apaf-1 appears to have a much lower affinity to GTP than to adenine nucleoside triphosphates and therefore forms a lesser amount of apoptosome within a short time span. This notion is supported by the observation that equal amounts of apoptosome indeed are equally active regardless of the bound triphosphate (Fig. 2C).
Recently, an even less pronounced specificity for the base in the activating nucleotide was reported for human NALP1 (38), which is structurally and mechanistically related to Apaf-1 (39). Like Apaf-1, NALP1 possesses an NOD, which is flanked by an N-terminal effector domain and a C-terminal regulatory domain. Unless an external signal in the form of a pathogen-associated molecular pattern binds to the regulatory domain, NALP1 stays in an autoinhibited conformation. In the presence of a pathogen-associated molecular pattern and a nucleoside triphosphate, NALP1 undergoes a conformational change and oligomerizes into the so-called inflammasome (38, 40). Faustin et al. (38) reconstituted NALP1 inflammasomes in vitro and demonstrated that a broad range of purine- and pyrimidine-containing nucleoside triphosphates induces NALP1 assembly. In this light, it seems less surprising that Apaf-1 exhibits a certain promiscuity toward the base of the oligomerization-inducing nucleotide.
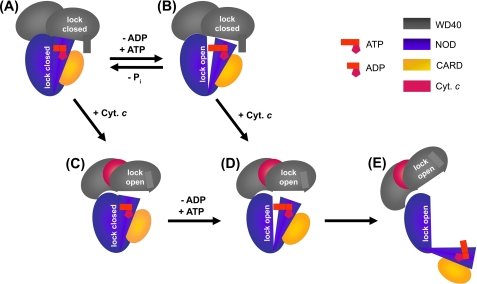
Based on our experimental data we propose a new model for the mechanism of apoptosome formation (Fig. 4). In our view, the autoinhibited conformation of Apaf-1 is secured by two different blocking fixtures as follows: one fastening the NOD at the WD40 domain and the other arising from interactions within the NOD with the ADP molecule as the organizing center (Fig. 4A). The latter is in line with the x-ray crystallographic model of the N-terminal half of Apaf-1 (30). Exchange of ADP for ATP releases the lock within the NOD (Fig. 4B). Binding of cytochrome c removes the interlock between the NOD and the WD40 domain (Fig. 4C). Only if both locks are open (Fig. 4D) is Apaf-1 ready to undergo the large conformational changes required for oligomerization (Fig. 4E). It is conceivable that cytochrome c facilitates the exchange of ADP for ATP and that the reaction preferably proceeds in the order A → C → D → E. In principle, a second route (via state B instead of state C) should be possible to yield assembly-competent Apaf-1. Because monomeric Apaf-1 hydrolyzes ATP at a low but steady level, at any given time a fraction of the Apaf-1 molecules in the cell will be loaded with ATP. This subpopulation would not need a further nucleotide exchange step but only binding of cytochrome c to be ready for the large conformational changes. Thus, apoptosome formation could either be initiated by binding of ATP to cytochrome c-bound Apaf-1 or by binding of cytochrome c to ATP-bound Apaf-1. A strict temporal order of these events therefore seems unlikely.
FIGURE 4.
Model of the mechanism of apoptosome formation. A, autoinhibited Apaf-1. Two blocking fixtures secure the closed state as follows: interactions between the WD40 propellers and the NOD (indicated by a gray rectangle protruding from one of the gray ovals) and interactions between subdomains of the NOD (indicated by tight arrangement of the blue oval and the blue triangle). B, Apaf-1 liganded with ATP. The presence of the γ-phosphate weakens subdomain interactions within the NOD (indicated by a gap between the blue oval and the blue triangle). Cyt. c, cytochrome c. C, Apaf-1 liganded with cytochrome c. Blocking interactions between WD40 propellers and the NOD are released. D, Apaf-1 liganded with cytochrome c and ATP. Both blocking fixtures are removed. E, open Apaf-1 ready to assemble into the apoptosomal heptamer.
Our results do not point toward a special need for dATP. At nucleotide concentrations higher than 10 μm, dATP and ATP are equally suited to induce apoptosome assembly as reflected in a similar level of caspase stimulation (Fig. 2D). Only at nucleotide concentrations of less than 10 μm do we find a moderately higher level of caspase activation if dATP is employed instead of ATP, which probably reflects a reported 10-fold higher affinity (20) of Apaf-1 for the deoxynucleotide. However, this difference seems to be more of academic than physiologic relevance. The ATP levels of healthy cells lie in the range of 2–5 mm, whereas the dATP concentration is about 3 orders of magnitude lower (Ref. 41 and references therein). In apoptotic cells, the ATP pool is decreased by a maximum of 60% (41), which corresponds to a minimal ATP concentration of several hundred micromolars in the cytosol at all times. Therefore, the nucleotide predominantly employed in vivo for apoptosome formation appears to be ATP.
Our findings well fit the recently proposed picture of Apaf-1 as a molecular switch with the “off” position corresponding to a monomeric ADP-bound state and the “on” position corresponding to an ATP-bound oligomeric state (42). Apaf-1 was classified to belong to the STAND (signal transduction ATPases with numerous domains) class of proteins (43), and our results confirm that Apaf-1 shares distinct mechanistic similarities with the STAND protein MalT (44), a bacterial transcription regulator. Neither protein needs ATP hydrolysis for oligomerization or downstream signaling. The ability of MalT to hydrolyze ATP serves to terminate signaling by transferring the protein back into the monomeric off state. Apaf-1 loses its hydrolytic competence upon oligomerization, which abrogates switching back and forth between active apoptosome and inactive monomer. This property appears to be reasonable considering the position of Apaf-1 in the death pathway and spotlights the release of cytochrome c as a decisive event for the progress of apoptosis. The hydrolytic competence of the monomer on the other hand is surprising because it is irrelevant for the progress of apoptosis. Whether this ability is just an inherited remnant or supports a recently proposed nonapoptotic function of Apaf-1 (45) cannot be answered with the present knowledge.
Supplementary Material
Acknowledgments
We thank Douglas R. Green for kindly providing the open reading frames of human Apaf-1-XL and human procaspase-9; Ernst Schonbrunn for valuable support in the early stage of the project; and Hans C. Mau for inspiring discussions.
This work was supported by European Union Grant 3D Repertoire LSHG-CT-2005-512028.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- CARD
- caspase recruitment domain
- AppNHp
- adenyl-5′-yl imidodiphosphate
- NOD
- nucleotide binding and oligomerization domain
- HPLC
- high pressure liquid chromatography
- AFC
- 7-amino-4-trifluoromethylcoumarin.
REFERENCES
- 1.Kumar S. (2007) Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 2.Taylor R. C., Cullen S. P., Martin S. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 3.Thornberry N. A., Lazebnik Y. (1998) Science 281, 1312–1316 [DOI] [PubMed] [Google Scholar]
- 4.de Almeida C. J., Linden R. (2005) Cell. Mol. Life Sci. 62, 1532–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 6.Opferman J. T. (2008) Cell Death Differ. 15, 234–242 [DOI] [PubMed] [Google Scholar]
- 7.Rathmell J. C., Thompson C. B. (2002) Cell 109, S97–S107 [DOI] [PubMed] [Google Scholar]
- 8.Friedlander R. M. (2003) N. Engl. J. Med. 348, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 9.Green D. R., Evan G. I. (2002) Cancer Cell 1, 19–30 [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 11.Nagata S. (2006) IUBMB Life 58, 358–362 [DOI] [PubMed] [Google Scholar]
- 12.Debatin K. M. (2004) Cancer Immunol. Immunother. 53, 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newmeyer D. D., Ferguson-Miller S. (2003) Cell 112, 481–490 [DOI] [PubMed] [Google Scholar]
- 14.Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 15.Kroemer G., Galluzzi L., Brenner C. (2007) Physiol. Rev. 87, 99–163 [DOI] [PubMed] [Google Scholar]
- 16.Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. (1997) Cell 90, 405–413 [DOI] [PubMed] [Google Scholar]
- 17.Hu Y., Ding L., Spencer D. M., Núñez G. (1998) J. Biol. Chem. 273, 33489–33494 [DOI] [PubMed] [Google Scholar]
- 18.Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. (1997) Cell 91, 479–489 [DOI] [PubMed] [Google Scholar]
- 19.Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. (1998) Mol. Cell 1, 949–957 [DOI] [PubMed] [Google Scholar]
- 20.Jiang X., Wang X. (2000) J. Biol. Chem. 275, 31199–31203 [DOI] [PubMed] [Google Scholar]
- 21.Yu X., Acehan D., Ménétret J. F., Booth C. R., Ludtke S. J., Riedl S. J., Shi Y., Wang X., Akey C. W. (2005) Structure 13, 1725–1735 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez J., Lazebnik Y. (1999) Genes Dev. 13, 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvesen G. S., Dixit V. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10964–10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Q., Park H. H., Chung J. Y., Lin S. C., Lo Y. C., da Graca L. S., Jiang X., Wu H. (2006) Mol. Cell 22, 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genini D., Budihardjo I., Plunkett W., Wang X., Carrera C. J., Cottam H. B., Carson D. A., Leoni L. M. (2000) J. Biol. Chem. 275, 29–34 [DOI] [PubMed] [Google Scholar]
- 26.Kim H. E., Du F., Fang M., Wang X. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leoni L. M., Chao Q., Cottam H. B., Genini D., Rosenbach M., Carrera C. J., Budihardjo I., Wang X., Carson D. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9567–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 29.Bao Q., Lu W., Rabinowitz J. D., Shi Y. (2007) Mol. Cell 25, 181–192 [DOI] [PubMed] [Google Scholar]
- 30.Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005) Nature 434, 926–933 [DOI] [PubMed] [Google Scholar]
- 31.John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., Goody R. S. (1990) Biochemistry 29, 6058–6065 [DOI] [PubMed] [Google Scholar]
- 32.Shi Y. (2008) Methods Enzymol. 442, 141–156 [DOI] [PubMed] [Google Scholar]
- 33.Bao Q., Shi Y. (2007) Cell Death Differ. 14, 56–65 [DOI] [PubMed] [Google Scholar]
- 34.Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. (1999) J. Biol. Chem. 274, 8359–8362 [DOI] [PubMed] [Google Scholar]
- 35.Hu Y., Benedict M. A., Ding L., Núñez G. (1999) EMBO J. 18, 3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou H., Li Y., Liu X., Wang X. (1999) J. Biol. Chem. 274, 11549–11556 [DOI] [PubMed] [Google Scholar]
- 37.Saleh A., Srinivasula S. M., Acharya S., Fishel R., Alnemri E. S. (1999) J. Biol. Chem. 274, 17941–17945 [DOI] [PubMed] [Google Scholar]
- 38.Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 39.Proell M., Riedl S. J., Fritz J. H., Rojas A. M., Schwarzenbacher R. (2008) PLoS ONE 3, e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tschopp J., Martinon F., Burns K. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 95– 104 [DOI] [PubMed] [Google Scholar]
- 41.Chandra D., Bratton S. B., Person M. D., Tian Y., Martin A. G., Ayres M., Fearnhead H. O., Gandhi V., Tang D. G. (2006) Cell 125, 1333–1346 [DOI] [PubMed] [Google Scholar]
- 42.Danot O., Marquenet E., Vidal-Ingigliardi D., Richet E. (2009) Structure 17, 172–182 [DOI] [PubMed] [Google Scholar]
- 43.Leipe D. D., Koonin E. V., Aravind L. (2004) J. Mol. Biol. 343, 1–28 [DOI] [PubMed] [Google Scholar]
- 44.Marquenet E., Richet E. (2007) Mol. Cell 28, 187–199 [DOI] [PubMed] [Google Scholar]
- 45.Zermati Y., Mouhamad S., Stergiou L., Besse B., Galluzzi L., Boehrer S., Pauleau A. L., Rosselli F., D'Amelio M., Amendola R., Castedo M., Hengartner M., Soria J. C., Cecconi F., Kroemer G. (2007) Mol. Cell 28, 624–637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.