Abstract
PGRL1 RNA and protein levels are increased in iron-deficient Chlamydomonas reinhardtii cells. In an RNAi strain, which accumulates lower PGRL1 levels in both iron-replete and -starved conditions, the photosynthetic electron transfer rate is decreased, respiratory capacity in iron-sufficient conditions is increased, and the efficiency of cyclic electron transfer under iron-deprivation is diminished. Pgrl1-kd cells exhibit iron deficiency symptoms at higher iron concentrations than wild-type cells, although the cells are not more depleted in cellular iron relative to wild-type cells as measured by mass spectrometry. Thiol-trapping experiments indicate iron-dependent and redox-induced conformational changes in PGRL1 that may provide a link between iron metabolism and the partitioning of photosynthetic electron transfer between linear and cyclic flow. We propose, therefore, that PGRL1 in C. reinhardtii may possess a dual function in the chloroplast; that is, iron sensing and modulation of electron transfer.
Introduction
Iron is used as a cofactor in numerous biochemical pathways. It is present in heme- and iron-sulfur proteins that are required for energy-transducing pathways like respiration and photosynthesis. Hence in cyanobacteria and plants, photosynthetic membranes are severely impacted by iron deficiency. Low iron availability causes changes in photosynthetic capacity and the composition of thylakoid membranes (1, 2). Cyanobacteria respond to iron deficiency by decreasing the ratio of photosystem I to photosystem II (PSI/PSII)3 and by degradation of light harvesting phycobilisomes (3). The PSI complex is a specific target of iron starvation, as it contains 12 iron atoms per monomer. In some marine Synechococcus (e.g. WH 8102), electron rerouting to the plastoquinone terminal oxidase is enhanced to counterbalance over-reduction of the plastoquinone pool (4). As an additional response, the “iron-stress-induced” gene isiA is expressed. The isiA protein has significant sequence similarity with CP43, a chlorophyll a-binding protein of PSII (PSII (5, 6)), and forms a ring of 18 molecules around a PSI trimeric reaction center, as revealed by electron microscopy (7, 8). Likewise a light-harvesting protein of the chlorophyll a/b type that associates with PSI is induced by iron deficiency in the green alga Dunaliella (9). In Chlamydomonas reinhardtii, iron deficiency leads not only to a pronounced degradation of PSI but also to a remodeling of the PSI-associated light-harvesting antenna (LHCI), which precedes iron limitation (10). Interestingly, a similar phenomenon is observed in red algae (11, 12). Long term adaptation to iron deficiency causes constitutive differences in the photosynthetic architecture as demonstrated for coastal and oceanic diatoms (13). Similar phenomena have also been proposed in marine cyanobacteria (4) and green algae, like Ostreococcus (14).
Other metabolic changes also occur in iron-starved cells. Transcriptomics and functional analyses revealed that the iron-deficiency response in the diatom Phaeodactylum tricornutum involves alterations in chlorophyll biosynthesis and pigment metabolism, removal of excess electrons by mitochondrial alternative oxidase, and an increase in capacity of non-photochemical quenching (15). Similar responses in regard to chlorophyll accumulation (10) were also observed for C. reinhardtii. In acetate-grown iron-deficient C. reinhardtii cells, removal of excess electrons is probably facilitated by the mitochondrial respiratory chain, which is significantly more resistant to iron starvation than is the photosynthetic electron transfer chain (16). Previously, the application of SILAC (stable isotopic labeling of amino acids in cell culture) in conjunction with quantitative proteomics indicated that several stress-related proteins as well as proteins with so far unknown function were induced under iron deprivation (18). Among the induced proteins are a 2-Cys peroxiredoxin, which functions in the detoxification of reactive oxygen species, and a stress-related light harvesting protein, LhcSR3, which is involved in regulation of photosynthesis and required particularly for the qE component of non-photochemical quenching in C. reinhardtii).4 Another interesting candidate protein that was found to be up-regulated in response to iron deficiency is PGRL1 (proton gradient regulation, earlier indicated as TEF3 or C_420064), which has an iron/zinc binding motif (16). Phylogenomic analysis (18) sorted the PGRL1 gene to a class of genes called the “Plastid Cut,” which contains 90 proteins that are absolutely conserved in all organisms with a photosynthetic plastid, from diatoms to vascular plants. The Arabidopsis thaliana orthologs of PGRL1, PGRL1a and PGRL1b, are likely to be involved in the switch between cyclic (CEF) and linear photosynthetic electron transfer (LEF) (19).
In LEF, ferredoxin is reduced by PSI and donates its electron to ferredoxin-NADP-oxidoreductase, generating NADPH, which is consumed along with ATP in the Calvin-Benson-Bassham cycle (hereafter called Calvin cycle) and other biosynthetic reactions (20). Conversely, both ferredoxin and NADPH have the capacity to independently return electrons to the plastoquinone pool in CEF, leading to ATP synthesis without net distribution of reducing equivalents (see Ref. 21 for further discussion). In plants and algae, the commutation between LEF and CEF is thought to be modulated primarily by the redox poise of the stromal electron carriers (e.g. Ref. 22). However, the finding that PGR5 (23, 24) and its partner, PGRL1, affect the efficiency of cyclic electron flow has led to the hypothesis that the PGR5-PGRL1 complex may also participate in regulation of LEF versus CEF through a mechanism that has not been elucidated yet (e.g. Ref. 25).
In C. reinhardtii, CEF is thought to have a relevant contribution to photosynthesis, particularly under reducing and/or stress conditions (for review, see Ref. 26). Thus, we investigated the function of PGRL1 in C. reinhardtii under iron-sufficient and iron-deficient growth conditions using a knockdown strategy. Our data suggest that PGRL1 participates in CEF and also plays a role in modulating acclimation to iron nutrition. This likely includes (i) binding, sensing, or distribution of this iron, (ii) remodeling of photosynthetic electron transfer by changing reaction center stoichiometries, and (iii) partitioning of electron flow between LEF, CEF, and respiration to adapt the ATP synthesis capacity to the overall cellular demand.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Cells were grown in the light (60 μE m−2s−1) in Tris acetate-phosphate (TAP) medium (27) at 25 °C and shaken at 120 rpm. The ΔpsaB (28) and the nac2 (29) mutant strains were kind gifts from K. Redding (Arizona State University, Tempe, AZ) and J. Nickelsen, (Ludwig-Maximilians University, Muenchen, Germany), respectively. The arginine auxotrophic strain 388 was a kind gift of T. Happe (Ruhr Universtity, Bochum, Germany). For growth of the strain 388, medium was supplemented with 100 μg/ml arginine. For experiments involving transition from iron-replete to iron-depleted conditions, cells were maintained in standard TAP medium and transferred to iron-free TAP as described previously (10). For experiments involving growth in different iron concentrations, cells were transferred from TAP plates into TAP medium at the indicated iron concentrations, and starter cultures were diluted at least 2–3× before the onset of each experiment. Initial cell density was set at 1 × 106 cells/ml, and cultures were allowed one doubling and subsequently collected or used for physiological measurements.
Plasmid Construction
Total RNA of C. reinhardtii cells was isolated with TRI® reagent (Sigma) according to the supplier's manual. The subsequent cDNA synthesis was performed with the reverse transcription system (Promega) according to the manual.
For generation of the inverted repeat PGRL1 cassette, a 694-bp fragment corresponding to the coding region of PGRL1 was amplified by using cDNA isolated from iron-deficient cells as template and the primers 42-FORS-P (5′-aactgcaggttccttgcggcgatgac-3′, the underline indicates the PstI site) and 42-REV-E (5′-cggaattccagcttctcctgcttgacct-3′, the underline indicates the EcoRI site), adding the respective restriction sites. A longer 1000-bp fragment, additionally containing 306 bp of adjacent coding sequence that functions as a spacer in the inverted repeat construct, was amplified using the primer 42-FORL-P (5′-aactgcagctgcgcataaggtcttccat-3′, the underline indicates the PstI) and 42-REV-E (5′-cggaattccagcttctcctgcttgacct-3′, the underline indicates the EcoRI site). Both fragments were cloned in antisense orientation into pBluescriptSK (Stratagene) using the added restriction sites. The resulting inverted repeat cassette was excised with EcoRI and cloned into the unique EcoRI site of the RNAi vector pNE537 (30). Design and construction of the artificial microRNA vector was done as described in Molnar et al. (31). The 90-mer oligonucleotides amiFor_TEF3UTR (5′-ctagtCGTCGCGCGTTTTTGTCAATAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTATTGACAAAAACGCGCGACGg-3′) and amiRev_TEF3UTR (5′-ctagcCGTCGCGCGTTTTTGTCAATAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTATTGACAAAAACGCGCGACGa-3′), which target the 3′-untranslated region of PGRL1, were annealed and cloned into the SpeI site of the pChlamyRNA2 vector, giving rise to the vector pHUN5.
Nuclear Transformation and Selection
Biolistic transformation of the RNAi construct into a cell wall containing the wild-type strain and the subsequent screening was carried out as described in Naumann et al. (32). The cell wall-deficient arginine auxotroph strain 388 was transformed with the amiRNA construct pHUN5 using glass beads according to (33). Subsequent selection of the mutants was done on TAP plates screening for complementation of the arginine auxotrophy. Several mutants (two RNAi mutant strains pgrl1–28 and pgrl1–23 and an artificial micro-RNA mutant, pHUN5–9) were isolated. A PGRL1-depleted RNAi strain was thoroughly characterized and is presented in Figs. 1–6. An independent amiRNA mutant, which also possesses diminished levels of PGRL1, is also more prone to iron deficiency than its respective wild type (supplemental Fig. 1), underscoring the importance of PGRL1 for the adaptation to iron deficiency (see below).
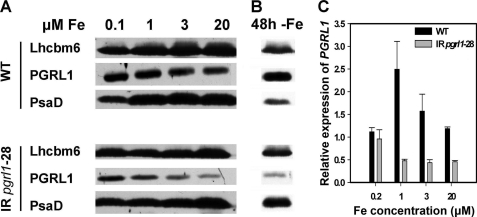
FIGURE 1.
PGRL1 expression is induced under iron deficiency in C. reinhardtii and is down-regulated in pgrl1–28 RNAi mutant strain in both iron-replete and -starved conditions. A, 80 μg of whole cell protein extracts from wild-type and pgrl1–28 mutant cells grown in TAP medium containing 0.1, 1, 3, and 20 μm iron were separated on a 13% SDS-PAGE and PGRL1, PsaD, and Lhcbm6 abundance were analyzed by immunoblot. Lhcbm6 was used as loading control. B, the same analyses were performed in wild-type and mutant cells grown for 48 h in iron-replete and iron-depleted TAP medium. C, PGRL1 expression was analyzed by quantitative real-time PCR. RNA was isolated from wild-type and pgrl1–28 mutant strains grown in TAP medium containing the indicated iron concentrations. Gene expression was normalized to CBLP expression. Relative expression was normalized to a mix of all samples and calculated by the 2−ΔΔCT method (17). Black bars indicate PGRL1 expression in wild type, and gray bars PGRL1 indicate expression in pgrl1. The values are the means ± S.D. of three independent experiments.
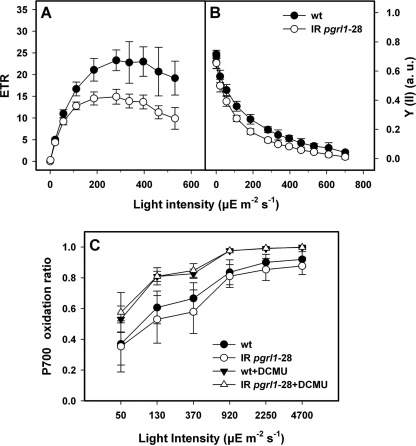
FIGURE 2.
In iron-replete conditions the pgrl1 RNAi mutant shows decreased photosynthetic electron flow as compared with the wild type but possesses an unchanged efficiency to perform cyclic electron flow. Irradiance dependence of relative electron transport through PSII (ETR, A) of quantum yield of PSII (Y(II); B) and of P700+ oxidation ratio (C) for wild-type and mutant cells. The values are the means ± S.D. of six independent experiments. a.u., arbitrary units.
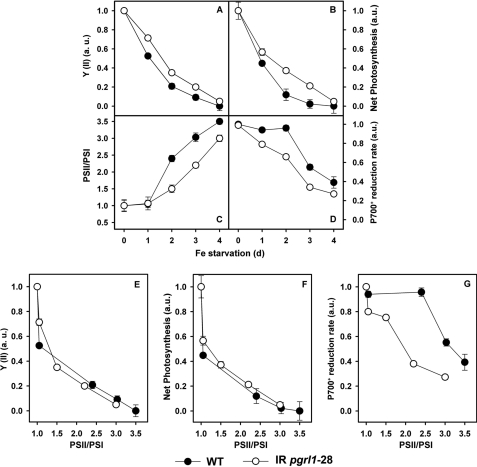
FIGURE 3.
PGRL1 is required for cyclic electron flow under iron-limited conditions. Wild-type and pgrl1 RNAi strains were grown with and without iron TAP, and quantum yield of PSII (Y(II); A), net photosynthesis (B), the PSII/PSI ratio (C), and P700 reduction rate (D) were measured over a 4-day period. The changes in these photosynthetic parameters are also presented as a function of PSII/PSI ratio (E–G). a.u., arbitrary units.
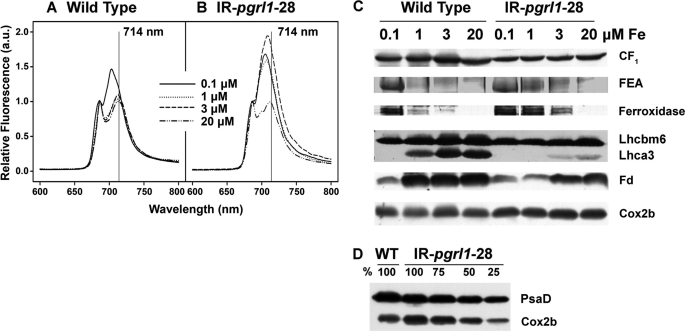
FIGURE 4.
The pgrl1 RNAi mutant strain shows a different response to iron as compared with the wild type. Cells were grown in TAP medium containing 0.1, 1, 3, and 20 μm iron to a cell density of 2 × 106 cells/ml. A, and B, shown are low temperature fluorescence emission spectra of wild-type and mutant cells, respectively. C, 80 μg of whole cell protein extracts from wild-type and mutant cells were fractionated on a 13% SDS-PAGE, and CF1, ferroxidase, Lhcbm6, Lhca3, and ferredoxin abundance were analyzed by immunoblot. Lhcbm6 and CF1 were used as loading controls. FEA1 abundance was analyzed by Coomassie Blue staining. D, Cox2b abundance in wild-type and mutant cells were grown in iron-replete TAP. 100% corresponds to 80 μg of protein. PsaD was used as loading control. a.u., arbitrary units.
FIGURE 5.
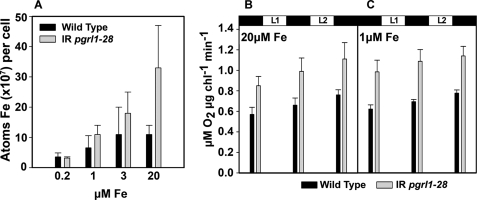
The pgrl1 RNAi mutant strain has an increased cellular iron content and enhanced respiration as compared with the wild-type. A, cells were grown in TAP medium supplemented with iron to the indicated concentration, and total iron content was measured by inductively coupled plasma-mass spectrometry. Means ± S.D. of three independent experiments are shown. Oxygen uptake in wild-type and mutant cells were grown in TAP medium containing 1 (C) and 20 μm (B) iron is shown. Measurements were made after 15 min of dark adaptation and after 5 min of illumination at 600 (L1) and 1200 (L2) μE m−2 s−1. The values are the means ± S.D. of three independent experiments.
FIGURE 6.
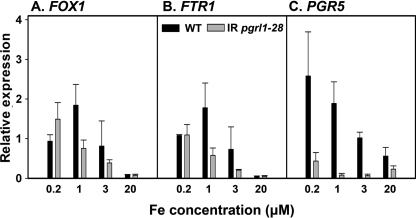
Expression of FOX1, FTR1, and PGR5 in response to iron nutrition. RNA was isolated from wild-type and pgrl1 knockdown strain grown under the indicated concentrations of iron. Gene expression was analyzed by quantitative real-time PCR. Gene expression was normalized to CBLP expression. Relative expression was normalized to a mix of all samples and calculated by the 2−ΔΔT method (17) for FOX1 (A), FTR1 (B), and PGR5 (C). Black bars indicate gene expression in wild-type, and gray bars indicate gene expression in pgrl1. The values are the means ± S.D. of three independent experiments.
Isolation of Thylakoids and the PSI Complexes
Thylakoids were isolated as described in Hippler et al. (34), breaking the cell wall-containing cells by passing them two times through a self-made bio-nebulizer at a pressure of 35 p.s.i. The thylakoids were solubilized with β-dodecyl-maltoside and loaded on a linear sucrose gradient as described in Hippler et al. (34).
Protein Analysis
Whole cells, thylakoids, or sucrose gradient fractions were analyzed by SDS-PAGE according to Laemmli (35). The gels were either stained with Coomassie Blue or blotted on nitrocellulose membrane. After incubation with specific antibodies, the signal was visualized by enhanced chemical luminescence (ECL). FOX1, Fd, and Cox2b antibodies, all used with a 1:1000 dilution, were purchased from Agrisera (Vännäs, Sweden). All other antibodies are described in Moseley et al. (10) and Naumann et al. (32).
Quantitative Real-time PCR
Total RNA was extracted from C. reinhardtii cells, and quantitative real-time PCR was performed as in Allen et al. (36). The data are presented as the -fold change in mRNA abundance and normalized to the endogenous reference gene (CBLP), relative to a mixture of all the samples. The abundance of CBLP did not change under the conditions tested in this work.
Measurement of Iron Content
Cells were grown in the indicated concentrations of iron to a cell density of 2 × 106 cells/ml. Cells (1 × 108) were collected, washed, and digested as in Allen et al. (36). Digested cell paste was diluted with 9 ml of deionized water before measurement. Total iron content was measured by inductively coupled plasma-mass spectrometry (Agilent 7500 ICP-MS, detection limit 100 ppb) in helium mode at the University of California, Los Angeles Molecular Instrumentation Center using the standard addition method. Manganese, magnesium, copper, and zinc were also measured (data not shown).
Measuring the in Vivo Thiol Redox Status of PGRL1
Whole cells from iron-replete or iron-depleted cultures were resuspended in solution A (0.1% formic acid, 5% acetonitrile). Samples containing 60 μg of protein were incubated with 100 mm DTT or 100 mm H2O2 at 37 °C for 20 min in solution A and were analyzed by non-reducing SDS-PAGE. Control samples did not undergo the 37 °C incubation.
For 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid (AMS) modification, samples were precipitated with 10% ice-cold trichloroacetic acid after their oxidation or reduction and centrifuged at 16,000 × g for 15 min. The pellet was washed with 10% trichloroacetic acid and then acetone, dried, and resuspended in freshly prepared solution B (solution B (1% SDS, 50 mm Tris-HCl, pH 7.5) containing 20 mm AMS (Invitrogen). The reaction mixture was incubated at room temperature for 30 min. Upon completion of the reaction, 2-mercaptoethanol (50 mm final concentration) was added to remove excess thiol-reactive reagent, and samples were analyzed by SDS-PAGE. Control samples containing no DTT or H2O2 were not incubated at 37 °C but were directly trichloroacetic acid-precipitated.
Alternatively, samples were incubated in solution B (described above) in the presence of ferrozine at room temperature for 20 min and were allowed to react with AMS without trichloroacetic acid precipitation. Control samples containing no ferrozine were directly AMS-modified without room temperature incubation.
Low Temperature (77 K) Fluorescence Emission Spectroscopy
Whole cells (2 × 106 cells/ml) were resuspended in 60% glycerol, 10 mm HEPES, pH 7.5, and frozen in liquid nitrogen. Low temperature fluorescence emission spectra were recorded with the FP-6500 spectrofluorometer (Jasco, Gross-Umstadt, Germany). The obtained data were normalized to the photosystem II emission peak at 685 nm.
Spectroscopic Measurements
For spectroscopic measurements, algal samples were resuspended in a HEPES-KOH 20 mm buffer, pH 7.2, with the addition of 20% Ficoll (w/v) to avoid cell sedimentation during measurements. Measurements were performed at room temperature using a JTS spectrophotometer (Biologic). Continuous light was provided by a red source (630 nm), which was switched off transiently while measuring P700 absorption changes at 705 nm, as required to estimate cyclic electron flow. PSI and PSII content was estimated spectroscopically from changes in the amplitude of the fast phase (100 μs) of the electrochromic shift signal (at 520–545 nm) upon excitation with a saturating laser flash. The electrochromic shift spectral change follows linearly the number of light-induced charge separations within the reaction centers (37). Thus, PSII contribution can be calculated from the decrease in the signal amplitude upon the addition of DCMU (20 μm) and hydroxylamine (1 mm) that irreversibly block PSII charge separation once the sample has been preilluminated. Conversely, PSI was estimated as the fraction of the signal that was insensitive to these inhibitors.
Fluorescence changes, as required to assess the Y(II) parameter, were measured using a home-built fluorometer, where excitation was provided by a green LED source (520 nm), and fluorescence was detected in the near IR region. Alternatively, fluorescence was measured using a Maxi-Imaging PAM chlorophyll fluorometer (Heinz Walz GmbH, Effeltrich, Germany). Samples were set at a density of 2 × 106 cells/ml in TAP and were dark-adapted for 15 min before each measurement. The effective photochemical quantum yield of photosystem II was measured as PSII yield (Y(II) = (Fm′ − F)/Fm′), where Fm′ is the maximum fluorescence emission level in the presence of actinic light and F is the steady-state level of fluorescence emission. Measurement of photosynthetic electron transfer rates (ETR) were based on fluorescence analysis (ETR = 0.5 × yield × 0.84 × PAR μE m−2 s−1), where PAR is the photosynthetically active radiation.
Oxygen Exchanges
Oxygen uptake and evolution rates were measured using an OxyLab Hansatech Clark-type oxygen electrode (Norfolk, UK) in the presence of 5 mm NaHCO3 at 20 °C. The electrode was calibrated using dithionite. Whole cells were centrifuged for 4 min at 4000 rpm at room temperature. The supernatant was removed, and the cells were resuspended in TAP medium in the indicated iron concentrations to a 20 μg/ml chlorophyll concentration suspension. Measurements were made in 1.5 ml of sample volume after 15 min of dark adaptation and after 5 min of illumination at 600 and 1200 μE m−2 s−1. Illumination was provided by light red LEDs centered on 650 nm.
RESULTS
The Cellular PGRL1 Content Is Controlled by Iron Availability in C. reinhardtii
PGRL1 protein and RNA abundance is increased under iron-deficient conditions in wild-type C. reinhardtii cells (16) (Fig. 1), suggesting a role of this protein in acclimation to nutrient starvation. To understand the function of PGRL1, an RNAi approach was taken. Immunoblot analysis showed that pgrl1–28 contained less PGRL1 protein as compared with the wild-type cells in iron-replete conditions (20 μm iron) (Fig. 1A, upper and lower panel). In WT cells, PGRL1 abundance increased when cells were grown on replete (20 μm iron) compared with iron-limited (0.1 μm iron) conditions. A less pronounced response was seen in the mutant. To assess whether the lower PGRL1 abundance in the mutant was due to decreased gene expression, we analyzed PGRL1 RNA abundance by real time PCR. RNA was isolated from mutant and wild-type cells grown to 2 × 106 cells/ml at 20, 3, 1, or 0.2 μm iron. Fig. 1C shows that PGRL1 expression in the wild-type cell is iron-dependent with a maximum expression at 1 μm iron. Expression is reduced in the mutant at 20, 3, and 1 μm iron compared with the wild-type cell. Surprisingly, PGRL1 mRNA abundance is similar as shown by the comparison between wild-type and pgrl1–28 at 0.2 μm iron, although the protein abundance in pgrl1–28 is reduced.
In parallel to the effect on PGRL1, a significant decrease in PSI was seen at 0.1 μm iron in the wild-type cells as compared with iron-sufficient conditions. This was expected because this iron concentration is sufficient to trigger the iron deficiency acclimation process in C. reinhardtii. A similar effect was observed in the mutant. However, the decrease of PSI in the WT was not as pronounced as in pgrl1–28, when the PsaD levels were normalized to those of Lhcbm6, a protein that is insensitive to iron starvation (16), and therefore used as a loading control. This result was confirmed in a parallel experiment where mutant and wild-type cells were collected from iron-sufficient conditions as well as after 48 h of iron deficiency, fractionated by SDS-PAGE, and further investigated by immunoblot analyses (Fig. 1B). We also observed that the level of cytochrome f, a marker for the presence of the cytochrome b6/f complex, was similar in WT and pgrl1–28 under iron-sufficient conditions as assessed by immunoblotting. Cytochrome f levels decrease upon a shift to iron deficiency, where the abundance of cytochrome f is maintained longer in the mutant as compared with WT (data not shown).
Acclimation of Photosynthesis to Iron Is Modified in the pgrl1-kd Mutant
To address the functional consequences of decreased pgrl1 expression, photosynthetic parameters were recorded for wild-type and mutant cells in iron-sufficient and -deficient conditions (Figs. 2 and 3, respectively). Dark-adapted WT and pgrl1–28 cells showed a similar PSII quantum efficiency in iron-replete conditions (Figs. 2B and 3A). However, the PSII quantum efficiency (Y(II)) of the knockdown line decreases faster with increasing light intensity as compared with the WT (Fig. 2B), resulting in 2-fold smaller ETRs at saturating light (Fig. 2A). This indicates that the electron flow chain was more reduced in the mutant, likely because of kinetic limitation by a dark reaction. To further elucidate this phenomenon, we measured the redox changes of P700, the primary electron donor of PSI, in the presence or absence of DCMU. This parameter provides information on the balance between PSII- and PSI-driven electron transfer and can be used to assess the capacity of the photosynthetic apparatus to perform CEF when measured under conditions where PSI turnover is enhanced with respect to PSII (i.e. in the presence of a PSII inhibitor (38)).
In the absence of DCMU, very similar P700+ levels were observed in both strains as a function of the light intensity. This is in stark contrast to previous reports from research with A. thaliana showing a large difference in the P700+ ratios in wild-type cells and pgr5 or pgrl1-deficient mutants (19, 23). Moreover, no differences were seen in the presence of DCMU (Fig. 2C). This suggests that the efficiency of CEF was unchanged in prgl1–28 cells in iron-replete conditions.
Because PGRL1 levels are increased 2–3-fold by iron deprivation (Fig. 1), we decided to explore its possible role in acclimation of photosynthesis to a changing iron environment. We, thus, shifted C. reinhardtii WT and pgrl1–28 cells from an iron-sufficient to a medium that did not contain iron and measured changes in photosynthetic parameters during the first 4 days of acclimation to reduced iron. Major changes were seen in PSII quantum efficiency (Y(II)), net photosynthesis, PSII/PSI ratios, and CEF (Fig. 3, A–D). The PSII/PSI ratio (estimated from the contribution of PSII and PSI photochemistry to the generation of the light driven electrochromic shift of membrane pigments (37)) remained constant during the first day of iron starvation and then increased progressively in both genotypes, as expected because PSII complexes are more stable than PSI under iron deficiency. The changes in PSII/PSI ratio were more pronounced in WT cells. In contrast, a very large decrease in the quantum efficiency of PSII and net photosynthesis was seen already at day one. This was followed by a smaller, more progressive change, which again was more pronounced in WT cells. The P700+ reduction rate in the presence of DCMU, which is indicative of CEF, also declined during iron starvation. However, this parameter was more affected in pgrl1–28 than in the WT, where no significant changes where seen until day 2.
When changes in the different photosynthetic parameters (net photosynthesis, CEF, Y(II)) were plotted as a function of changes in the PSII/PSI ratio, i.e. of the iron-induced degradation of PSI, all the photosynthetic parameters related to linear electron flow showed a marked initial decrease, which was not related to PSI decrease (Fig. 3, E–G). Then, a slower decline was seen, which follows the changes in the PSII/PSI ratio with a nearly identical slope in both genotypes. Conversely, the P700+ reduction rate (i.e. CEF) remained almost unchanged in WT cells until a PSII/PSI ratio of 2.5 was reached. At that point, CEF decreases along with PSI with a much steeper slope than the one observed in the case of Y(II) and net photosynthesis. In pgrl1–28 cells, CEF declined almost linearly with PSI, showing a similar slope as the one measured in the case of Y(II) and net photosynthesis in the whole range of iron acclimation explored in this experiment. These results indicate that PGRL1 is required for CEF under iron-limited conditions, i.e. when PSI levels are reduced.
This view was further confirmed by measurements of the same photosynthetic parameters (net photosynthesis, PSII quantum efficiency, and P700 reduction) in cells grown at 20, 1, and 0.1 μm iron (supplemental Figs. 2). Indeed, the P700+ reduction rate was about 1.5 times slower in pgrl1 than in the WT at 0.1 μm iron, whereas the decline in net photosynthesis and PSII quantum efficiency at 0.1 μm iron were almost identical.
Besides affecting the PSI cellular content, iron starvation is known to modify its light-harvesting apparatus. In particular, detachment of PSI antenna from the core complex has been reported, which precedes the degradation of PSI (10). Thus, we decided to investigate the role of PGRL1 in iron deficiency dependent antenna remodeling. Wild-type and pgrl1–28 cells were grown at 20, 3, 1, and 0.1 μm iron, and the status of PSI antenna complexes was investigated by measuring low temperature fluorescence emission spectra as well as Coomassie Blue staining and immunoblot analyses of total cellular proteins fractionated by SDS-PAGE (Fig. 4). In iron-replete conditions (20 μm iron), WT and pgrl1–28 cells showed fluorescence emission maxima of 686 and 713 nm. The first fluorescence peak is associated with PSII emission, whereas the second one is ascribable to the presence of energetically coupled PSI-LHCI supercomplex. Upon acclimation to iron-starved conditions (0.1 μm iron), the PSI emission peak was blue-shifted to 703 and 705 nm in WT and mutant cells, respectively, confirming that PSI and LHCI have become functionally uncoupled in iron-deficient conditions (10). However, pgrl1–28 displayed modified fluorescence properties already at 3 μm iron. Specifically, we note an increase in the PSI fluorescence emission band and a small blue shift to 709 nm. In contrast, the fluorescence emission peaks of PSI-LHCI in WT cells remains unaltered at iron concentrations of 1, 3, and 20 μm iron, pointing to the fact that the knockdown strain pgrl1–28 has a higher sensitivity to iron limitation than does the wild type. Overall, it appears that pgrl1–28 cells respond to limited iron starvation by maintaining their PSI and increasing LHCIs uncoupling and degradation. This interpretation is supported by biochemical analysis, showing that the amount of Lhca3 was decreased 5–10-fold in pgrl1–28 compared with wild type. At 1 μm iron, Lhca3 was not detectable in the RNAi strain pgrl1–28, whereas it was still present in the WT. The larger LHCI degradation seen in pgrl1–28 would, therefore, represent a typical strategy to photoprotect PSI, which is not degraded under limited iron starvation in this strain.
Cellular Iron Metabolism Is Modified in pgrl1 Mutants
The modified response of the photosynthetic apparatus to iron starvation in pgrl1–28 is paralleled by other changes in cellular energetic metabolism. For example, the Cox2b protein, a typical marker for respiratory electron protein complexes, was more abundant in the mutant as compared with the WT (Fig. 4, C and D). This suggests that the respiratory capacity is increased in the mutant under iron-sufficient conditions. Consistent with this, oxygen consumption was ∼1.5 times higher in pgrl1–28 than in the wild-type cells in iron-sufficient and -deficient conditions (Fig. 5, B and C). We found that Cox2b levels did not decline in both wild-type and mutant cells under iron deficiency, consistent with the notion that the mitochondrial respiratory chain is more resistant to iron starvation than is the photosynthetic electron transport chain in acetate-grown C. reinhardtii (16).
The increase in respiratory electron transfer complexes should translate into a higher demand of iron in the mutant when compared the WT. Measurements of the iron content of the RNAi pgrl1–28 mutant and wild-type cells using inductively coupled plasma-mass spectrometry are consistent with this idea. Fig. 5A shows that the amount of iron per cell in the mutant is significantly higher than that of the wild type. This difference can be partially explained by the fact that the pgrl1–28 mutant cells are slightly larger than WT cells (especially in replete iron conditions, see supplemental Fig. 3). Still, the differences measured in iron-replete conditions support the notion that the overall iron metabolism is modified in the mutant. In line with this conclusion, although the iron content per cell declines in both WT and pgrl1–28 cells in both iron-deficient (3 and 1 μm iron) and iron-limited (0.2 μm iron) conditions, the effect was more pronounced in the mutant. Its iron content is still higher than that of the wild type at 1 μm iron, but at 0.2 μm iron the difference between wild type and pgrl1–28 is no longer evident.
The Iron Uptake Machinery Is Up-regulated in pgrl1–28
Consistent with the notion that iron cellular content is higher in pgrl1 cells, higher FEA1 and ferroxidase were found in the mutant (Fig. 4C). FEA1 and ferroxidase are part of the high affinity iron uptake system in C. reinhardtii and are induced by low iron availability (36, 39). FEA is enormously induced at the protein level after the onset of iron deficiency. The FEA protein can be seen in a Coomassie Blue-stained SDS-PAGE after fractionation of mutant cells grown at 1 μm iron, whereas an iron concentration of 0.1 μm iron is needed to clearly observe the protein in the wild type. In contrast, the levels of ferredoxin, the electron acceptor of PSI, were largely decreased by iron starvation (40). Again, differences were seen between the two genotypes; although Fd was already diminished at 1 μm iron in the mutant, this protein only decreased when cells when were grown in 0.1 μm iron in the wild type. This finding further supports the conclusion that the mutant is iron-deficient at 1 μm iron, whereas the wild type is not.
The different response to iron in WT and pgrl1–28 cells was confirmed while looking at the pattern of expression of genes encoding for components of the C. reinhardtii iron-uptake system in WT and mutant cells (Fig. 6). RNA was isolated from mutant and wild-type cells grown to 2 × 106 cells/ml at 20, 3, 1, or 0.2 μm iron. The mRNA expression levels for ferroxidase FOX1 and the Fe3+-specific transporter FTR1 clearly increase from 20 to 0.2 μm iron with a maximal expression at 1 and 0.2 μm iron for the wild type and the mutant, respectively (Fig. 6, A and B). Notably, the abundance of the ferroxidase is already visibly induced at 1 μm iron in the RNAi line (Fig. 4C), indicating that there is maybe a post-transcriptional component to the regulation of its abundance in addition to transcriptional control (36). This can also be seen for the wild type where the mRNA abundance is maximal at 1 μm iron, whereas the FOX1 protein is primary detectable at 0.1 μm iron (Fig. 4C).
Because PGRL1 is part of a complex including PGR5, we also tested the levels of the PGR5 gene (23, 24) upon iron starvation. The abundance of PGR5 mRNA was about 6-fold increased by iron deficiency with a maximal abundance evident at 0.2 μm (Fig. 6C). This iron deficiency-dependent induction of PGR5 mRNA expression is largely suppressed in the pgrl1-RNAi background. The suppression is even more pronounced than the one observed for FOX1 and FTR1, pointing to a regulation of PGR5 expression by the PGRL1 levels.
PGRL1 Is Part of a High Molecular Mass Complex and Is Possibly an Iron-binding Protein
Our data clearly suggest a role of PGRL1 in modulating iron acclimation in C. reinhardtii. To obtain further information on the molecular mechanism underlying this function of PGRL1, we undertook further biochemical analysis of this protein. Previous work in A. thaliana has shown that PGRL1 interacts not only with PGR5, but also with PSI, leading to the formation of a multimeric complex (19). Still, the presence of PSI is not required for stable accumulation of PGRL1 in the thylakoid membranes. In C. reinhardtii, we confirmed that PGRL1 can accumulate in the absence of PSI and PSII (supplemental Fig. 4A). Moreover, fractionation of detergent-solubilized iron-deficient thylakoids isolated from WT or PSI-lacking cells on sucrose density gradients confirmed the presence of a high molecular mass complex, which contained PGRL1. This complex was observed in mutants lacking PSI, indicating that the formation of the high molecular mass complex is not dependent on the presence of PSI in this alga either. In line with this, the integrity of PSI core as well as the subunit composition of the PSI-LHCI complex is comparable between WT and pgrl1–28 (supplemental Fig. 5) in iron-replete cells.
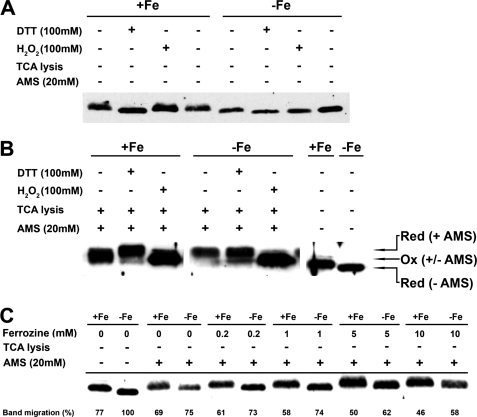
Another interesting feature of PGRL1 is the finding that its apparent molecular mass decreases when the iron concentration in the medium is diminished (Figs. 1A and 7A). The decrease in molecular mass can also be observed when broken cells are treated with 100 mm DTT. On the other hand, a slight up-shift of the PGRL1 band is seen when iron-replete and -deficient cells are incubated with hydrogen peroxide (Fig. 7A). This observation suggested that the migration of PGRL1 could be dependent on the redox state of cysteines in PGRL1, prompting us to further investigate the biochemical properties of this protein using the cysteine-specific alkylation reagent AMS (41). AMS alkylation adds 0.5 kDa to the molecular mass of a protein for each modified cysteine. The rationale behind this is that reduced cysteines can be alkylated, whereas oxidized cysteines cannot. PGRL1 contains six cysteines; four of those are found in two highly conserved CXXC motifs (19). Broken iron-sufficient and -deficient cells were either directly trichloroacetic acid-precipitated or incubated with DTT or H2O2 for 30 min at 37 °C and then trichloroacetic acid-precipitated. The trichloroacetic acid-precipitated samples were subsequently resolved in a Tris-SDS buffer containing AMS, leading to the alkylation of cysteines. The alkylated samples were then fractionated by SDS-PAGE and further analyzed by immunoblotting using anti-PGRL1 antibodies (Fig. 7B). The immunodetection revealed that DTT treatment led to the highest molecular mass bands of PGRL1. An increase in molecular mass of PGRL1 of about 3–5 kDa was observed, suggesting that all six cysteines were reduced by DTT and alkylated. In contrast, incubation with AMS in H2O2-treated extracts did not change the molecular mass of PGRL1, indicating that hydrogen peroxide led to oxidation of cysteines, thereby preventing alkylation of PGRL1. Altogether, these data support the notion that the molecular mass difference between PGRL1 from iron-sufficient and iron-deficient samples (Fig. 7A) is due to a different redox state of the cysteines. In the thiol-trapping experiment (Fig. 7B), PGRL1 from iron-deficient cells showed a higher molecular mass band, which runs at the position of the DTT-AMS samples. This band likely presents PGRL1 with all cysteines alkylated, as suggested by its increase in molecular mass. PGRL1 from iron-sufficient cells possessed, on the other hand, a broad protein band after cysteine alkylation, which is lower in molecular mass than PGRL1 from iron-deficient conditions or from the DTT-AMS samples, pointing to the fact that PGRL1 cysteines were less reduced and, therefore, less alkylated when stemming from iron-replete growth conditions. Next we questioned whether iron binding impacts the alkylation of PGRL1 cysteines. For these experiments we lysed iron plus or minus cells directly in an AMS-containing Tris-SDS buffer in the presence or absence of the Fe2+ chelator ferrozine (42). In the absence of ferrozine, the Tris-SDS AMS treatment caused an alkylation of PGRL1 from iron-sufficient and -deficient cells in respect to the non-alkylated samples. The shift in molecular mass of PGRL1 from iron-deficient cells was more pronounced as compared with the one in iron-replete conditions. Incubation of Tris-SDS-treated cells with AMS and increasing ferrozine concentrations from 0.2 to 10 mm resulted in a clear increase in the molecular mass of PGRL1 from iron-replete cells. The increase in molecular mass was also observable for PGRL1 from iron-deprived cells, although higher ferrozine concentrations of 5–10 mm were required. These changes can be visualized by expressing the migration of PGRL1 with respect to the molecular mass found for PGRL1 from iron-deficient conditions without AMS treatment, which is set to 100%. In line with a successful alkylation of PGRL1, the molecular mass increases, and the percent of band migration decreases, which is clearly observable for experiments with rising ferrozine concentrations. These data strongly suggest that during the Tris-SDS treatment in the absence of ferrozine, PGRL1 cysteines were partially protected from AMS alkylation. Given the fact that ferrozine is a Fe2+ chelator, we propose that iron withdrawal from PGRL1 by this chelator makes the cysteines of this protein available for alkylation. Therefore, we suggest that PGRL1 is involved in iron binding and that iron-loaded cysteines in PGRL1 are reduced but protected from AMS-dependent alkylation.
FIGURE 7.
PGRL1 is redox-regulated via the iron status of the cells. Cells from iron replete and iron-depleted cultures were resuspended in solution A, and then 60 μg of protein-containing samples were used in the different treatments described below. 13% SDS-PAGE electrophoresis and PGRL1 immunodetection followed each different treatment. A, shown is the effect of 100 mm H2O2 (oxidation) and 100 mm DTT (reduction) in the migration of PGRL1 band in plus-iron and minus-iron cell samples. B, plus-iron and minus-iron samples were oxidized or reduced as in A and were subsequently lysed in 10% (w/v) trichloroacetic acid to “freeze” the redox state of PGRL1 cysteines. The samples were then allowed to react with AMS, a 0.5-kDa chemical reagent that conjugates selectively to reduced cysteines. Plus-iron and minus-iron samples in solution A with were also run in the same gel (last 2 lanes). C, plus-iron and minus-iron samples were treated with ferrozine (0.2, 1, 5, and 10 mm) were subsequently alkylated with AMS without trichloroacetic acid precipitation. Control samples without ferrozine or AMS were also run in the same gel, which shows that the presence of ferrozine increases the availability of PGRL1 cysteines to AMS alkylation.
DISCUSSION
In this work we have addressed the function of PGRL1 in the green alga C. reinhardtii. PGRL1 expression is increased under iron deprivation (16), suggesting a possible role of this protein in the response to availability of this metal. To test this hypothesis, we took advantage of reverse genetic approaches and produced mutant strains having diminished levels of PGRL1. Comparative analysis of the wild-type and pgrl1–28 strains with respect to iron-deficiency responses suggests that PGRL1 is a player in the response of the photosynthetic apparatus to changes in iron nutrition. We suggest that this occurs via iron-dependent changes in thiol-disulfide chemistry of PGRL1. Given the proposed role of PGRL1 in CEF, this may provide a mechanism for controlling the partitioning between LEF and CEF.
Iron Acclimation Is Perturbed in pgrl1 RNAi Mutants of C. reinhardtii
Our data show that PGRL1 depletion under replete growth conditions leads to several related phenotypes, which all reflect a modified response to iron. (i) The cellular iron content is increased, likely reflecting an augmented iron cellular demand. This may stem from a higher respiratory activity, as indicated by the increased levels of respiratory complex protein marker Cox2b in pgrl1–28 (Fig. 4, C and D) and by the enhanced oxygen consumption observed in these cells (Fig. 5, B and C). At the same time (ii) PSII-driven electron transfer rate, as estimated from fluorescence parameters (ETR, Fig. 2A), was reduced in pgrl1–28. This suggests that electron flux in the cells is subjected to different partitioning between the photosynthetic and respiratory electron flow chains. Consistent with this, net photosynthesis (i.e. oxygen consumption plus evolution) is identical in WT and mutant strains (Fig. 3). Furthermore, (iii) the levels of ferredoxin and Lhca3, two proteins that are very sensitive to iron deficiency (32, 40), were already diminished at 20 μm iron in the mutant as compared with wild type (Fig. 4C).
In addition, PGRL1-depleted cells showed iron-deficiency symptoms at higher iron concentrations (3 and 1 μm) than did WT cells when exposed to iron starvation (Fig. 5, A, B, and C). This is typically evidenced by the overexpression of the FEA1 and FOX1 proteins, two proteins that participate in the high affinity iron uptake system of C. reinhardtii (36, 39) as well as by the higher decrease in Lhca3 and ferredoxin. Thus, our data suggest that PGRL1 plays a role in modulating acclimation of the two major bioenergetic pathways in the cell (the photosynthetic and respiratory ones) to iron starvation. Previous results have shown that a hierarchy exists with respect to iron requirement and distribution in C. reinhardtii cells, which favors maintenance and/or distribution of iron in the mitochondria under iron-deficient and photo-heterotrophic growth conditions (1). C. reinhardtii switches from photo-heterotrophic to heterotrophic metabolism under iron deficiency (16), whereas iron-rich photosynthetic proteins are degraded, and iron-containing respiratory proteins remained or even increased in abundance. Thus, it is likely that the enhanced respiration observed in pgrl1–28 cells most likely reflects an indirect effect of PGRL1 on the respiratory apparatus, namely the enhanced recycling of chloroplast-released iron into the respiratory chain, as required to compensate for impaired photosynthetic activity (see below).
Altogether, our findings suggest a very complex function for PGRL1 during acclimation to iron starvation, which can be rationalized assuming that the iron-sensing process is modified in pgrl1–28. This would explain not only the cell overloading with iron under replete conditions but also the chlorotic phenotype of pgrl1 A. thaliana mutant plants when grown under normal iron-sufficient conditions (19). The expression of some iron-responsive genes, such as FOX1 and FTR1, which is decreased in the mutant, is also consistent with this hypothesis. Also, the observation that expression of both PGR5 and PGRL1 (which results in the formation of the PGR5-PGRL1 complex) is tightly regulated by the cellular iron concentration (Figs. 1C and 6C) in another argument in favor of a wider role of this protein in iron metabolism. In all these cases, the iron deficiency-induced expression is decreased. However pgrl1-depleted mutants possess more ferroxidase at 1 and 3 μm iron as compared with wild type. This strongly suggests that there may be a post-transcriptional component to determine ferroxidase abundance in C. reinhardtii (36).
What is then (if any) the molecular mechanism relating PGRL1 to iron sensing in C. reinhardtii? As shown in Figs. 1A and 7A, the PGRL1 protein migrates more rapidly under low iron when fractionated by SDS-PAGE from C. reinhardtii cells. The shift from lower to higher mobility can be triggered by reducing conditions (DTT) and is related to the number of reduced cysteines in this protein as revealed by AMS-dependent alkylation experiments (Fig. 7). In addition, the redox state of PGRL1 cysteines appears to be linked to the iron binding status of the protein, as cysteines in iron-loaded PGRL1 are reduced but protected from alkylation (Fig. 7C). AMS-dependent alkylation can only take place when the iron-loaded PGRL1 is unloaded by the Fe2+ chelator ferrozine. This suggests that PGRL1 may have the capacity to bind iron in iron-sufficient as well as iron-deficient growth conditions. PGRL1 possesses two CXXC motifs that are known to enable binding of mono- or divalent metal ions (43). This motif has been shown to modulate iron binding by thioredoxin reductase (Trr1p) from Saccharomyces cerevisiae, where a role in intracellular iron transport has been suggested for this protein (44). Thus, the CXXC motif in PGRL1 could be important for iron binding. It is known that the iron-binding ferritin protein is largely up-regulated after the onset of iron deprivation (45, 46) in C. reinhardtii, where they participate in rapid remodeling of the photosynthetic apparatus in response to iron availability (45) and in protection against photo-oxidative damage. It is possible that iron binding of PGRL1 under iron deficiency is related to ferritin and/or to a general mechanism of iron loading into iron storage protein. Likewise it is possible that iron binding to PGRL1 is an independent defense line to take up iron released due to degradation of PSI and to minimize photo-oxidative damage. In this respect the increase of PGRL1 under iron deficiency would enhance the capacity of the thylakoid membrane to bind iron. Obviously direct binding of iron to PRGL1 would also provide this protein with a very efficient system to detect changes in the chloroplast iron concentration.
Role of PGRL1 in Energy Metabolism in C. reinhardtii
Our comparative analysis of photosynthesis in WT and pgrl1–28 cells points to the existence of a tight relationship between PSI levels, PGRL1 levels, and CEF in C. reinhardtii. Indeed, increasing PGRL1 in the presence of reduced PSI levels leads to constant CEF rates (WT cells). Conversely low PGRL1 levels result in CEF inhibition and delayed PSI degradation (pgrl1–28 cells). The possible rationale for this relationship has to be searched in the different response of photosynthetic and respiratory metabolism in the two genotypes. As seen in Fig. 3, net photosynthesis (i.e. O2 evolution + O2 consumption) and the efficiency of photosynthetic electron flow in steady state (Y(ΙΙ)) show an initial drop upon exposure to iron starvation, which largely precedes PSI degradation in both genotypes. After this initial decrease, a slower decline is seen that follows linearly the decrease in PSI. This suggests that a PSI-independent reaction step limits electron flow from H2O to CO2 at the beginning of iron limitation (Fig. 3, day 0 to day 1). This step is substituted by a second one (likely the decrease of PSI itself) that controls photosynthesis efficiency in fully iron-starved cells. Our data, showing a significant decrease of ferredoxin during iron deficiency (see also Ref. 40), suggest that degradation of this protein is probably responsible for the initial inhibition of linear electron flow. Indeed, a similar phenotype has been reported when ferredoxin 2, the most abundant leaf ferredoxin in A. thaliana, was down-regulated via RNAi (47). On the other hand, changes in the Fd levels cannot fully explain the different CEF capacity observed in WT and pgrl1 cells in low iron conditions. Indeed, although FDX1 levels are very similar at 0.1 μm iron. CEF is more affected in prgl1 than in the WT.
In principle, this finding suggests that other ferredoxin isoforms, which are induced under iron starvation (2, 40), could support cyclic electron transfer under low iron. In this case, the different response of CEF in WT and pgrl1–28 iron-starved cells would simply reflect the different capacity to induce the alternative ferredoxins in the two genotypes. However, an alternative possibility can be conceived which directly links CEF capacity to the levels of PGRL1 in the thylakoids. Previous data have suggested that this protein modulates CEF by enhancing the PSI affinity for soluble electron carriers involved in cycling, at the expense of those implicated in linear flow (48–50). Overexpression of PGRL1 in WT cells upon reduction of Fd levels would, therefore, maintain high CEF levels by recruiting the residual ferredoxin for this pathway at the expense of a linear one. In agreement with this, CEF is almost constant in WT during the first 2 days of starvation, whereas Y(II) and net photosynthesis show a larger decrease when compared with pgrl1 (Fig. 3). Compensation of ferredoxin deficiencies by increased PGRL1 accumulation would take place until PSI would be degraded to a threshold concentration, below which both CEF and LEF would be equally limited by the amount of this complex. In mutant cells, where PGRL1 levels remain low even under iron starvation, no preferential partitioning of CEF would occur, leading to a very similar decrease of LEF and CEF.
It is acknowledged that the Calvin cycle requires ATP and NADPH in a stoichiometry of 1.5, i.e. a ratio that cannot be entirely fulfilled by the sole operation of LEF, even in iron replete conditions. This process has an insufficient proton to electron balance when compared with the stoichiometry of H+ required to fuel ATP synthesis by the chloroplast CFo − Fi complex (e.g. Ref. 21; see, however, Ref. 51). In particular, no more than 1.4 ATP should be synthesized per NADPH in C. reinhardtii (≈1.39, e.g. Ref. 52). Thus, alternative processes must contribute to the generation of additional ATP synthesis to re-equilibrate the ATP/NADPH stoichiometry for proper carbon assimilation. All these processes require “extra” PSI activity when compared with PSII. Synthesis of ATP can be fuelled by cyclic photophosphorylation (53) and by reducing molecular oxygen in the Mehler reaction (for review, see Ref. 54). Mitochondria can also provide extra ATP while consuming PSI-generated reducing equivalents exported from the chloroplast via the malate or the triose phosphate transporter (for review, see Ref. 55). It is acknowledged that the chloroplast-mitochondria energetic interaction is extremely efficient in C. reinhardtii when compared with vascular plants (for review, see Ref. 52). Therefore, it is tempting to speculate that in prgl1–28 cells, where cyclic flow cannot be maintained, mitochondria activity would be enhanced to counterbalance the decreased CEF-driven ATP production. Consistent with this, enhanced respiration in pgl1–28 cells prevents over-reduction of the PSI-reducing side, at variance with previous observations in pgrl1 mutants of A. thaliana (19). This results in similar levels of P700+ generation in this strain when compared with the WT (Fig. 2), in contrast to the phenotype of pgrl1 mutant plants in A. thaliana, where no light-induced P700 oxidation can be seen (19).
A Dual Role of PGRL1 in Iron Homeostasis and Energy Metabolism in C. reinhardtii
What is the link between iron-sensing and cyclic electron flow? As discussed above, consequences of PGRL1 removal on photosynthesis do not necessarily reflect a direct involvement of this protein in photosynthesis. Still, our data confirm the existence of a link between PGRL1 and CEF. The conformational changes observed in PGRL1 in relationship to iron starvation may provide a molecular mechanism to explain this link. The reduction state of PGRL1 cysteines is clearly related to the iron status of the cells and increases as the cells become more and more iron-deficient (Figs. 1A, 7A, and B). At the same time PGRL1 is likely to bind iron, as suggested by the enhanced AMS alkylation observed upon addition of ferrozine (Fig. 7C). Higher ferrozine concentrations are required to alkylate PGRL1 cysteines from iron-deficient cells, indicating a stronger iron binding affinity or an enhanced iron load of the protein in respect to PGRL1 from iron-replete conditions. Thus, it is tempting to speculate that the iron-loading capability of PGRL1 may represent a molecular mechanism to control iron distribution and CEF at the same time. Iron-dependent molecular switches are well known and have been e.g. described for IRP1 and IRP2, which control the binding of ribosomes to the ferritin mRNA in mammals in relation of the iron status of the cell (56). In addition, metal release from CXXC motifs upon oxidation has been shown for E. coli Trx2 (57). In this case zinc unloading by hydrogen peroxide induces major conformational changes, which might act as a redox switch for Trx2 protein function (57). In this scenario, PGRL1 could affect CEF (e.g. by modulating the affinity of the linear and cyclic pathways for their soluble electron carriers) via its iron-dependent conformational changes. Alternatively the redox state of PGRL1 cysteines, independent of metal binding, might control the conformational state of the protein and function as a redox switch that directly impacts CEF without a direct implication of iron. Obviously, the two hypotheses are not mutually exclusive, and a model where the iron-loading and redox state of cysteines would be synergistic to impact CEF and iron-sensing is also conceivable. In this context it is of note that for IRP2 also, cysteine oxidation besides iron availability regulates the RNA binding activity of the protein, thereby demonstrating that iron- and redox-dependent regulation may occur in conjunction (58). In general oxidoreduction of protein thiols in redox regulation is a common (59) and in particular a well known mechanism in chloroplasts (60, 61).
In conclusion we propose a dual role of PGRL1 in C. reinhardtii, including iron sensing or distribution and modulation of cyclic electron transfer. Furthermore, due to redox-induced conformational changes in PGRL1, CEF would be stimulated under reducing conditions, in agreement with previous data indicating increased CEF efficiency in anaerobic C. reinhardtii cells (62).
Supplementary Material
Acknowledgments
We thank Dr. J. Alric for support with spectroscopic measurements and Dr. F. A. Wollman for fruitful discussions.
This work was supported by the German Science Foundation (to M. H.), an Institutional Ruth L. Kirschstein National Research Service Award GM070104 and a Dissertation Year Fellowship from the UCLA graduate division (to A. M. T.) and by Department of Energy Grant DE-FD02-04ER15529 (to S. S. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
G. Peers, T. B. Truong, E. Ostendorf, E. A. Busch, D. Elrad, A. Grossman, M. Hippler, and K. K. Niyogi, manuscript in preparation.
- PSI and PSII
- photosystems I and II, respectively
- LHCI
- light-harvesting antenna of PSI
- CEF
- cyclic electron transfer
- DCMU
- 3-(3′,4′-dichlorophenyl)-1,1-dimethylurea
- LEF
- linear photosynthetic electron transfer
- TAP
- Tris acetate-phosphate
- AMS
- 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid
- ETR
- electron transfer rate
- μE
- microeinsteins
- DTT
- dithiothreitol
- WT
- wild type.
REFERENCES
- 1.Clemens S., Naumann B., Hippler M. (2009) Front. Biosci. 14, 1955–1969 [DOI] [PubMed] [Google Scholar]
- 2.Merchant S. S., Allen M. D., Kropat J., Moseley J. L., Long J. C., Tottey S., Terauchi A. M. (2006) Biochim. Biophys. Acta 1763, 578–594 [DOI] [PubMed] [Google Scholar]
- 3.Guikema J. A., Sherman L. A. (1984) Plant Physiol. 74, 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey S., Melis A., Mackey K. R., Cardol P., Finazzi G., van Dijken G., Berg G. M., Arrigo K., Shrager J., Grossman A. (2008) Biochim. Biophys. Acta 1777, 269–276 [DOI] [PubMed] [Google Scholar]
- 5.Laudenbach D. E., Straus N. A. (1988) J. Bacteriol. 170, 5018–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnap R. L., Troyan T., Sherman L. A. (1993) Plant Physiol. 103, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boekema E. J., Hifney A., Yakushevska A. E., Piotrowski M., Keegstra W., Berry S., Michel K. P., Pistorius E. K., Kruip J. (2001) Nature 412, 745–748 [DOI] [PubMed] [Google Scholar]
- 8.Bibby T. S., Nield J., Barber J. (2001) Nature 412, 743–745 [DOI] [PubMed] [Google Scholar]
- 9.Varsano T., Wolf S. G., Pick U. (2006) J. Biol. Chem. 281, 10305–10315 [DOI] [PubMed] [Google Scholar]
- 10.Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doan J. M., Schoefs B., Ruban A. V., Etienne A. L. (2003) FEBS Lett. 533, 59–62 [DOI] [PubMed] [Google Scholar]
- 12.Desquilbet T. E., Duval J. C., Robert B., Houmard J., Thomas J. C. (2003) Plant Cell Physiol. 44, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 13.Strzepek R. F., Harrison P. J. (2004) Nature 431, 689–692 [DOI] [PubMed] [Google Scholar]
- 14.Cardol P., Bailleul B., Rappaport F., Derelle E., Béal D., Breyton C., Bailey S., Wollman F. A., Grossman A., Moreau H., Finazzi G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7881–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen A. E., Laroche J., Maheswari U., Lommer M., Schauer N., Lopez P. J., Finazzi G., Fernie A. R., Bowler C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 17.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 18.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., Leister D. (2008) Cell 132, 273–285 [DOI] [PubMed] [Google Scholar]
- 20.Taiz L., Zeiger E. (2006) Plant Physiology, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 21.Allen J. ( 2002) Cell 110, 273– 276 [DOI] [PubMed] [Google Scholar]
- 22.Johnson G. N. (2005) J. Exp. Bot. 56, 407–416 [DOI] [PubMed] [Google Scholar]
- 23.Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002) Cell 110, 361–371 [DOI] [PubMed] [Google Scholar]
- 24.Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004) Nature 429, 579–582 [DOI] [PubMed] [Google Scholar]
- 25.Shikanai T. (2007) Annu. Rev. Plant Biol. 58, 199–217 [DOI] [PubMed] [Google Scholar]
- 26.Finazzi G. (2005) J. Exp. Bot. 56, 383–388 [DOI] [PubMed] [Google Scholar]
- 27.Harris E. H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- 28.Redding K., MacMillan F., Leibl W., Brettel K., Hanley J., Rutherford A. W., Breton J., Rochaix J. D. ( 1998) EMBO J. 17, 50– 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchka M. R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J. D. (1989) Cell 58, 869–876 [DOI] [PubMed] [Google Scholar]
- 30.Rohr J., Sarkar N., Balenger S., Jeong B. R., Cerutti H. (2004) Plant J. 40, 611–621 [DOI] [PubMed] [Google Scholar]
- 31.Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009) Plant J. 58, 165–174 [DOI] [PubMed] [Google Scholar]
- 32.Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 33.Kindle K. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hippler M., Drepper F., Farah J., Rochaix J. D. (1997) Biochemistry 36, 6343–6349 [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 36.Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007) Eukaryot. Cell 6, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witt H. T. (1979) Biochim. Biophys. Acta 505, 355–427 [DOI] [PubMed] [Google Scholar]
- 38.Maxwell P. C., Biggins J. (1976) Biochemistry 15, 3975–3981 [DOI] [PubMed] [Google Scholar]
- 39.Chen J. C., Hsieh S. I., Kropat J., Merchant S. S. (2008) Eukaryot. Cell 7, 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terauchi A. M., Lu S. F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J. N., Knaff D. B., Farmer P. J., Lemaire S. D., Hase T., Merchant S. S. (2009) J. Biol. Chem. 284, 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagamori S., Nishiyama K., Tokuda H. (2002) J. Biochem. 132, 629–634 [DOI] [PubMed] [Google Scholar]
- 42.Stookey L. (1970) Anal. Chem. 42, 779–781 [Google Scholar]
- 43.Romero-Isart N., Vasák M. (2002) J. Inorg. Biochem. 88, 388–396 [DOI] [PubMed] [Google Scholar]
- 44.Kang H. J., Choi S. W., Heo D. H., Jeong M. Y., Sung H. C., Yun C. W. (2008) Biochem. Biophys. Res. Commun. 371, 63–68 [DOI] [PubMed] [Google Scholar]
- 45.Busch A., Rimbauld B., Naumann B., Rensch S., Hippler M. (2008) Plant J. 55, 201–211 [DOI] [PubMed] [Google Scholar]
- 46.Long J. C., Sommer F., Allen M. D., Lu S. F., Merchant S. S. (2008) Genetics 179, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanke G. T., Hase T. (2008) Photochem. Photobiol. 84, 1302–1309 [DOI] [PubMed] [Google Scholar]
- 48.Breyton C., Nandha B., Johnson G. N., Joliot P., Finazzi G. (2006) Biochemistry 45, 13465–13475 [DOI] [PubMed] [Google Scholar]
- 49.Joliot P., Joliot A. (2006) Biochim. Biophys. Acta 1757, 362–368 [DOI] [PubMed] [Google Scholar]
- 50.Nandha B., Finazzi G., Joliot P., Hald S., Johnson G. N. (2007) Biochim. Biophys. Acta 1767, 1252–1259 [DOI] [PubMed] [Google Scholar]
- 51.Steigmiller S., Turina P., Gräber P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3745–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eberhard S., Finazzi G., Wollman F. A. (2008) Annu. Rev. Genet 42, 463–515 [DOI] [PubMed] [Google Scholar]
- 53.Arnon D. I. (1959) Nature 184, 10–21 [DOI] [PubMed] [Google Scholar]
- 54.Ort D. R., Baker N. R. (2002) Curr. Opin. Plant Biol. 5, 193–198 [DOI] [PubMed] [Google Scholar]
- 55.Noguchi K., Yoshida K. (2008) Mitochondrion 8, 87–99 [DOI] [PubMed] [Google Scholar]
- 56.Rouault T. A. (2006) Nat. Chem. Biol. 2, 406–414 [DOI] [PubMed] [Google Scholar]
- 57.Collet J. F., D'Souza J. C., Jakob U., Bardwell J. C. (2003) J. Biol. Chem. 278, 45325–45332 [DOI] [PubMed] [Google Scholar]
- 58.Zumbrennen K. B., Wallander M. L., Romney S. J., Leibold E. A. (2009) Mol. Cell. Biol. 29, 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghezzi P. (2005) Biochem. Soc. Trans. 33, 1378–1381 [DOI] [PubMed] [Google Scholar]
- 60.Pesaresi P., Schneider A., Kleine T., Leister D. (2007) Curr. Opin. Plant Biol. 10, 600–606 [DOI] [PubMed] [Google Scholar]
- 61.Oelze M. L., Kandlbinder A., Dietz K. J. (2008) Biochim. Biophys. Acta 1780, 1261–1272 [DOI] [PubMed] [Google Scholar]
- 62.Finazzi G., Furia A., Barbagallo R. P., Forti G. (1999) Biochim. Biophys. Acta 1413, 117–129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.