Abstract
We have examined function of the bacterial β replication clamp in the different steps of methyl-directed DNA mismatch repair. The mismatch-, MutS-, and MutL-dependent activation of MutH is unaffected by the presence or orientation of loaded β clamp on either 3′ or 5′ heteroduplexes. Similarly, β is not required for 3′ or 5′ mismatch-provoked excision when scored in the presence of γ complex or in the presence of γ complex and DNA polymerase III core components. However, mismatch repair does not occur in the absence of β, an effect we attribute to a requirement for the clamp in the repair DNA synthesis step of the reaction. We have confirmed previous findings that β clamp interacts specifically with MutS and MutL (López de Saro, F. J., Marinus, M. G., Modrich, P., and O'Donnell, M. (2006) J. Biol. Chem. 281, 14340–14349) and show that the mutator phenotype conferred by amino acid substitution within the MutS N-terminal β-interaction motif is the probable result of instability coupled with reduced activity in multiple steps of the repair reaction. In addition, we have found that the DNA polymerase III α catalytic subunit interacts strongly and specifically with both MutS and MutL. Because interactions of polymerase III holoenzyme components with MutS and MutL appear to be of limited import during the initiation and excision steps of mismatch correction, we suggest that their significance might lie in the control of replication fork events in response to the sensing of DNA lesions by the repair system.
Introduction
Mismatch repair is a conserved genetic stabilization system that in bacteria corrects DNA replication errors and ensures the fidelity of genetic recombination (1–3). In Escherichia coli, the strand specificity necessary for removal of DNA biosynthetic errors from the daughter strand is based on the transient absence of d(GATC) methylation in newly synthesized DNA (4). A methyl-directed reaction that can account for replication error correction has been reconstituted using purified E. coli activities (5–8). Mismatch recognition by MutS initiates the E. coli mismatch repair, leading to recruitment of MutL. Assembly of a MutS-MutL-heteroduplex complex activates MutH endonuclease, which incises the unmethylated strand at a hemimethylated d(GATC) site (9). The resulting strand break, which may reside 3′ or 5′ to the mismatch, serves as the entry site for an excision system comprised of DNA helicase II and an appropriate single-strand exonuclease. Excision directed by a 3′ strand break depends on the 3′ to 5′ hydrolytic activity of exonuclease I, exonuclease VII, or exonuclease X, whereas the 5′ to 3′ activity of either RecJ exonuclease or exonuclease VII is sufficient to support hydrolysis directed by a 5′ strand break (6–8). Excision in this manner removes that portion of the incised strand spanning the two DNA sites. The ensuing gap is repaired by DNA synthesis, in a reaction that depends on the integrity of dnaX (formerly called dnaZ) (5). dnaX encodes the γ and τ subunits of DNA polymerase (Pol)3 III holoenzyme, which can be resolved into three components: core polymerase, the β processivity clamp, and the γ complex clamp loader (10).
Direct interactions of the β clamp with MutS and MutL have been demonstrated (11), suggesting the possibility that this component of the replication apparatus could be involved in early steps of mismatch repair. Two MutS motifs have been implicated in this interaction, one located near the N terminus and the second near the C terminus. Deletion of the C-terminal motif (812QMSLL816) abolishes in vitro MutS interaction with β in the absence of DNA but does not confer hypermutability in vivo (12). By contrast, Ala substitution mutagenesis within the N-terminal MutS motif (15QQYLRL20) is without effect in MutS-β interaction in solution but confers strong hypermutability in the E. coli cell. The latter finding has led to the proposal that the N-terminal β interaction motif of MutS might play an important role in the interaction of the two proteins, but only when they are DNA-bound.
To clarify the role(s) of the β clamp in mismatch repair, we have evaluated the effects of the E. coli β, γ complex, and Pol III core on individual steps of the reconstituted E. coli methyl-directed reaction. Our results indicate that β is dispensable for MutH activation and excision steps of the reaction but essential for overall repair.
EXPERIMENTAL PROCEDURES
Proteins and DNAs
MutS (13), MutSN containing four Ala substitutions within the N-terminal β interaction motif (15QQYLRL20 to 15AAYAAL20) (12), MutL (14), MutH (15), DNA helicase II (16), β clamp (17), βPK (a β clamp variant containing a 6-residue C-terminal cAMP-dependent protein kinase recognition motif) (18), γ complex (19), and Pol III core (20) were isolated by published methods. Exonuclease I, SSB, NheI, ClaI, XhoI, and HindIII, cAMP-dependent protein kinase were obtained from commercial sources. RecJ exonuclease was a gift from Susan Lovett (Brandeis University). All of the protein concentrations are expressed as monomer equivalents. For Pol III core and γ complex, these equivalents correspond to the αϵθ and δδ′γψχ assemblies, respectively (10).
Supercoiled closed circular 6,444 bp heteroduplex or homoduplex DNAs contained a G-T mismatch or A·T base pair located at position 5636 and a single hemimethylated d(GATC) site 1,024 bp from the mismatch at position 216. These molecules, which contained d(GATC) methylation on either the complementary (3′ heteroduplex) or viral DNA strand (5′ heteroduplex), were prepared using phage DNAs derived from f1MR65 and f1MR66 as described previously (21). DNAs used to score nick-directed excision were prepared using phages f1MR70 and f1MR71 (22) and contained a G-T mismatch (or A·T base pair) at position 5632 and a single-strand break on the complementary strand located either 3′ or 5′ to the mispair at a separation distance of 128 bp. Hemimethylated heteroduplex/homoduplex DNAs that also contained a 3′ strand break (see Fig. 3) were prepared in a similar manner except that the d(GATC) site located 1,024 bp from the mismatch was hemimodified as indicated.
FIGURE 3.
Orientation of β loading does not affect MutH activation. Open circular G-T heteroduplex (or A·T homoduplex) DNAs contained a single strand break within the complementary DNA strand 128 bp 3′ to the mismatch (shorter path), as well as d(GATC) methylation on the complementary DNA strand (A, 3′ heteroduplex/homoduplex) or viral strand (B, 5′ heteroduplex/homoduplex). DNAs were preincubated with β clamp and γ complex (G-T, open circles; A·T, open triangles) or mock preincubated (G-T, closed circles; A·T, closed inverted triangles) as described under “Experimental Procedures.” The products were used immediately for MutH activation assays. The error bars represent one standard deviation for three independent experiments.
Assembly of β Clamp onto DNA
Loading of β clamp by γ complex was performed as described (23). Briefly, 15-μl reactions contained 20 mm Tris-HCl, pH 7.5, 4% glycerol, 0.1 mm EDTA, 40 μg/ml bovine serum albumin (BSA), 5 mm dithiothreitol, 2 mm ATP, 8 mm MgCl2, 9.6 nm supercoiled or nicked DNA, 150 nm β clamp, 12 nm γ complex, and when indicated, 320 nm Pol III core. After incubation for 10 min at 37 °C, β·DNA assemblies were used immediately for MutH activation and excision assays as described below.
Quantitation of clamp loading employed a modified form of β containing a C-terminal cAMP-dependent protein kinase recognition motif, which allows the protein to be 32P-labeled (23). The assembly of β onto DNA was performed as described above except that 20 mm HEPES-NaOH, pH 7.5, was substituted for Tris-HCl, and the 15-μl reactions contained 0 or 30 mm NaCl as indicated. After 10 min at 37 °C the reactions were supplemented with 45 μl of buffer A (60 mm HEPES-NaOH, pH 7.5, 6.7 mm Tris-HCl, pH 7.5, 8 mm potassium phosphate, pH 7.4, 0.9% glycerol, 0.04 mm EDTA, 0.4 mm dithiothreitol, 400 μg/ml BSA, 2 mm ATP, 4 mm MgCl2) containing 66.7 mm NaCl (0 mm NaCl assembly reaction) or 56.7 mm NaCl (30 mm NaCl assembly reaction), and β was cross-linked to DNA by the addition of 6 μl of 1% glutaraldehyde and incubation for 10 min at room temperature. Cross-linking was quenched by the addition of 1 m Tris-HCl, pH 7.5, to a final concentration of 100 mm, and protein-DNA complexes were analyzed by electrophoresis through 1% agarose in 40 mm Tris acetate, 1 mm EDTA, pH 8.0, at 7.1 volts/cm for 2 h. DNA was visualized by ethidium bromide staining. [32P]β clamp was visualized and quantified using a Typhoon phosphorimaging device and ImageQuant 5.2 software (GE Healthcare). For analysis of β·DNA complex lifetimes, β assembly reactions were performed in a similar manner in the absence or presence of 100 mm NaCl. After 10 min of incubation at 37 °C, the reactions were supplemented with 45 μl of buffer A containing 133 mm NaCl (or 100 mm NaCl for the assembly reaction performed in the presence of 100 mm NaCl) and incubation continued at 37 °C. Ten-μl samples were removed as a function of time and cross-linked, and β·DNA complexes were determined as described above.
MutH Activation
d(GATC) incision by activated MutH (9) was performed in 60-μl reactions containing buffer B (50 mm HEPES-KOH, pH 8.0, 5 mm Tris-HCl, pH 7.5, 6 mm potassium phosphate, pH 7.4, 50 mm KCl, 1.7% glycerol, 0.055 mm EDTA, 1.6 mm dithiothreitol, 310 μg/ml BSA, 2 mm ATP, 5 mm MgCl2), 2.4 nm 3′ or 5′ hemimethylated heteroduplex/homoduplex DNA (preincubated with β clamp and γ complex as described above; preincubated with β, γ complex, and Pol III core; or mock preincubated (Pol III components omitted)), 1 nm MutH, 25 nm MutL, and MutS or MutSN (37 nm or as indicated). Incubation was at 37 °C. The samples (10 μl) were removed as a function of time and quenched with 90 μl of 22 mm EDTA, and DNA products were isolated by phenol extraction and ethanol precipitation. d(GATC) incision was scored by indirect end labeling (9). DNA products were cleaved with ClaI, subjected to electrophoresis through 1% alkaline agarose, transferred to a Hybond-XL membrane (Amersham Biosciences) after depurination and strand breakage to ensure effective transfer of large fragments (24), and probed with an excess of 5′ 32P-labeled probe C2527 (d(AGCAGCACCGTAATCAGTAGCG)) or V2505 (d(CGCTACTGATTACGGTGCTGCT)). V2505 hybridizes to the complementary strand adjacent to the ClaI site and was used to map incision on the 5′ hemimethylated substrate; C2527 hybridizes to the viral strand adjacent to the ClaI site and was used to score incision on the 3′ hemimethylated DNA. The results were quantified by phosphorimaging analysis as described above.
3′ and 5′ Directed Excision
Mismatch-provoked excision on 3′ or 5′ hemimethylated supercoiled heteroduplex/homoduplex DNA (preincubated with β clamp, γ complex, and Pol III core; or mock preincubated) was performed in 60-μl reactions containing buffer B, 2.4 nm DNA, 0.1 mm ddTTP, 0.1 mm each dATP, dCTP, and dGTP, 1 nm MutH, 25 nm MutL, and MutS or MutSN (37 nm or as indicated), 1000 nm SSB, 12 nm DNA helicase II, and 1.8 nm exonuclease I (3′-directed excision) or 7.8 nm RecJ exonuclease (5′-directed hydrolysis) (6). Incubation was at 37 °C, and samples (10 μl) were removed as a function of time and quenched by the addition of 90 μl of 22 mm EDTA, followed by phenol extraction and ethanol precipitation. Recovered DNA was digested with ClaI and NheI and subjected to electrophoresis through 1% agarose to score for excision (25). Excision end points were mapped by indirect end labeling using 5′-32P-labeled oligonucleotide probe C2552 (d(GAAACGTCACCAATGAAACCAT)) (for 3′ directed excision) or V2531 (d(ATGGTTTCATTGGTGACGTTTC)) (for 5′ directed excision).
Excision on circular DNAs with a pre-existing strand break was performed in a similar manner, except MutH was omitted from the reactions. The extent of excision was determined by digestion with ClaI and NheI as described above.
3′ and 5′ Directed Repair
Methyl-directed repair of 2.4 nm 3′ or 5′ hemimethylated supercoiled heteroduplex DNAs was performed in 60-μl reactions containing buffer B, 37 nm MutS, 25 nm MutL, and 1 nm MutH, 12 nm DNA helicase II, 1000 nm SSB, and 1.8 nm exonuclease I (3′-directed repair) or 7.8 nm RecJ exonuclease (5′-directed repair), 37 nm β clamp as indicated, 2.9 nm γ complex, 79 nm Pol III core, and 0.1 mm each dATP, dCTP, dGTP and dTTP. Incubation was at 37 °C, and the samples (10 μl) were removed as a function of time and quenched by the addition of 90 μl of 22 mm EDTA, followed by phenol extraction and ethanol precipitation. Recovered DNA products were digested with ClaI and HindIII or ClaI and XhoI to score 5′- or 3′-directed repair, respectively (5). Recovered DNA was also mapped by indirect end labeling after ClaI digestion as described for excision reactions.
Western Analysis
pET-MutSwt or pET-MutSN (12) were transformed into E. coli BT199ΔmutS2K6. The cultures were grown from single colonies overnight at 37 °C in LB medium supplemented with ampicillin (100 μg/ml). One ml of cells was spun down and resuspended in 100 μl of 20 mm potassium phosphate, pH 7.4, 1 mm EDTA, 0.5 mm dithiothreitol. SDS-denatured lysate (90 μg protein) was subjected to electrophoresis on an 8% SDS-PAGE and transferred onto the polyvinylidene difluoride membrane, which was blocked overnight at 4 °C with 0.01 m Tris-HCl, pH 8.0, 0.15 m NaCl, 1 mm EDTA, 0.1% Triton X-100 containing 5% milk solids (all washes were performed in the same buffer). The membrane was then incubated with rabbit anti-MutS for 2 h at room temperature, washed, and then incubated 1 h with peroxidase-conjugated goat anti-rabbit antibody (Amersham Biosciences). The data were visualized using ECL+ system (Amersham Biosciences).
Far Western
β clamp, γ complex, Pol III core, BSA, MutL, MutS, and DNA helicase II (0.25–4 pmol) as indicated were spotted onto Protran nitrocellulose membranes (Whatman). Alternatively, 4 pmol of each protein was resolved by electrophoresis on a 10% SDS-polyacrylamide gel, followed by transfer to a Protran nitrocellulose membrane. The membranes were blocked for 1 h at room temperature in the blocking buffer described above and then incubated with 0.6 μm of MutL or MutS in the same buffer overnight at 4 °C. After wash, the membranes were incubated with rabbit anti-MutL or anti-MutS antibodies for 2 h at room temperature, washed, and then incubated for 1 h with peroxidase-conjugated goat anti-rabbit antibodies (Amersham Biosciences). The data were visualized using the ECL+ system (Amersham Biosciences).
Strain Construction
BT199ΔmutS2K6 was constructed using the method of de Lorenzo and Timmis (26). A BglII fragment of pMS312 (27) that includes mutS and flanking genomic sequences was cloned into the BamHI site pUC18. A precise deletion of the mutS open reading frame was obtained by PCR using primers oriented to replicate the entire plasmid except the MutS coding sequence. Each primer (5′-GGG GCG GCC GCG GGG TTA TGT CCG GTT CCC TG and 5′-GGG GCG GCC GCT AAT AAC AAT TCC CGA TAG TC) contained a NotI site at its 5′ terminus. The resulting plasmid was digested with NotI and ligated to a 2.1-kb NotI fragment from pCK155 (28) containing the Tn5 npt gene. The resulting kanamycin-resistant, ampicillin-resistant plasmid was transformed into JC7623 (recB recC sbcB sbcC). Kanamycin-resistant, ampicillin-sensitive colonies were found to have the appropriate deletion of mutS. The deletion was transferred into BT199 by P1virA-mediated transduction. The resulting strain BT199ΔmutS2K6 is ΔmutS::npt thr-1 leuB6 thi-1 lacY1 galK2 ara14 xyl5 mtl-1 Δ(gpt-proA2) rpsL31 tsx33 supE44 rac rfbD1 mgl-51 kdgK51.
Mutation Rates
E. coli strain BT199ΔmutS2K6 was transformed with pET-MutS or pET-MutSN (12) and plated onto LB plates (1% tryptone, 0.5% yeast extract, 1% NaCl, 1.5% agar) containing ampicillin (100 μg/ml). Ten to twenty cultures of BT199ΔmutS2K6 or BT199ΔmutS2K6-pET-MutSN were grown overnight from single colonies at 37 °C. Mutation rates to rifampicin resistance (100 μg/ml) were calculated according to the method of the median (29) based on two to five independent experiments.
RESULTS
Lifetimes of the β Processivity Clamp on Supercoiled or Nicked Heteroduplex Substrates
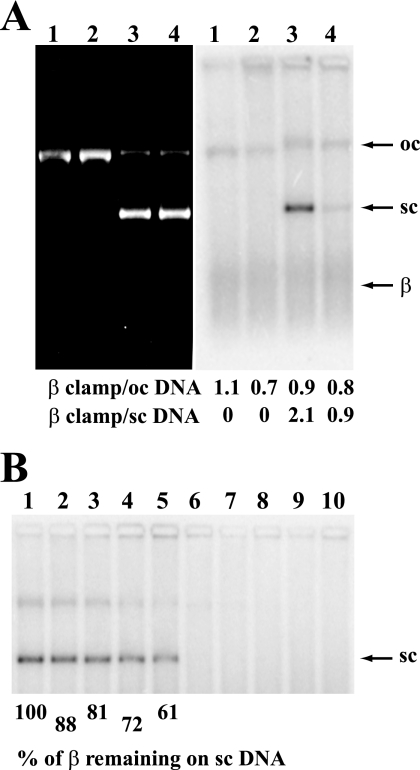
Methyl-directed mismatch repair has been routinely scored in vitro using supercoiled closed circular hemimethylated heteroduplexes (30), whereas open circular DNA can be used to study the MutH-independent reaction (5). A 32P-labeled form of β (23) was used to determine whether the clamp is maintained in the loaded form on such DNAs under conditions of repair assay. As shown in Fig. 1A and consistent with previous findings, γ complex efficiently loads β onto nicked or supercoiled DNA provided that the ionic strength is low (lanes 1 and 3). However, loading of the clamp onto supercoiled DNA is severely reduced if NaCl is present at 30 mm (Fig. 1A, lane 4) (23) and abolished at 100 mm salt (Fig. 1B, lanes 6–10). The latter effect was exploited to estimate the lifetime of β·DNA complexes by preloading β onto a superhelical G-T heterduplex in the absence of NaCl, followed by the addition of NaCl to 100 mm to prevent reloading. Dissociation of preloaded complexes at 100 mm NaCl occurred with a t½ of ∼45 min (Fig. 1B, lanes 1–5), somewhat less than the 72-min half-life for β·DNA complexes determined previously after γ complex removal by gel filtration (31). Because the mismatch repair assays used here range from 5 to 15 min, the 45 min half-life of the β·DNA complex is sufficient to assess possible involvement of the preloaded clamp in various steps of the reaction.
FIGURE 1.
Assembly of β clamp in the presence of γ complex onto supercoiled or nicked DNA. A, 3′ nicked G-T heteroduplex (lanes 1 and 2) or 3′ G-T supercoiled heteroduplex (lanes 3 and 4) were incubated with 32P-labeled β clamp and γ complex in the absence (lanes 1 and 3) or presence of 30 mm NaCl (lanes 2 and 4), β·DNA complexes cross-linked, and the products were resolved by electrophoresis through 1% agarose (“Experimental Procedures”). DNA was visualized after ethidium staining (left panel) and β·DNA complexes scored by phosphorimaging (right panel). The positions of the nicked open circular (oc) DNA, supercoiled (sc) DNA, and free β are shown on the right. The stoichiometry of β loading was determined from percentage of the total signal in each lane. B, lifetime of β·DNA complexes on supercoiled G-T heteroduplex DNA was determined by loading [32P]β in the absence of NaCl, followed by the addition of NaCl to 100 mm to prevent further loading. Incubation was continued, and the samples were removed and scored for β·DNA complexes (“Experimental Procedures”). Lanes 1–5, samples removed 0, 5, 10, 20, and 30 min after 100 mm NaCl addition. Lanes 6–10, assembly reaction was performed in the presence of 100 mm NaCl, and the samples were taken at 0, 5, 10, 20, and 30 min. β·DNA complexes were visualized as in A.
β Is Not Required for Mismatch-dependent MutH Activation
β clamps that remain on DNA after replication fork passage could associate with MutS and/or MutL and hence be involved in early steps of mismatch repair. However, as shown in Fig. 2, preloaded β clamp does not significantly affect the rate or extent of incision of a hemimethylated d(GATC) site by activated MutH without regard to a d(GATC) target sequence location 3′ or 5′ to the mismatch, and identical results were obtained when preincubation included β, γ complex, and Pol III core (not shown). Significant incision of hemimodified homoduplex control DNA also occurs under these conditions. As shown previously, this effect, which varies from preparation to preparation (compare Figs. 2 and 3), is likely due to natural variation in the DNA populations that are used for substrate preparation (9).
FIGURE 2.
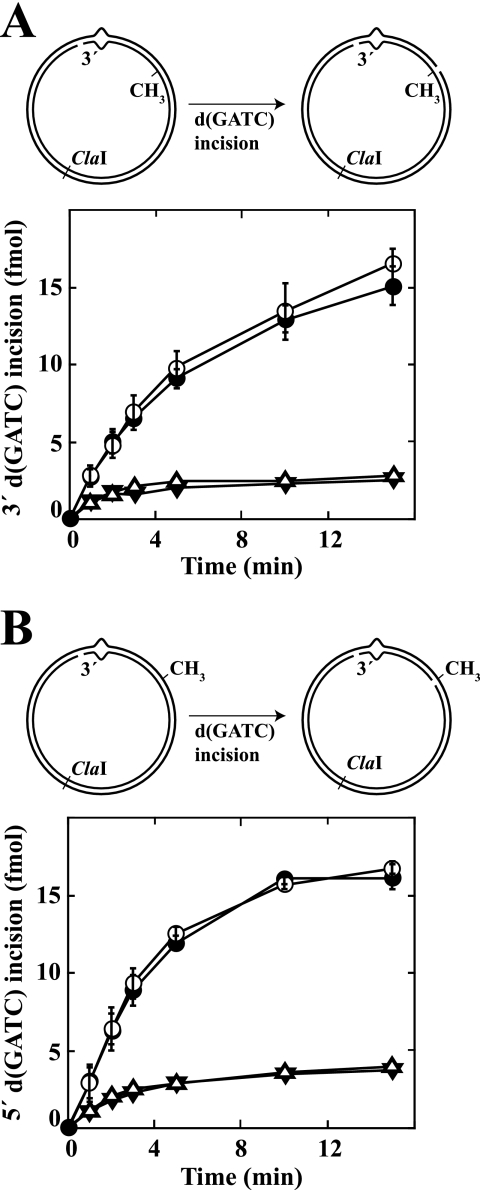
Loaded β clamp is not required for MutH activation. A, diagram depicts incision of a 3′ G-T supercoiled hemimethylated heteroduplex by activated MutH. Substrates contained a G-T mismatch (A·T base pair in homoduplex control) and d(GATC) methylation on the complementary DNA strand (unmethylated d(GATC) sequence 3′ to mismatch as viewed along the shorter path between the two sites). The 3′ G-T heteroduplex was preincubated with 150 nm β clamp and 12 nm γ complex (open circles), preincubated with 300 nm β clamp and 16 nm γ complex (diamonds), or mock preincubated (β clamp and γ complex omitted) (closed circles) at low ionic strength to ensure a proper assembly of β clamp onto DNA (“Experimental Procedures”). Control A·T homoduplex was preincubated with 300 nm β and 16 nm γ complex (open triangles) or mock preincubated (inverted closed triangles) in a similar manner. The products were used immediately for MutH activation assays (“Experimental Procedures”). The error bars represent one standard deviation for three independent experiments. B, experimental procedures and symbols are as in A except the substrate was a 5′ G-T supercoiled hemimethylated heteroduplex with d(GATC) modification on the viral DNA strand.
Because the β clamp is assembled at a double strand-single strand junction with a preferred orientation (32), the potential for interaction could depend on the relative orientations with which β and the repair components of interest are loaded onto the helix. For example, previous studies (11) have indicated that the MutS-binding motif of β is located on one face of the clamp, implying that the orientation of loaded β could affect the potential for MutS·β complex formation. To evaluate possible β orientation effects on MutH activation, we loaded the clamp onto hemimethylated heteroduplex/homoduplex DNAs that contained a single strand break 3′ to the mismatch, substrates onto which β is expected to be assembled with an orientation that is unique relative to the absolute orientation of the DNA molecule. Possible effects of preloaded β on MutH activation were then evaluated. As shown in Fig. 3, the efficiency of mismatch-dependent MutH activation was unaffected by the preloaded clamp, regardless of which heteroduplex strand was subject to MutH incision. These results indicate that the β clamp is not required for the initiation step of methyl-directed mismatch repair.
β Is Required for Methyl-directed Mismatch Repair
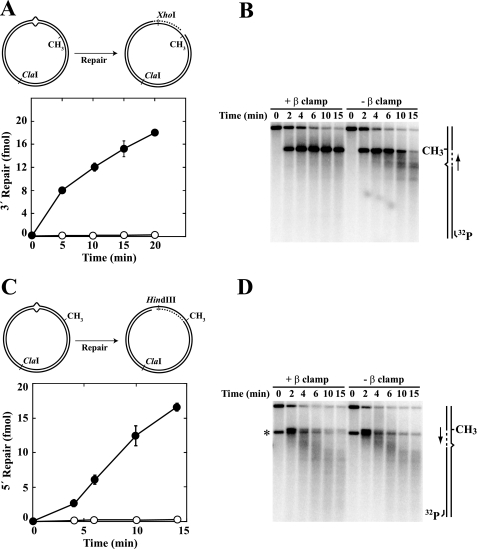
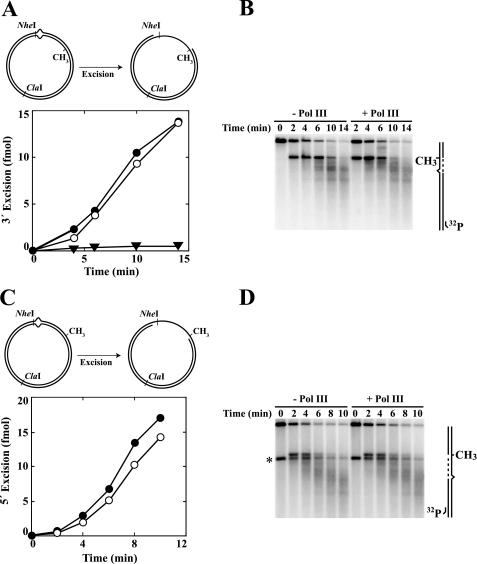
Although β is dispensable for MutH activation, omission of the clamp from reconstituted repair reactions abolished correction of supercoiled G-T heteroduplexes directed by an unmethylated d(GATC) sequence located either 3′ or 5′ to the mispair (Fig. 4, A and C). Failure to repair these DNAs in the absence of β could be due to a defect in excision, repair DNA synthesis, or both. To distinguish between these possibilities, we used indirect end labeling to visualize DNA termini produced during the course of incubation in the presence or absence of the clamp. These experiments revealed the presence of excision intermediates when β was omitted from repair reactions (Fig. 4, B and D). Furthermore, the patterns of excision intermediates produced with a 5′ heteroduplex were identical in the absence or presence of β clamp (Fig. 4D). A similar comparison is not possible for the 3′ heteroduplex (Fig. 4B) because repair DNA synthesis that occurs in the presence of β on this substrate restores the excision products to their original length.
FIGURE 4.
β clamp is essential for the repair synthesis step of mismatch correction. Supercoiled 3′ (A and B; dGATC methylation on complementary strand) or 5′ (C and D; methylation on viral strand) G-T heteroduplexes were incubated with MutS, MutL, MutH, DNA helicase II, SSB, Pol III core, γ complex, dNTPs, and exonuclease I (3′ heteroduplex) or RecJ (5′ heteroduplex) in the presence or absence of β clamp (“Experimental Procedures”). The G-T mismatch in substrate DNAs resides within overlapping HindIII and XhoI sites, rendering the heteroduplexes resistant to both enzymes (30). Repair of 3′ (A) or 5′ (C) heteroduplexes confers XhoI or HindIII sensitivity, respectively (closed circles, + β clamp; open circles, β omitted from reactions). The error bars correspond to one standard deviation for three independent measurements. The reaction products were also digested with ClaI and analyzed by indirect end labeling to visualize excision repair end points produced during the reaction (“Experimental Procedures”) as illustrated in the diagrams on the right. Excision on the unmethylated DNA strand proceeds 3′ to 5′ (3′ heteroduplex) or 5′ to 3′ (5′ heteroduplex) toward the mismatch from the MutH-produced strand break at the d(GATC) site (33). The arrows adjacent to the diagrams indicate the direction of DNA synthesis on the repaired strand. The asterisk in D indicates a DNA terminus that was present in a subset of 5′ heteroduplex molecules because of incomplete ligation of a strand break during substrate preparation.
Pol III Components Are Dispensable for Mismatch-provoked Excision
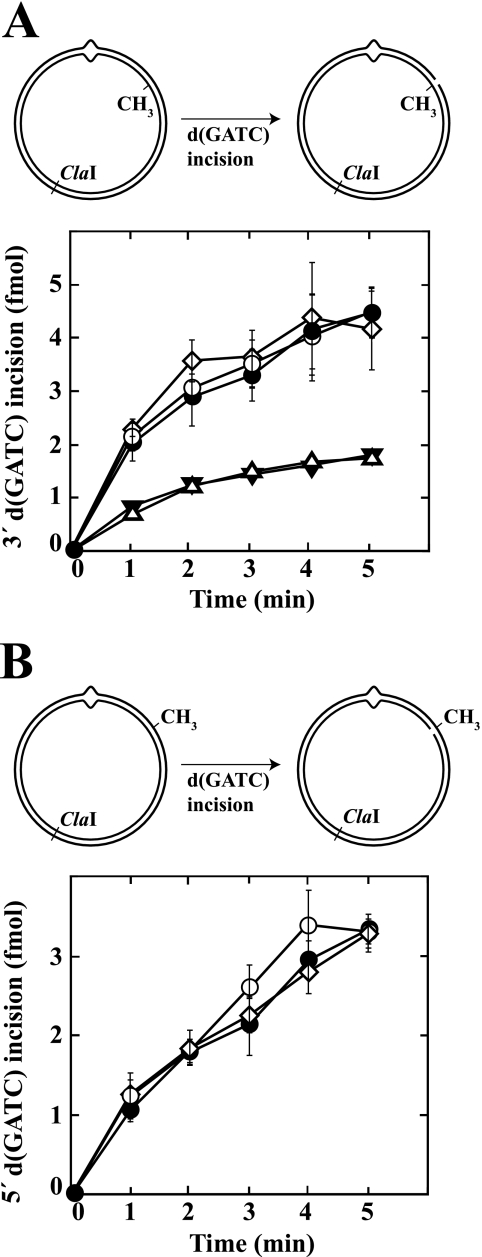
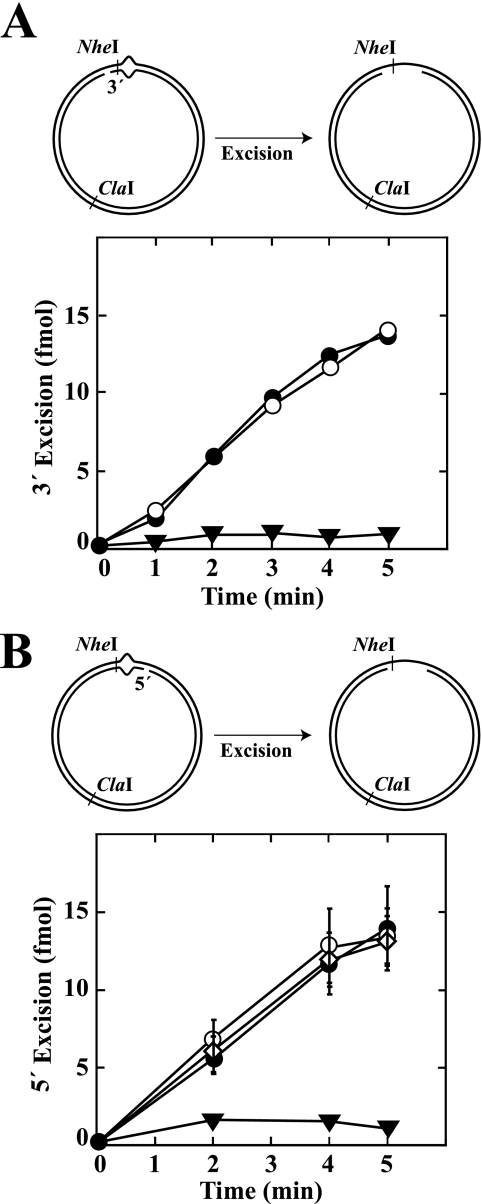
To further assess potential involvement of Pol III holoenzyme components in the excision step of repair, hemimethylated supercoiled 3′ or 5′ G-T heteroduplex substrates were preincubated with β, γ complex, and Pol III core to permit clamp loading or mock preincubated. The products were used immediately for excision assay in the presence of dATP, dGTP, dCTP, and ddTTP (“Experimental Procedures”). The latter nucleotide has been shown to inhibit the repair DNA synthesis step of methyl-directed repair (33). As shown in Fig. 5 (A and C), the rates and extents of excision determined on 3′ and 5′ heteroduplexes were similar whether Pol III components were present or not. Furthermore, the distributions of excision tract end points as determined by indirect end labeling were essentially identical in the absence or presence of Pol III activities (Fig. 5, B and D).
FIGURE 5.
β clamp does not influence the excision step of methyl-directed mismatch repair. Supercoiled 3′ (A and B; dGATC methylation on complementary strand) or 5′ (C and D; methylation on viral strand) G-T heteroduplexes were preincubated with β clamp, γ complex, and Pol III core (A and C, open circles) or mock preincubated (A and C, closed circles). Supercoiled 3′ A·T homoduplex control DNA was also subjected to mock preincubation (A, inverted triangles). The products were used immediately for mismatch-provoked excision assays, which contained MutS, MutL, DNA helicase II, SSB, dATP, dGTP, dCTP, ddTTP, and exonuclease I (3′ heteroduplex/homoduplex) or RecJ exonuclease (5′ heteroduplex). Substrates contain an NheI site 5 bp from the location of the mispair, which is rendered resistant to cleavage by mismatch-provoked excision (“Experimental Procedures”). Extents of excision scored by this assay are shown in A and C. Heteroduplex reaction products were also digested with ClaI and analyzed by indirect end labeling to visualize excision end points produced in the absence or presence of Pol III components (B and D). The asterisk in D indicates a DNA terminus that was present in a subset of 5′ heteroduplex molecules because of incomplete ligation of a strand break during substrate preparation.
We also examined the kinetics of excision in the presence of ddTTP using open circular heteroduplexes that had been preincubated with β and γ complex; preincubated with β, γ complex, and Pol III core; or mock preincubated. On such DNAs, β is expected to load in orientation-dependent fashion (32), and excision is MutH-independent. However, as shown in Fig. 6, no significant effects of Pol III components on excision kinetics were observed. Thus, Pol III holoenzyme or the loaded form of the β clamp does not play a significant role in the excision step of the mismatch repair as judged by the assays used here.
FIGURE 6.
β clamp does not influence the nick-directed mismatch excision. Open circular G-T heteroduplex DNAs (closed circles, open circles, and open diamonds), or control A·T homoduplexes (inverted triangles) contained a single-strand break in the complementary strand 128 bp 3′ (A) or 128 bp 5′ (B) to the location of the mismatch. Nicked DNAs were preincubated with β clamp and γ complex (open circles); preincubated with β clamp, γ complex and Pol III core (open diamonds); or mock preincubated (closed circles and inverted triangles). Products were used immediately for mismatch-provoked excision assays, which contained MutS, MutL, DNA helicase II, SSB, dATP, dGTP, dCTP, ddTTP, and exonuclease I (3′ strand break) or RecJ exonuclease (5′ strand break). Excision was scored by NheI resistance assay (Fig. 5 and “Experimental Procedures”). The error bars for the heteroduplex with a 5′ strand break correspond to one standard deviation for three independent experiments. The results shown for the heteroduplex with a 3′ strand break are the averages of two determinations.
MutSN Mutants
Two putative β binding motifs have been identified within MutS: one located near the C terminus and a second in N-terminal part of the protein. Deletion of the C-terminal motif has shown it to be nonessential as judged by mutability assay, but multiple Ala substitutions within the N-terminal motif (15QQYLRL20 to 15AAYAAL20; designated MutSN) confer a strong in vivo defect in mismatch repair (12). We have confirmed this finding by determining mutation rates to rifampicin resistance of a mutSΔ strain and an otherwise isogenic strain harboring mutSΝ on a plasmid vector, which do not differ significantly (1.8 × 10−7 and 1.1 × 10−7/generation, respectively).
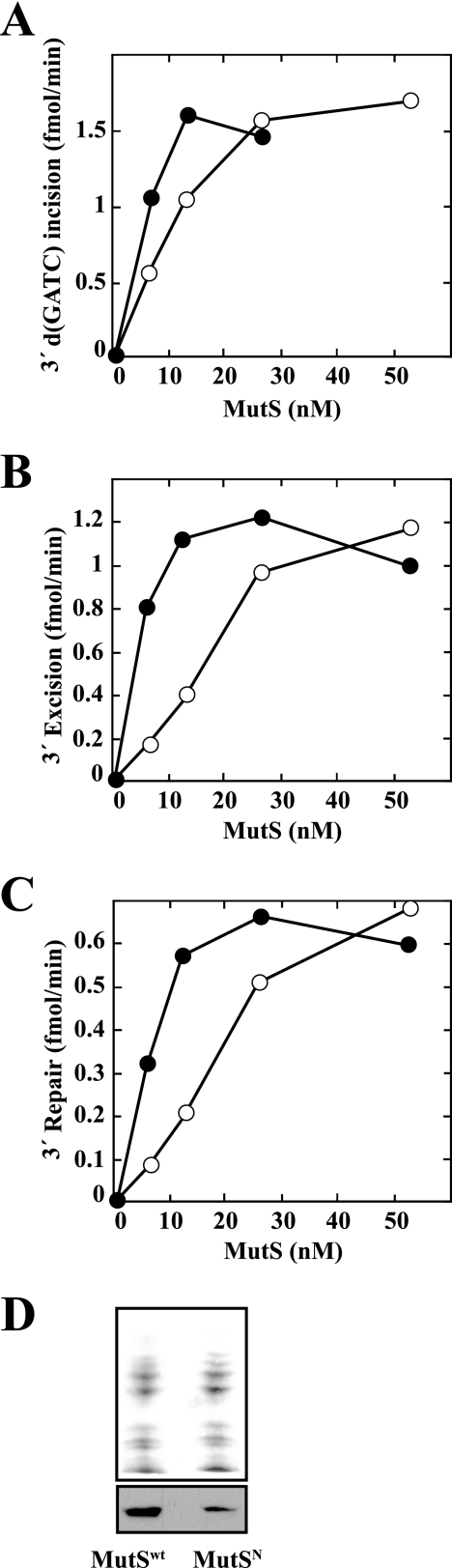
We compared MutS and MutSN for their activities in MutH activation, excision, and mismatch repair. As judged by initial rate measurements, the specific activity of MutSN is ∼65% of that of native MutS in MutH activation assay and ∼35% of that of the wild type protein in mismatch-provoked excision and repair assays (Fig. 7, A–C). We have also found that isolated MutSN is unstable to prolonged storage. The mutant protein also appears to be unstable in the cell as judged by Western blot of extracts derived from otherwise isogenic cells expressing MutS or MutSN. Such experiments have indicated levels of MutSN ranging from 20 to 50% of those observed for the wild type protein (Fig. 7D). The mutSN phenotype may thus be due to instability coupled with a general activity defect.
FIGURE 7.
Activity comparisons for wild type MutS or MutSN. A, supercoiled 3′ G-T hemimethylated heteroduplex (Fig. 2A) was incubated with MutH, MutL, and variable concentrations of wild type MutS (closed circles) or MutSN (open circles) in the absence of Pol III components. The reactions were sampled as a function of time and initial rates of d(GATC) incision determined (“Experimental Procedures”). B, supercoiled 3′ G-T hemimethylated heteroduplex was incubated with MutH, MutL, DNA helicase II, SSB, exonuclease I, and variable concentrations of wild type MutS (closed circles) or MutSN (open circles) under excision conditions (“Experimental Procedures”) except that Pol III components, ddTTP, and dNTPs were omitted. The reactions were sampled as a function of time to determine initial rates of excision, which were quantified by NheI resistance assay (Fig. 5). C, supercoiled 3′ G-T hemimethylated heteroduplex was incubated under repair conditions (“Experimental Procedures”) with MutL, MutH, DNA helicase II, SSB, exonuclease I, Pol III core, β clamp, γ complex, and MutS (closed circles) or MutSN (open circles) as indicated. The samples were taken as a function of time and repair quantified by XhoI-sensitivity assay (Fig. 4A) to determine initial rates of repair. D, extracts of E. coli BT199ΔmutS2K6 (harboring pET-mutSwt or pET-mutSN) were analyzed for MutS protein by Western blot as described under “Experimental Procedures.” Ponceau S staining of the Western membrane was used as a loading control.
Protein-Protein Interactions That Link Replication Machinery and the Mismatch Repair System
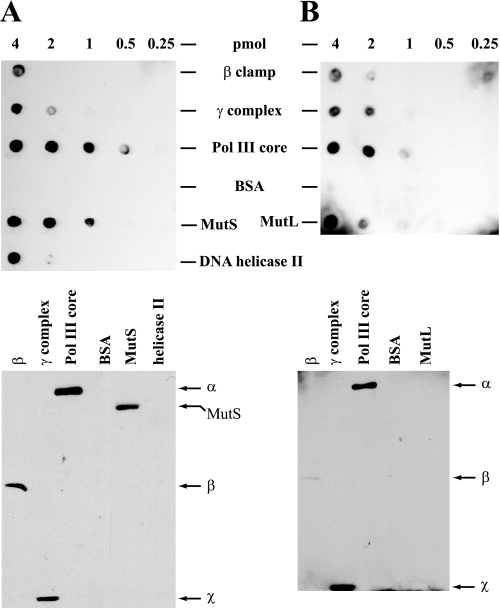
In view of our failure to observe any significant effects of β on MutH activation or excision steps of methyl-directed mismatch repair, we re-evaluated interactions of MutS and MutL with β clamp and with the other components of Pol III holoenzyme, using a method distinct from those employed in the previous study (12). As shown in Fig. 8 (upper panels), Far Western analysis confirmed the ability of MutS and MutL to interact with β, as well as several other interactions that have been previously documented in this system including the interaction of MutS with MutL (34), MutL with DNA helicase II (35, 36), and MutL with γ complex (37). In addition, this analysis also indicated interaction of MutS with γ complex and strong interaction of both MutS and MutL with Pol III core.
FIGURE 8.
Interactions of MutL and MutS with components of Pol III holoenzyme. β clamp, γ complex, Pol III core, BSA, MutS, MutL, and DNA helicase II were applied as indicated to nitrocellulose membranes (upper panels). Alternatively, proteins samples were subjected to SDS-PAGE on 10% gels (lower panels; 4-pmol sample load) followed by transfer to nitrocellulose membranes. The membranes were used for far Western analysis by incubation with MutL (A) or MutS (B), followed by immunochemical visualization of membrane-bound MutL and MutS (“Experimental Procedures”). SDS gel species that bind MutL and MutS are indicated to the right of each gel transfer, with identification based on parallel gels that were stained with Coomassie Blue. No signals were observed with otherwise identical membranes when MutS or MutL incubation steps were omitted (not shown).
To clarify subunit involvement in these interactions, β, γ complex, Pol III core, MutS, MutL, and helicase II were subjected to SDS gel electrophoresis, and resolved components were transferred to nitrocellulose membranes, which were used as templates for Far Western incubation with MutL or MutS (Fig. 8, lower panels). Strong interaction of both MutL and MutS was observed with β and with the α catalytic subunit of Pol III core. However, several interactions that were observed when native proteins were spotted on nitrocellulose (Fig. 8, upper panels) were not seen or were only weakly evident in the SDS gel transfers, possibly because of the denatured state of transferred proteins. The possible significance of this set of interactions will be considered below.
DISCUSSION
Sliding clamp involvement in mismatch repair has been the subject of extensive analysis. Popular models for clamp function in mismatch repair have invoked clamp-based recruitment of a MutS protein to sites of replication (11, 12, 38), stabilization of a MutS·mismatch complex (39–41), or clamp-based linkage of excision and repair DNA synthesis steps of the reaction (42). Nevertheless, inactivation of the proliferating cell nuclear antigen interaction motif of yeast MutSα results in only a modest mutability increase (39, 43), and deletion of this motif from human MutSα is without significant effect on the initiation or excision steps of in vitro mismatch correction, although it does confer a partial defect in 5′-directed repair when the complete reaction is scored (44).
As judged by in vitro assay using covalently closed or nicked circular model heteroduplexes, we have found that the β sliding clamp has little if any effect on the initiation or excision steps of methyl-directed mismatch repair. However, because β is required for overall repair as scored using the same DNAs, we infer that the clamp plays an essential role during the repair DNA synthesis stage of mismatch correction. It is noteworthy that our findings do not exclude a role for β in providing a link between the excision and repair synthesis steps of mismatch correction, particularly in the case of 5′ heteroduplex repair where excision and repair synthesis could occur in a concerted manner.
As discussed above, two potential β interaction motifs have been identified within E. coli MutS (11, 12). A C-terminal MutS element is a bona fide β interaction motif, conferring strong interaction with the clamp in vitro. Deletion of this motif (MutSC) abolishes in vitro interaction of the two proteins but does not result in elevated mutability in vivo (12). Although synthetic peptides containing the N-terminal motif interact with β, Ala substitution mutagenesis of this motif (MutSN) does not alter in vitro interaction of β with native MutS in the absence of DNA. However, the finding that the mutSN mutation results in hypermutability has led to the proposal that this sequence element may support β interaction, but only when the two proteins are DNA-bound. Our results suggest that this is not the case and that the biological phenotype associated with the mutSN mutation is the probable result of instability coupled with a general activity defect. It therefore seems likely that the MutS-β interaction is largely mediated via the C-terminal MutS motif and that this interaction is of limited significance during the early steps of methyl-directed mismatch repair.
Thus, as in the MutSα-dependent eukaryotic pathway where genetic and biochemical studies have indicated a limited contribution of the MutSα-proliferating cell nuclear antigen clamp interaction to mismatch repair (39, 43, 44), interaction of E. coli MutS with β clamp is not required for methyl-directed mismatch correction as judged by in vitro assay. Our biochemical experiments have also failed to reveal significant contributions of the γ complex clamp loader or Pol III core components to the initiation and excision steps of the methyl-directed mismatch correction. Despite these observations, we have confirmed previous findings that β interacts specifically with both MutS and MutL (11, 12) and that γ complex interacts with MutL (37). In addition we have shown that the γ complex interacts specifically with MutS and that Pol III core interacts with both MutS and MutL. In view of the role of mismatch repair in replication fidelity, such interactions may not be surprising, but our failure to identify the functional consequences of such interactions was unexpected.
We have considered several explanations for these puzzling findings. Because the biochemical assays that we use to score mismatch repair rely on nonreplicating model heteroduplexes, our in vitro experiments may not accurately reproduce repair events that occur at the replication fork insofar as such interactions are concerned. An alternate view posits that our biochemical results are providing an accurate indication of the significance of these interactions in mismatch repair. As noted above, the limited in vitro consequences of inactivation of the proliferating cell nuclear antigen-binding site of human MutSα are consistent with the genetic consequences of inactivation of this motif within the yeast protein, and a similar consistency of biochemical and genetic findings applies if the MutS C-terminal β interaction motif is solely responsible for MutS-clamp interaction, as has been indicated by experiments performed in the absence of DNA (12). In this case, interactions that have limited implications for repair as deduced by biochemical methods could nevertheless be sufficiently significant in biological terms to confer selective advantage.
There is, however, another way to think about the interaction of mismatch repair and replication activities. Like the study described here, previous work on this issue has been restricted to the significance of these interactions in the context of mismatch repair. A plausible alternative is that the functional consequences of these interactions are primarily manifested at the level of replication, providing a regulatory mechanism whereby mismatch repair proteins modulate fork activity in response to DNA lesions. Such a mechanism could be valuable for several reasons. It would permit replication error removal to be integrated into the context of replication fork events, an effect that might be particularly useful in those organisms where termini at the fork may provide the strand signals that direct mismatch repair (1, 45). It could in principle also function to regulate fork movement in response to a transient elevation of DNA biosynthetic errors such as those that may arise from transient nucleotide pool imbalances or chemical damage to exposed bases within template strands. The possibility of mismatch repair modulation of fork activities in response to chemical lesions may be particularly worth considering in the context of the mammalian mismatch repair system, which is known to participate in checkpoint and apoptotic responses to certain classes of DNA damage (1, 3, 46, 47).
This work was supported, in whole or in part, by National Institutes of Health Grant GM23719.
- Pol
- polymerase
- BSA
- bovine serum albumin
- SSB
- single-strand binding protein.
REFERENCES
- 1.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 2.Li L. S., Morales J. C., Hwang A., Wagner M. W., Boothman D. A. (2008) J. Biol. Chem. 283, 21394–21403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh P., Yamane K. (2008) Mech. Ageing Dev. 129, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. (1983) Genetics 104, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahue R. S., Au K. G., Modrich P. (1989) Science 245, 160–164 [DOI] [PubMed] [Google Scholar]
- 6.Cooper D. L., Lahue R. S., Modrich P. (1993) J. Biol. Chem. 268, 11823–11829 [PubMed] [Google Scholar]
- 7.Burdett V., Baitinger C., Viswanathan M., Lovett S. T., Modrich P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanathan M., Burdett V., Baitinger C., Modrich P., Lovett S. T. (2001) J. Biol. Chem. 276, 31053–31058 [DOI] [PubMed] [Google Scholar]
- 9.Au K. G., Welsh K., Modrich P. (1992) J. Biol. Chem. 267, 12142–12148 [PubMed] [Google Scholar]
- 10.Kelman Z., O'Donnell M. (1995) Annu. Rev. Biochem. 64, 171–200 [DOI] [PubMed] [Google Scholar]
- 11.López de Saro F. J., O'Donnell M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8376–8380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López de Saro F. J., Marinus M. G., Modrich P., O'Donnell M. (2006) J. Biol. Chem. 281, 14340–14349 [DOI] [PubMed] [Google Scholar]
- 13.Blackwell L. J., Bjornson K. P., Allen D. J., Modrich P. L. (2001) J. Biol. Chem. 276, 34339–34347 [DOI] [PubMed] [Google Scholar]
- 14.Spampinato C., Modrich P. (2000) J. Biol. Chem. 275, 9863–9869 [DOI] [PubMed] [Google Scholar]
- 15.Welsh K. M., Lu A. L., Clark S., Modrich P. (1987) J. Biol. Chem. 262, 15624–15629 [PubMed] [Google Scholar]
- 16.Runyon G. T., Wong I., Lohman T. M. (1993) Biochemistry 32, 602–612 [DOI] [PubMed] [Google Scholar]
- 17.Kong X. P., Onrust R., O'Donnell M., Kuriyan J. (1992) Cell 69, 425–437 [DOI] [PubMed] [Google Scholar]
- 18.Naktinis V., Onrust R., Fang L., O'Donnell M. (1995) J. Biol. Chem. 270, 13358–13365 [PubMed] [Google Scholar]
- 19.Onrust R., Finkelstein J., Naktinis V., Turner J., Fang L., O'Donnell M. (1995) J. Biol. Chem. 270, 13348–13357 [DOI] [PubMed] [Google Scholar]
- 20.Studwell-Vaughan P. S., O'Donnell M. (1993) J. Biol. Chem. 268, 11785–11791 [PubMed] [Google Scholar]
- 21.Pluciennik A., Modrich P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12709–12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genschel J., Modrich P. (2009) J. Biol. Chem. 284, 21536–21544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao N., Leu F. P., Anjelkovic J., Turner J., O'Donnell M. (2000) J. Biol. Chem. 275, 11440–11450 [DOI] [PubMed] [Google Scholar]
- 24.Dzantiev L., Constantin N., Genschel J., Iyer R. R., Burgers P. M., Modrich P. (2004) Mol. Cell 15, 31–41 [DOI] [PubMed] [Google Scholar]
- 25.Genschel J., Bazemore L. R., Modrich P. (2002) J. Biol. Chem. 277, 13302–13311 [DOI] [PubMed] [Google Scholar]
- 26.de Lorenzo V., Timmis K. N. (1994) Methods Enzymol. 235, 386–405 [DOI] [PubMed] [Google Scholar]
- 27.Su S. S., Modrich P. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 5057–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen C. S., Eberl L., Sanchez-Romero J. M., Givskov M., Molin S., De Lorenzo V. (1995) J. Bacteriol. 177, 52–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lea D. E., Coulson C. A. (1949) J. Genet. 49, 264–285 [DOI] [PubMed] [Google Scholar]
- 30.Su S. S., Lahue R. S., Au K. G., Modrich P. (1988) J. Biol. Chem. 263, 6829–6835 [PubMed] [Google Scholar]
- 31.Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. (1996) Genes Cells 1, 101–113 [DOI] [PubMed] [Google Scholar]
- 32.Georgescu R. E., Kim S. S., Yurieva O., Kuriyan J., Kong X. P., O'Donnell M. (2008) Cell 132, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grilley M., Griffith J., Modrich P. (1993) J. Biol. Chem. 268, 11830–11837 [PubMed] [Google Scholar]
- 34.Grilley M., Welsh K. M., Su S. S., Modrich P. (1989) J. Biol. Chem. 264, 1000–1004 [PubMed] [Google Scholar]
- 35.Hall M. C., Jordan J. R., Matson S. W. (1998) EMBO J. 17, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi M., Dao V., Modrich P. (1998) J. Biol. Chem. 273, 9197–9201 [DOI] [PubMed] [Google Scholar]
- 37.Li F., Liu Q., Chen Y. Y., Yu Z. N., Zhang Z. P., Zhou Y. F., Deng J. Y., Bi L. J., Zhang X. E. (2008) Mutat. Res. 637, 101–110 [DOI] [PubMed] [Google Scholar]
- 38.Kleczkowska H. E., Marra G., Lettieri T., Jiricny J. (2001) Genes Dev. 15, 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flores-Rozas H., Clark D., Kolodner R. D. (2000) Nat. Genet. 26, 375–378 [DOI] [PubMed] [Google Scholar]
- 40.Lau P. J., Kolodner R. D. (2003) J. Biol. Chem. 278, 14–17 [DOI] [PubMed] [Google Scholar]
- 41.Simmons L. A., Davies B. W., Grossman A. D., Walker G. C. (2008) Mol. Cell 29, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson R. E., Kovvali G. K., Guzder S. N., Amin N. S., Holm C., Habraken Y., Sung P., Prakash L., Prakash S. (1996) J. Biol. Chem. 271, 27987–27990 [DOI] [PubMed] [Google Scholar]
- 43.Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A. (2000) J. Biol. Chem. 275, 36498–36501 [DOI] [PubMed] [Google Scholar]
- 44.Iyer R. R., Pohlhaus T. J., Chen S., Hura G. L., Dzantiev L., Beese L. S., Modrich P. (2008) J. Biol. Chem. 283, 13310–13319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlov Y. I., Mian I. M., Kunkel T. A. (2003) Curr. Biol. 13, 744–748 [DOI] [PubMed] [Google Scholar]
- 46.Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 47.Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]