Abstract
Amyloid β-protein (Aβ) assemblies are thought to play primary roles in Alzheimer disease (AD). They are considered to acquire surface tertiary structures, not present in physiologic monomers, that are responsible for exerting toxicity, probably through abnormal interactions with their target(s). Therefore, Aβ assemblies having distinct surface tertiary structures should cause neurotoxicity through distinct mechanisms. Aiming to clarify the molecular basis of neuronal loss, which is a central phenotype in neurodegenerative diseases such as AD, we report here the selective immunoisolation of neurotoxic 10–15-nm spherical Aβ assemblies termed native amylospheroids (native ASPDs) from AD and dementia with Lewy bodies brains, using ASPD tertiary structure-dependent antibodies. In AD patients, the amount of native ASPDs was correlated with the pathologic severity of disease. Native ASPDs are anti-pan oligomer A11 antibody-negative, high mass (>100 kDa) assemblies that induce degeneration particularly of mature neurons, including those of human origin, in vitro. Importantly, their immunospecificity strongly suggests that native ASPDs have a distinct surface tertiary structure from other reported assemblies such as dimers, Aβ-derived diffusible ligands, and A11-positive assemblies. Only ASPD tertiary structure-dependent antibodies could block ASPD-induced neurodegeneration. ASPDs bind presynaptic target(s) on mature neurons and have a mode of toxicity different from those of other assemblies, which have been reported to exert their toxicity through binding postsynaptic targets and probably perturbing glutamatergic synaptic transmission. Thus, our findings indicate that native ASPDs with a distinct toxic surface induce neuronal loss through a different mechanism from other Aβ assemblies.
Introduction
Neurodegenerative diseases, such as Alzheimer disease (AD),2 Parkinson disease, prion diseases, and the polyglutamine diseases, arise from abnormal protein interactions in the central nervous system (1). In these diseases, complex multistep processes of protein conformational change and accretion produce various nonfibrillar assemblies, leading finally to fibrils (1–5). Recent studies have suggested that the early assemblies in this process might be the most toxic, possibly through the exposure of buried moieties and the formation of surface tertiary structures not present in physiologic monomers (6). These surface tertiary structures could mediate abnormal interactions with other cellular components (1).
In AD, extensive studies have suggested that accumulation of amyloid β-protein (Aβ), a physiologic derivative of amyloid precursor protein (APP), plays a primary pathogenic role (7–9). Various forms of assemblies ranging in mass from dimers up to multimers of ∼1 MDa have been reported as neurotoxins (10–13) as follows: protofibrils (14); dimers/trimers (natural low-n oligomers) (15); 3–24-mer Aβ-(1–42) assemblies termed Aβ-derived diffusible ligands (ADDLs) (16); 12-mers termed globulomers (17) or Aβ*56 (18); 15–20-mer Aβ assemblies termed Aβ oligomers (AβOs) (19); and 150-mer or higher assemblies termed β-sheet intermediates (20). Whether or not they share a common surface, the tertiary structure responsible for toxicity remains unsettled; some of these assemblies are detected by specific antibodies (17, 21), whereas others are detected by a polyclonal A11 antibody (18, 19) that is reported to recognize epitopes associated with a certain oligomer state of amyloids regardless of their amino acid sequence (22). However, these assemblies, which differ in origin, mass, and toxic activity, mostly bind to postsynapses, leading to synaptic impairment (17–19, 23, 24). They are also suggested to play a role in synaptic impairment in AD model mice carrying human APP (17, 18, 25), which retain early features of AD such as amyloid plaques, synaptic loss, and mild memory deficits (26, 27). These observations collectively suggest that these assemblies play a role in AD pathogenesis by causing synaptic impairment. On the other hand, it remains largely unknown how, after the synaptic impairment, these assemblies cause subsequent neuronal loss in human AD brains. One reason is that no overt neuronal cell loss has been observed in most APP transgenic mice (except APP23 mice (28, 29)), even in the presence of these assemblies (26, 27). Another reason is that, as for the nonfibrillar Aβ assemblies actually present in human AD brains, Aβ dimers that induce synaptic impairment and not neuronal loss were recently isolated (30), but Aβ assemblies that directly cause neuronal loss have not yet been isolated either from AD patients or from the mice. Because soluble fractions of brains from humans with AD have been reported to contain Aβ assemblies ranging in size from dimers to polymers larger than 100 kDa (31), which appear to correlate with dementia (32, 33), Aβ assemblies responsible for neuronal loss might be present in the soluble fractions of AD brains. As has recently been shown clinically and diagnostically (34–37), neuronal loss plays an important role in cognitive deterioration of AD patients, so we aimed to isolate toxic Aβ assemblies from the soluble fractions of AD brains.
As a first step to isolate such Aβ assemblies in vivo, we have previously prepared highly toxic spherical Aβ assemblies termed “amylospheroids” (ASPDs) in vitro (38). Notably, ASPDs are considered not to be intermediates in the pathway leading to fibrils, because ASPDs were not incorporated into mature fibrils and continued to exist after fibril formation (13, 38). They also differ from protofibrils and ADDLs in morphology and size (11, 13, 38).
Here, we generated ASPD tertiary structure-dependent antibodies and used them to selectively immunoisolate a human counterpart of ASPDs (native ASPDs) from patients with AD or dementia with Lewy bodies (DLB). To distinguish native ASPDs from in vitro-produced ASPDs, the latter is hereafter referred to as synthetic ASPDs. Native ASPDs are A11-negative, high mass Aβ assemblies that induce degeneration of human neuronal cells in vitro, particularly those with mature character, and they differ in mass, surface tertiary structure, and neurotoxicity mechanism from other reported nonfibrillar Aβ assemblies (summary in supplemental Table S1).
EXPERIMENTAL PROCEDURES
Aβ Source
Aβ-(1–40) peptides were synthesized using N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry on an Applied Biosystems model 433A peptide synthesizer and purified (38). Their structure and purity were confirmed using quantitative amino acid analysis, analytic high pressure liquid chromatography, and matrix-assisted laser desorption/ionization-time-of-flight/mass spectrometry (MALDI-TOF/MS; Ultraflex II, Bruker Daltonics). The purified Aβ-(1–40) was lyophilized, dissolved in 35% (v/v) acetonitrile in 0.1% (v/v) trifluoroacetic acid (∼50 nmol/tube), and lyophilized. This step was repeated twice. Aβ-(1–42) peptides (25 mg/ampoule; Bachem lots 0552992 and 1000255) were completely dissolved in ∼54 ml of 1,1,1,3,3,3-hexafluoro-2-propanol (Aldrich) by incubating the peptide solution overnight at 4 °C and then for 3 h at 37 °C and finally lyophilized (∼40 nmol/tube). This step was repeated two or three times. The lyophilized peptides were kept at −20 °C.
Preparation and Purification of Synthetic ASPDs
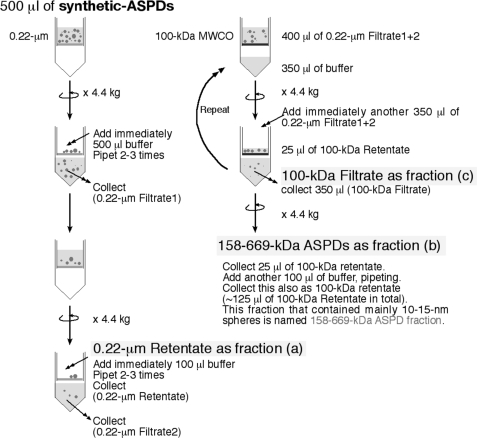
Synthetic ASPDs were prepared in vitro either from 50 μm solution of Aβ-(1–40) (0.5× Dulbecco's phosphate-buffered saline without Ca2+ and Mg2+ (PBS); Nissui Pharmaceutical Co. Ltd.) or of Aβ-(1–42) (either 0.5× PBS or F12 buffer without l-glutamine and phenol red) by slowly rotating the solution (5–7 days for Aβ-(1–40); 14 h for Aβ-(1–42)), as described previously (38). At concentrations below a critical fibril-forming concentration (∼100 μm) (39), spherical Aβ assemblies (5–20 nm in diameter for Aβ-(1–40); 5–25 nm for Aβ-(1–42); >85% 10–15 nm spheres), with rare fibril-like structures, were usually produced. The most toxic ASPDs (prepared either from Aβ-(1–40) or Aβ-(1–42)) were previously identified as 10–15-nm spheres recovered by glycerol gradient centrifugation in the fraction migrating near the thyroglobulin (669 kDa) standard (38). Further analysis of standard proteins using this glycerol gradient sedimentation assay revealed that the mass of the most toxic ASPDs is approximately equal to that of aldolase (158 kDa) but does not exceed that of thyroglobulin (669 kDa).3 Therefore, the most toxic ASPDs were purified as retentates by using 100-kDa molecular mass cutoff filters (Ultrafree-MC, Millipore) to remove lower mass Aβ assemblies. In some experiments, including mature neuron-binding assays, the most toxic ASPD fraction was also purified by two-step filtrations (see Scheme 1). Studies using transmission electron microscopy (TEM) revealed that 10–15-nm spheres were predominantly recovered in the most toxic ASPD fraction (termed “158–669-kDa ASPDs”) that passed through 0.22-μm filters but were retained on 100-kDa molecular mass cutoff filters (data not shown). Although these 10–15-nm spheres were hardly detectable in 100-kDa filtrates, smaller particles with a diameter of 5–6 nm were present in 100-kDa filtrates. A very small amount of 10–15-nm ASPDs was also present in 0.22-μm retentates because they remained bound to the filter (data not shown). These TEM observations were in good agreement with the results of dot blots and toxicity assays shown in Fig. 1A. Quantitative amino acid analysis revealed that generally ∼25% of total Aβ was recovered as 158–669-kDa ASPDs. Synthetic ASPDs were prepared every week, and their quality was confirmed using dot blotting, TEM, and toxicity assays. Aβ concentration of each preparation was determined every week by quantitative amino acid analysis (Waters AccQ-Tag system) (38).
SCHEME 1.
Fractionation of the most toxic 158–669-kDa ASPDs by two-step filtrations.
FIGURE 1.
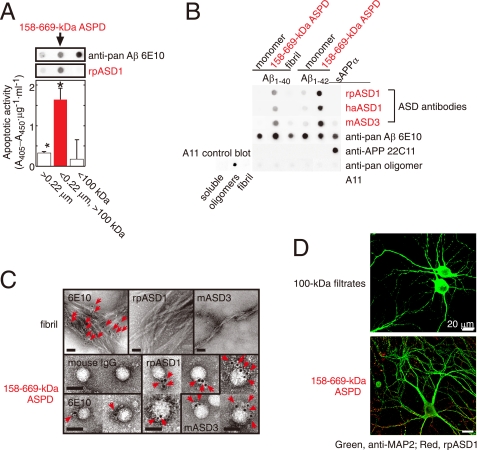
Characterization of ASD antibodies. A, evaluation of two-step filtered fractions (0.22-μm retentates, the 158–669-kDa ASPDs, and 100-kDa filtrates; see Scheme 1 under “Experimental Procedures”) by dot blotting using rpASD1 and 6E10 (upper panel) and by toxicity assays using rat primary septal cultures (lower panel; mean ± S.D.; Games-Howell post hoc test, *, p < 0.001, n = 6). B, dot blotting of Aβ and APP (5 ng/dot). Synthetic ASPDs were prepared in vitro either from Aβ-(1–40) or Aβ-(1–42) as described (7). Purified 158–669-kDa ASPD fraction was recovered in 100-kDa retentates as in A. Unlike anti-APP-(66–81) (22C11), anti-Aβ-(1–16) (6E10), or A11 antibody, ASD antibodies selectively detected synthetic ASPDs and the 158–669-kDa ASPDs. The control blot membrane for A11 was provided by Invitrogen (supplemental Experimental Procedures). C, immuno-TEM analysis. Arrows show the secondary antibody-conjugated immunogold. 6E10 detected the 158–669-kDa ASPDs weakly, probably because of its low affinity for synthetic ASPDs. rpASD1 and mASD3 showed little reactivity to fibrils but clearly detected the 158–669-kDa ASPDs. Bar, 20 nm. D, rpASD1 detected intense signals in 27-DIV mature rat hippocampal neurons treated with the 158–669-kDa ASPDs (in A) for 30 min but did not detect signals in those treated with the 100-kDa filtrates containing monomers and Aβ-(1–42) assemblies with mass <100 kDa. Z-stack images are shown (supplemental Experimental Procedures).
Preparation of Aβ Monomers and Fibrils
To prepare monomers, Aβ-(1–40) or Aβ-(1–42) lyophilizates were solubilized to 50 μm in 1,1,1,3,3,3-hexafluoro-2-propanol. The solution was incubated for 30 min at room temperature and then centrifuged at 20,400 × g for 30 min at 4 °C, and the upper 90% of the supernatant volume was collected. The monomer concentration was determined by means of quantitative amino acid analysis (38). To produce fibrils, Aβ-(1–40) was dissolved at a concentration of 100 μm in 0.5× PBS, pH 3.5. This solution was incubated without agitation at 37 °C for 2 days, after which fibrils were separated from the monomer and low mass Aβ assemblies by filtration through 100-kDa molecular mass cutoff filters. Large amounts of fibrils without ASPDs were detected reproducibly by TEM. The fibril concentration was determined by means of quantitative amino acid analysis (38). To obtain different types of fibrils for immuno-TEM analysis, fibrils were also produced by slowly rotating the above Aβ-(1–40) solution or by dissolving Aβ-(1–40) at a concentration of 350 μm in PBS, pH 7.5, followed by incubation for 5–7 days at 37 °C, with or without slow rotation (38).
Preparation of Aβ Oligomers for A11 Antibody
A test membrane, on which soluble Aβ oligomers (termed AβOs) and Aβ fibrils (1–3 μg/dot) were spotted, was produced by Dr. C. Glabe (University of California, Irvine) according to reported methods (22, 40). This test membrane was kindly provided by Invitrogen as a positive control for A11 antibody.
Preparation of ADDLs
ADDLs were produced as described previously (16). Aβ-(1–42) lyophilizates were solubilized to 5 mm in DMSO, diluted to 100 μm with F12, and incubated for 24 h at 4 °C. The solution was centrifuged at 14,000 × g for 10 min at 4 °C, and the supernatant was collected. These ADDL preparations were further purified by obtaining the flow-through fraction of 100-kDa molecular mass cutoff filters as described (21).
Human Brain Extracts
The Bioethics Committees and the Biosafety Committees of Mitsubishi Kagaku Institute of Life Sciences, Niigata University, and Kyoto University approved all experiments using human subjects. Freshly frozen brains obtained at autopsy were homogenized to 0.15 g/ml in an ice-cold extraction buffer (either 20 mm Tris-HCl, pH 7.6, 137 mm NaCl, or F12 buffer without l-glutamine and phenol red, containing 1 mm EDTA, 1 mg/ml pepstatin, and complete protease inhibitor mixture (Roche Diagnostics)) using a Potter Teflon/glass homogenizer. Soluble fractions were collected as the supernatant following centrifugation at 104,300 × g (TLA100.4) at 4 °C for 1 h. SDS-extractable insoluble fractions were obtained from the pellet by homogenizing in 2% (w/v) SDS and by 1-h gentle shaking at 37 °C, followed by centrifugation for 1 h at 10 °C. Formic acid-extractable fractions were obtained by homogenizing the SDS pellet in 70% formic acid, followed by centrifugation for 1 h. Approximate protein recoveries were 10% for soluble fractions and 90% for insoluble fractions. The possibility of artificial generation of ASPD-like structures from Aβ-(1–42) monomers or of destruction of ASPD-like structures during the extraction procedures was excluded (data not shown). More details are given in the supplemental Experimental Procedures.
Immunoprecipitations (IP)
To remove other assemblies (<100 kDa), soluble extracts from AD or NCI brains were concentrated using 100-kDa molecular mass cutoff filters (Millipore). This process was repeated until we obtained AD-derived 100-kDa retentates that contained native ASPDs at ∼10–20 μm; this was verified by dot blotting using rpASD1. IPs were performed using an immunocapturing kit 100 MB-IAC Prot G (Bruker Daltonics), according to the manufacturer's instructions, except that 3% (w/v) bovine serum albumin (Sigma A7030) was used to suppress nonspecific binding. Monoclonal ASD antibodies (haASD1 or mASD3) were used for the immunoisolation because of their high affinity for ASPDs. Captured proteins were eluted using Gentle Elution buffer (Pierce), because ASPDs retained their structure and toxicity (∼100%) after a 60-min exposure to this buffer. The amount of native ASPDs in eluates was immediately examined by dot blotting with polyclonal rpASD1, a suitable antibody for dot blot analysis. Details are given in supplemental Experimental Procedures.
Other Methods
Preparation and screening of ASD antibodies, dot blotting, Western blotting, TEM and immuno-TEM examinations, surface plasmon resonance by Biacore and competitive enzyme-linked immunosorbent assay experiments, pathologic examinations of human brains, Tg2576 mice experiments, toxicity assays, immunocytochemistry, human neuronal cells, and monkey neural progenitors and neurons, as well as statistics, are described in supplemental Experimental Procedures.
RESULTS
Production and Characterization of ASPD-specific Antibodies
To isolate native ASPDs from AD patients, we raised antibodies against ASPDs in 6 rabbits, 43 mice, and 10 hamsters. As an immunogen, synthetic ASPDs were prepared in vitro from 50 μm solutions of Aβ-(1–42) by slowly rotating the solutions for 14 h (38); they included spherical Aβ assemblies of 5–25 nm (>85% of them were 10–15-nm spheres). IgG-class antibodies were purified and named “ASD antibodies,” with prefixes indicating the source (rabbit polyclonal as rpASD1; hamster monoclonal as haASD1; mouse monoclonal as mASD3).
We examined the reactivity of ASD antibodies against the most toxic synthetic ASPD fraction separated as follows. Because the mass of the most toxic 10–15-nm ASPDs is almost equal to that of 158-kDa aldolase but does not exceed that of 669-kDa thyroglobulin in sedimentation analysis (38), synthetic ASPDs were further size-separated by means of two-step filtrations (see Scheme 1 under “Experimental Procedures”) to concentrate the most toxic 158–669-kDa ASPDs in fraction b, the fraction that passed through 0.22-μm filters but was retained on 100-kDa molecular mass cutoff filters. As expected, 158–669-kDa ASPDs recovered in fraction b included 10–15-nm spheres, as determined by TEM observation (data not shown), and were confirmed to be the most toxic species in toxicity assays (Fig. 1A).
Importantly, rpASD1 specifically detected the 158–669-kDa ASPDs in fraction b in dot blotting but had little or no cross-reactivity to fraction a 0.22-μm retentates or fraction c 100-kDa filtrates containing monomers and 5–6-nm particles, which are strongly detected by anti-pan Aβ 6E10 antibody (Fig. 1A). We also confirmed that rpASD1 did not cross-react with ADDLs (supplemental Fig. S1A). These results indicated that rpASD1 recognizes an epitope that is associated with the most toxic ASPDs but not with ADDLs.
We further characterized rpASD1 and the other ASD antibodies using the most toxic 158–669-kDa ASPDs. As shown in Fig. 1B, all ASD antibodies detected primarily the 158–669-kDa ASPDs (prepared from either Aβ-(1–42) or Aβ-(1–40)) but had very low or no cross-reactivity to sAPPα (human secreted form of APP), Aβ monomers, or Aβ fibrils, whereas 6E10 equally detected all these Aβ species and sAPPα (Fig. 1B). Consistent with this, ASD antibodies detected 10–15-nm spheres in the 158–669-kDa ASPDs but did not react with fibrils as observed with immuno-TEM under mild fixation conditions (Fig. 1C). In accordance with their ASPD preference in dot blots and immuno-TEM, the ASD antibodies showed the highest affinity for the 158–669-kDa ASPDs (Kd 10−12–10−13 m; Table 1), rather than for Aβ monomers, fibrils, or sAPPα (supplemental Table S2). These results demonstrated ASPD specificity of all the ASD antibodies. As described above, A11 antibody is a pan-oligomer polyclonal antibody that recognizes epitopes associated with an oligomer state (18, 19, 22). To our surprise, this anti-pan oligomer A11 antibody failed to detect the 158–669-kDa ASPDs (Fig. 1B). These results strongly suggested that epitopes recognized by the ASD antibodies are associated with the tertiary structure of ASPDs, which differs from that of A11-positive oligomers, such as Aβ*56 and AβOs, and from that of fibrils. To further elucidate the epitope specificity, we performed epitope mapping by means of competitive enzyme-linked immunosorbent assay using a series of pentapeptides covering the entire Aβ-(1–42) sequence. As summarized in Table 1, no single pentapeptide could compete out binding of the ASD antibodies to the 158–669-kDa ASPDs, suggesting that different Aβ regions exist in close proximity within ASPDs to form ASPD tertiary structure-dependent epitopes that are not present in a single Aβ monomer (supplemental Fig. S1B). The ASD antibodies produced only weak or no bands in Western blots under denaturing conditions (data not shown), as would be expected from the fact that they recognize ASPD tertiary structure. We finally examined whether the ASD antibodies are available for detecting ASPDs bound to mature neurons, because we have previously shown that synthetic ASPDs directly induce neuronal cell death, possibly by binding to neuronal cell surfaces (38). As shown in Fig. 1D, the ASD antibodies clearly detected synthetic ASPDs bound on mature rat hippocampal neurons (see “Experimental Procedures”) when the neurons were briefly treated with the 158–669-kDa ASPDs and fixed under mild conditions, but they did not label neurons treated with the 100-kDa filtrates, which contained monomers and 5–6-nm particles of less than 100 kDa. The characteristics of the antibodies are summarized in Table 1. All of these results demonstrate that the ASD antibodies recognize epitopes that are specific to the surface tertiary structure of ASPDs, which differ from that of ADDLs, A11-positive prefibrillar oligomers, and fibrils.
TABLE 1.
Summary of characters of ASD antibodies and anti-pan Aβ antibodies
The characters of newly established anti-ASPD antibodies (upper three rows) and previously reported anti-pan Aβ antibodies (lower two rows) are summarized. The original epitope mapping data are shown in supplemental Fig. S1B; see Fig. 1A and supplemental Fig. S3A for dot blots and supplemental Table S2 for Kd values determined by Scatchard analysis of enzyme-linked immunosorbent assay data; see Fig. 1C (except haASD1) for immuno-TEM and supplemental Fig. S5A for toxicity blockade.
| Antibody | Preference among Aβ types in dot blotting | Kd for ASPDs | Epitope map | Response to APP in dot blotting | Response to fibrils in immuno-TEM | Blockade of ASPD toxicity |
|---|---|---|---|---|---|---|
| nm | ||||||
| rpASD1 | ASPD | 0.005 | Several regionsa | − | − | + |
| mASD3 | ASPD | 0.003 | Several regionsa | ± | − | + |
| haASD1 | ASPD | 0.0005 | Could not be determinedb | − | − | − |
| 6E10 | All types | 0.2 | Aβ5–9c | + | + | − |
| 82E1 | All types | NDd | Aβ1–5c | − | ND | − |
a The binding of these antibodies to synthetic-ASPDs was most strongly inhibited by N-terminal pentapeptides of Aβ. In addition, the binding was also inhibited by specific sets of non-N-terminal pentapeptides. The data suggest that different Aβ regions exist in close proximity to form the ASPD-specific epitope.
b The binding of haASD1 to synthetic-ASPDs was not inhibited by the addition of any pentapeptide, suggesting that haASD1 recognizes a nonlinear epitope formed by noncontiguous Aβ regions.
c For 6E10 and 82E1, which did not discriminate ASPDs from other types of Aβ, complete inhibition was attained with a single pentapeptide in each case.
d ND means not determined.
ASPD-specific Antibody-stained AD Brain
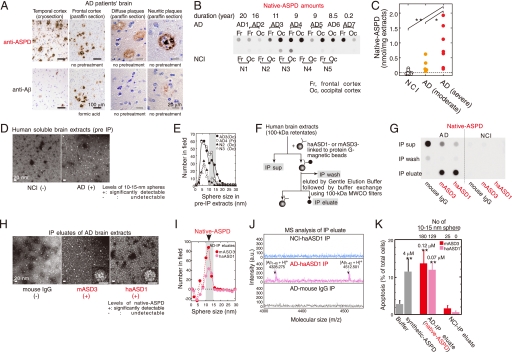
To elucidate whether synthetic ASPD-like assemblies are present in vivo, brain sections of patients with clinico-pathologically confirmed AD (41) (n = 10; age 80.4 ± 9.2 years, brain weight 964 ± 82 g, disease duration 10.1 ± 5.5 years) and those of NCI people (n = 7; age 71.3 ± 15.2 years, brain weight 1226 ± 96 g) were immunostained with ASD antibodies. The reactivity of the ASD antibodies in AD patients was strongly associated with brain regions where prominent neurodegeneration had occurred (e.g. temporal cortex, frontal cortex, and hippocampus) (Fig. 2A and supplemental Fig. S2) but was rarely observed in NCI brains (data not shown). This immunoreactivity in AD brains was associated mainly with plaques and occasionally with neurites and some microvessels and was eliminated by prior treatment of the ASD antibodies with the 158–669-kDa ASPDs (data not shown).
FIGURE 2.
Isolation of native ASPDs. A, AD brains were stained with rpASD1 (5 μg/ml) or anti-Aβ1–42 C-terminal antibody (0.5 μg/ml; 2 μg/ml for cryosections). B and C, dot blotting of 100-kDa retentates (>100 kDa) of AD or NCI brain extracts (1 μg of soluble extracts/dot) using rpASD1 (Scheffé post hoc test; **, p = 0.0011; *, p = 0.0388). Fr, frontal cortex; Oc, occipital cortex. D and E, TEM images (D) and particle analysis of 100-kDa retentates (n = 3; 10 randomly selected fields) (E). F and G, method for immunoprecipitation (IP) (F) and dot blotting (using rpASD1) of IP supernatants (sup), wash, and eluate fractions. IPs were performed using haASD1, mASD3, or mouse IgG (G). H and I, TEM images (inset, bar, 10 nm) (H) and particle analysis of IP eluates (n = 3; 15 randomly selected fields, background (a small amount of spheres <10 nm contained in eluate with buffers)-subtracted data are shown) (I). J, representative MALDI-TOF/MS data. Aβ-(1–40) and Aβ-(1–42) were detected only in native ASPDs at theoretical monoisotopic mass values (([Aβ-(1–40) + H]+, 4328 Da; [Aβ-(1–42) + H]+, 4512 Da) as observed in synthetic Aβ peptides. K, toxicity of isolated native ASPDs toward primary rat septal neurons (mean ± S.D.; Scheffé post hoc test, **, p < 0.0001, compared with buffer, n > 8) correlated with the 10–15-nm sphere number determined as in I. Neurons treated with NCI-IP eluates showed only background levels of apoptosis similar to those of neurons treated with buffers. Inset, synthetic or native ASPD amounts in Aβ monomer concentrations.
We next compared the reactivity to plaques under various conditions between the ASD antibodies and anti-pan Aβ antibodies, the most widely used antibodies for detecting fibrils in plaques. Although anti-pan Aβ antibodies labeled plaques only in formalin-fixed paraffin sections after pretreatments such as microwaving or formic acid, ASD antibody-specific reactivity was observed most strongly in cryosections and more weakly in formalin-fixed paraffin sections with or without pretreatments (except that haASD1 is available only for cryosections) (Fig. 2A). This difference in immunoreactivity to plaques suggested that anti-pan Aβ antibodies and ASD antibodies detect different structures in plaques; anti-pan Aβ antibodies detect fibrils when buried epitopes are exposed by protein-denaturing treatments, whereas ASD antibodies are considered to detect tertiary structure-dependent epitopes on putative human ASPD counterparts under conditions where the native structure of proteins is preserved. To confirm this, we performed biochemical fractionation of AD brains and examined whether these antibodies reacted with insoluble or soluble fractions. Consistent with previous data that fibrils include an insoluble core of plaques (42), anti-pan Aβ antibodies reacted mainly (>85%) with insoluble fractions of AD brains extracted with SDS or formic acid (supplemental Fig. S3A). Furthermore, this insoluble fraction produced broad smears in Western blots of Aβ, as is usually observed with fibrils (42) (supplemental Fig. S3B). In contrast, the ASD antibodies reacted only with soluble fractions of AD brains (supplemental Fig. S3A) in which the human ASPD counterpart was actually present, as described below (see under “Isolation of Native ASPD from Brains of AD Patients”). These results collectively indicate that the ASD antibodies detect a human ASPD counterpart, namely native ASPD, associated with plaques and neurites in AD brains. In subsequent work, we used monoclonal mASD3 and haASD1 for isolating ASPDs, because of their high affinity, and polyclonal rpASD1 for detecting ASPDs (except Fig. 3A; see also “Immunoprecipitations” under “Experimental Procedures”).
FIGURE 3.
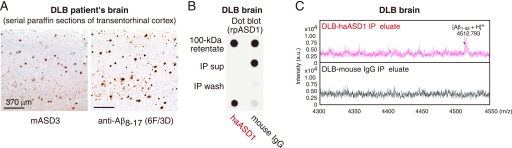
Native ASPDs exist in DLB brains. A, immunostaining using mASD3 (2.5 μg/ml) and anti-Aβ8–17 (pretreated with formic acid; 1:100; DAKO). B, IP was performed with haASD1 or mouse IgG as in Fig. 2F using 100-kDa retentates (4 μg of soluble brain extracts/IP). Dot blotting (0.04 μg/ml rpASD1) of 100-kDa retentates (2 μg of soluble brain extracts/dot), IP supernatants (sup), wash, and eluate is shown. C, representative MALDI-TOF/MS data.
Isolation of Native ASPD from Brains of AD Patients
The tissue fractionation study revealed that native ASPDs are recovered in soluble fractions of AD brains. To investigate the amount of native ASPD, we prepared soluble fractions of AD brains (n = 7; age 85.6 ± 3.1 years, brain weight 1025 ± 104 g) and NCI (n = 5; age 72.6 ± 9.5 years, brain weight 1236 ± 64 g) by means of a nondenaturing procedure using solutions of physiologic ionic strength and pH without detergents. We then obtained 100-kDa retentates of the soluble fractions to concentrate native ASPDs (larger than 100 kDa) and to eliminate other Aβ assemblies smaller than 100 kDa (as performed in Fig. 1A). The 100-kDa retentates of AD brains thus obtained had high levels of rpASD1-reactive substances, but those of NCI brains had very low or negligible reactivity (Fig. 2B). Consistent with the above data, much higher numbers of spheres sized 10–15 nm were present in 100-kDa retentates of AD patients than in those of NCI (Fig. 2, D and E). These results suggest that rpASD1-reactive 10–15-nm spheres in 100-kDa retentates of AD are native ASPD candidates. We then immunoisolated native ASPDs (Fig. 2F) from large amounts of AD-derived 100-kDa retentates using two monoclonal antibodies, haASD1 and mASD3 (Fig. 2, G–J). These antibodies were chosen for their extremely high affinity for ASPD (Kd < 10−12 m) and for their recognition of different epitopes (Table 1). Judging from the results of quantification of dot blots using rpASD1 (Fig. 2G), we obtained about 43 pmol of native ASPDs (expressed as Aβ monomer concentration) from 1 g of AD brain tissues (n = 6). The rpASD1 reactivity in the IP eluates was considered to be mostly due to the 10–15-nm spheres, because the number of spheres counted by TEM (Fig. 2H) (1.0 × 1010 10–15-nm sphere/μl estimated from the number of spheres in Fig. 2K, n = 6) was very similar to the amount of rpASD1-reactive ASPD obtained from dot blots (1.1 × 1010 native ASPD/μl based on the ASPD concentration in Fig. 2K, n = 8). This means that rpASD1-reactive 10–15-nm spheres were selectively isolated by a combination of 100-kDa retention and IP. Indeed, as shown by the TEM data (Fig. 2H), the non-ASPD small-sized spheres (<10 nm) that had been present in large amounts in 100-kDa retentates of AD and NCI were largely eliminated by the IP procedure (compare Fig. 2, I with E). Accordingly, we successfully isolated native ASPDs, consisting of 10–15-nm spheres (>95%; Fig. 2, H and I), from 100-kDa retentates of AD. In contrast, native ASPD-like assemblies were scarcely detected in IP eluates from 100-kDa retentates of NCI (Fig. 2, G and K). We next examined whether native ASPDs consisted of Aβ. Mass spectrometric analysis showed that singly charged ions corresponding to Aβ-(1–42) and Aβ-(1–40) were detected in native ASPDs (Fig. 2J). These results collectively demonstrate that 10–15-nm spherical Aβ assemblies isolated from AD brains are native ASPDs. Notably, anti-pan Aβ 6E10 could not immunoisolate native ASPDs (data not shown), probably because of its weak affinity for ASPDs (Kd ≈ 10−9 m) compared with ASD antibodies (Kd < 10−12 m) (Table 1). We also confirmed that anti-pan oligomer A11 antibody failed to detect native ASPDs (supplemental Fig. S4).
Having isolated native ASPDs selectively from human AD brains, we next examined whether they elicited neurodegeneration of rat primary neuronal cells. Surprisingly, AD-derived native ASPDs were even more toxic than synthetic ASPDs (Fig. 2K). These results collectively demonstrate that we have newly isolated A11-negative, high mass assemblies that cause neuronal cell death and that differ in mass and surface tertiary structure from other reported nonfibrillar Aβ assemblies.
Native ASPD Amount Correlates with the Pathologic Severity of AD
We next examined whether the amount of native ASPD correlated with the pathologic severity of AD brains. Larger amounts of native ASPD were present in AD patients with severe pathology (diagnosed “C” according to the CERAD criteria (43)) than in AD patients with moderate pathology (diagnosed “B”) (Fig. 2C). Furthermore, in AD patients with severe pathology, significantly higher amounts of native ASPD were detected in the frontal or temporal cortices (7.2 ± 1.5 nmol/g brain tissue, n = 3, Scheffé post hoc test p = 0.0012) than in the cerebellum (0.14 ± 0.1 nmol/g brain tissue). The result is consistent with previous findings that the cerebellum in AD is pathologically less affected (44, 45).
The above observations suggest the involvement of native ASPDs in neurodegeneration of AD brains. We therefore examined brains of patients suffering from DLB, the second most frequent cause of cognitive decline associated with neurodegeneration in the elderly (46, 47), because the majority of DLB brains have been shown to have AD-type pathology, including plaques (46–48). Interestingly, native ASPDs were also isolated from DLB brains (Fig. 3, A–C).
AD-derived Native ASPDs Cause Severe Degeneration of Human Neuronal Cells
To further elucidate the relationship between neuronal loss and native ASPDs, we first examined whether native ASPDs induce degeneration of human mature neuronal cells. Because studies using human primary neurons are problematic for ethical and practical reasons, cells with neuronal properties were induced from human bone marrow stromal cells (MSCs) (49). Initially, postmitotic neuronal cells were induced from human MSCs (>95% were neuron-specific MAP2ab-positive cells without glia) (49). Treatment of these cells with glial cell line-derived neurotrophic factor promoted their maturation into functional neuronal cells (49). We found that a 2-day treatment of the human MSC-derived functional neuronal cells with isolated native ASPDs caused severe degeneration, whereas IP eluates from NCI brains had no effect (Fig. 4A). In addition, pretreatment with mASD3 antibody (100 μg/ml) significantly blocked this toxicity (Fig. 4A), as observed in the case of the 158–669-kDa ASPDs (supplemental Fig. S5A), demonstrating that the observed neuronal cell death was caused by native ASPDs.
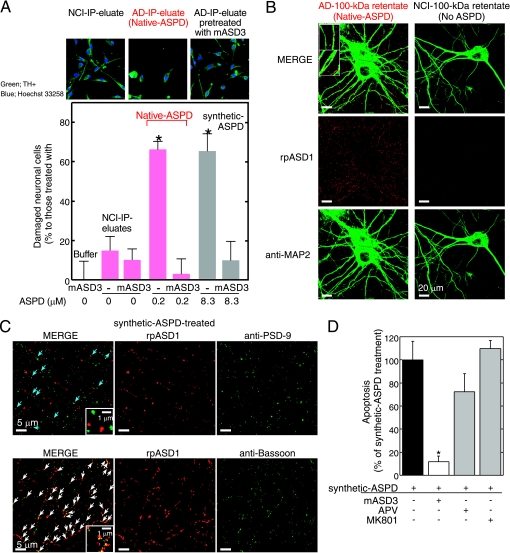
FIGURE 4.
Characterization of native and synthetic ASPD-induced toxicity. A, IP was performed using haASD1 as in Fig. 2F. Human neuronal cells were treated for 2 days with AD or NCI-IP eluates, with or without 2-h mASD3 (100 μg/ml) pretreatment. Nondamaged cells were counted after tyrosine hydroxylase (TH+) and Hoechst 33258 staining. The ratio of damaged cells to neuronal cells treated with buffer alone (mean ± S.D.) is shown (Scheffé post hoc test; *, p < 0.0001, n = 5). Neuronal cells treated with mASD3 alone or NCI-IP eluates showed only background levels of damaged cells similar to those in the case of cells treated with buffer. B and C, mature rat hippocampal neurons (24 DIV in B and 19 DIV in C) were incubated for 30 min either with 100-kDa retentates of AD (containing 0.8 μm native ASPDs) or NCI (no native ASPD detected) brain extracts in B or with 0.5 μm 158–669-kDa ASPDs (prepared from Aβ-(1–42); see Fig. 1A) in C. Bound ASPDs were detected by rpASD1, as in Fig. 1D. Punctate labeling was found primarily on neurites and surrounding cell bodies of neurons treated with native or synthetic ASPDs, but it was hardly detectable in neurons treated with the NCI retentates. A representative high power view is shown in the inset (B; bar, 5 μm). Neurons were co-stained with an antibody against anti-MAP2 in B, against a postsynaptic marker PSD-95 in C (upper panels), or against a presynaptic marker bassoon in C (lower panels). Z-stack images are shown (except lower panels in C) as in Fig. 1D. Bound ASPDs did not co-localize with PSD-95 but were concentrated with bassoon (white arrows in C), although they were occasionally localized in close proximity to PSD-95 (blue arrows in C). D, mature rat hippocampal neurons (21 DIV) were treated with 1 μm 158–669-kDa ASPDs for 2 days, with or without pretreatment (100 μg/ml mASD3 for 2 h; competitive (APV) or uncompetitive (MK801) NMDA-R antagonists (10 μm) for 30 min). Data represent mean ± S.D. (Scheffé post hoc test; *, p = 0.0039, compared with synthetic ASPDs; synthetic ASPDs, n = 7; synthetic ASPDs + APV or MK801, n = 5; synthetic ASPDs + mASD3, n = 4).
We then examined whether native ASPDs bind mature rat hippocampal neurons, as is observed in the case of synthetic ASPDs (Fig. 1D and supplemental Fig. S5B). Binding of AD-derived native ASPDs to 24-DIV mature rat hippocampal neurons was detected with rpASD1 most intensely in neurites and also to some extent in cell bodies (Fig. 4B). These results suggest that, despite the difference in dose dependence of neurotoxicity (Figs. 2K and 4A), native and synthetic ASPDs share essentially the same mechanism of neurotoxicity, i.e. they have the same surface tertiary structure that is responsible for exerting the toxicity. We speculate that the apparent difference in dose dependence might be attributed to differences in molecular compositions, but testing this idea will require further analyses using large amounts of isolated native ASPDs.
Mode of Native ASPD Neurotoxicity Is Different from That of Other Reported Aβ Assemblies
The above results (Fig. 4, A and B) show that native ASPDs cause neuronal cell death, possibly by binding to neuronal cell surfaces. We therefore examined ASPD-binding sites on mature neurons to elucidate the molecular basis of native ASPD neurotoxicity. As shown in the high power images in Fig. 4B (inset), bound native ASPDs appeared to protrude from the MAP2 staining of dendrites. Essentially the same results were obtained with the binding of synthetic ASPDs (supplemental Fig. S5B (inset)). Because of the limited availability of native ASPDs, we employed synthetic ASPDs for further analysis, as synthetic and native ASPDs share essential properties. Consistent with the above observation, the binding of synthetic ASPDs did not co-localize with a postsynaptic marker, PSD-95 (Fig. 4C, upper panel), although it was occasionally detected in close proximity to PSD-95 (blue arrows in C). Instead, ASPD-binding sites appeared to be concentrated at presynaptic sites stained by the antibody against a presynaptic marker, bassoon (white arrows in Fig. 4C, lower panel).
Although previous studies using cell or slice culture systems have found that Aβ assemblies such as dimers, ADDLs and AβOs bind postsynapses and depend on postsynaptic signaling mechanisms for exerting synaptotoxicity (23), the presynaptic binding of ASPDs apparent in Fig. 4B suggests that ASPD neurotoxicity would not require postsynaptic signaling mechanisms such as the N-methyl-d-aspartate glutamate receptor (NMDA-R) pathway. Indeed, neither a competitive (APV) nor an uncompetitive (MK801) NMDA-R antagonist inhibited synthetic ASPD-induced neurodegeneration (Fig. 4D). As noted above, native and synthetic ASPDs share the common surface tertiary structure responsible for exerting the toxicity. Therefore, the findings obtained with synthetic ASPDs (Fig. 4, C and D) strongly suggest that native ASPDs cause neuronal cell death through presynaptic target(s) on mature neurons. Furthermore, these observations are consistent with the findings indicating that native ASPDs have a distinct surface tertiary structure from other reported Aβ assemblies and support the hypothesis that native ASPDs have a different target(s) from other Aβ assemblies.
DISCUSSION
Aβ assemblies are considered to acquire surface tertiary structures that are not present in physiologic Aβ monomers and that induce synaptic impairment and neuronal loss through interactions with neuronal cells. Therefore, as recently suggested (12), it is reasonable to classify seemingly different Aβ assemblies in terms of their immunoreactivity to antibodies that recognize particular surface tertiary structure. Because the surface tertiary structure mediates the binding of Aβ assemblies to their target(s) and is therefore responsible for exerting the toxic effects, Aβ assemblies having distinct surface tertiary structures are likely to have distinct mechanisms of neurotoxicity and may contribute differently to the disease development. Here we have demonstrated the existence of patient-derived native ASPDs by selectively immunoisolating them from AD and DLB brains (Figs. 2 and 3) using ASPD tertiary structure-dependent antibodies (Fig. 1 and Table 1). The native ASPDs (>100 kDa) thus obtained are larger in mass than AD-derived Aβ dimers and other reported assemblies such as 12-mers (53∼60 kDa; ADDLs, globulomer, Aβ*56) or AβOs (∼90 kDa) (supplemental Table S1). More importantly, native ASPDs are considered to have a distinct surface tertiary structure from those other assemblies because they differ in immunospecificity, as illustrated by the fact that ASPD tertiary structure-dependent antibodies showed minimal reactivity with the 100-kDa filtrate containing monomers and dimers (Fig. 1A) or with ADDLs (supplemental Fig. S1A) (16) in dot blots. Additionally, anti-pan oligomer A11 antibody (22) recognized AβOs but not synthetic ASPDs (Fig. 1B) or native ASPDs (supplemental Fig. S4). Finally, anti-Aβ N-terminal antibodies such as 82E1 blocked the synaptotoxicity of AD-derived dimers (30) but failed to block synthetic ASPD-induced neurodegeneration (supplemental Fig. S5A). These results all indicate a difference in the surface tertiary structure between these assemblies and ASPDs.
As for the cellular basis of the Aβ-induced synaptic changes, previous studies have suggested the involvement of postsynaptic signaling mechanisms (23). For example, the binding of ADDLs and AβOs has been reported to co-localize with PSD-95 (19, 23). As expected from the postsynaptic locale of their binding, ADDLs bind close to or at NMDA-R (23), and NMDA-R antagonists inhibit ADDL-induced dendritic changes (23), reactive oxygen species formation (50), and insulin receptor impairment (51). NMDA-R antagonists have also been reported to inhibit Aβ dimer-induced synaptic loss (24, 30). Interestingly, cellular prion protein, which interacts with NMDA-R (52), has recently been reported to serve as a high affinity postsynaptic receptor mediating ADDL-induced synaptic dysfunction (53). Taken together, these studies are consistent with the idea that Aβ dimers, ADDLs, and AβOs perturb postsynaptic transmission (19, 23, 30).
We found that, unlike the above Aβ assemblies, ASPDs bind presynaptic target(s) on neurons to induce neurodegeneration (Fig. 4, A–C). This may be reasonable in view of the distinct ASPD surface tertiary structure. Although the actual targets of native ASPDs remain to be elucidated, native ASPDs seem to affect mature neuron-specific molecules or cellular pathways, as synthetic ASPD-induced neurotoxicity appeared to be confined to neurons, being especially active toward mature neurons, but sparing non-neuronal cells and immature neurons (supplemental Fig. S6, A–C). Together, the findings indicate that native ASPDs are patient-derived, A11-negative, high mass Aβ assemblies with a distinct toxic surface that binds presynaptic target(s) on mature neurons, leading to neuronal loss (supplemental Table S1). Although further studies are required to reveal how native ASPDs exert neurotoxicity in the brains of patients with AD, our findings indicate for the first time that presynaptic signaling mechanisms may play a critical role in Aβ-induced neurodegeneration in AD.
Recent in vivo as well as in vitro studies support the toxicity of nonfibrillar Aβ assemblies and their possible causative roles in the neuropathology of AD (54–56), which is consistent with the dissociation between fibril load and cognitive decline in patients with AD (32, 57, 58). Thus, Aβ assemblies other than fibrils have been considered to be the preferred therapeutic targets for AD (54). However, the nature of the Aβ species and the oligomer state responsible for the pathogenesis remain controversial because of the heterogeneity of Aβ assemblies in terms of Aβ species and oligomer size. It is also unknown how Aβ monomers assemble into oligomers in living human brains. Nevertheless, previous in vitro studies have shown that Aβ monomers develop into a variety of assemblies that might represent distinct structural variants (10–13). These studies suggest that assembly may not be a linear process but may be the result of a series of multiple processes involving intermediates from side paths. Taking all the results together, it seems reasonable to assume that the brains of patients with AD contain distinct types of Aβ assemblies with different surface tertiary structures that may play different roles in AD development. Therefore, identification and characterization of all types of Aβ assemblies actually present in brains from humans with AD will be important for understanding the molecular mechanisms underlying the AD progression from the initial step to the symptomatic phase and for the development of therapies based on this understanding. Fractionation studies using oligomer tertiary structure-dependent antibodies as shown here will help to elucidate the assembly process and to determine the Aβ assembly state causing the pathogenesis. We have isolated native ASPDs that cause degeneration of mature human neuronal cells in vitro (Fig. 4A) and have shown that the amount of native ASPD is correlated with the pathologic severity of clinically proven AD cases (Fig. 2C).
These findings suggest that native ASPDs might be a candidate for Aβ assemblies that directly cause neuronal loss in the brains of humans with AD. However, it remains to be elucidated whether or not ASPDs play a particular role in the onset or early stage of disease development. Braak and co-workers (59) have compared the expansion of Aβ pathology in whole brain regions between AD cases and nondemented cases with or without Aβ-related pathology. They found that patients with clinically proven AD exhibit late Aβ stages, although the nondemented cases with AD-related pathology show early Aβ stages. Their findings suggest that AD brains develop pathologic Aβ deposition before clinical symptoms become apparent, and this may start much earlier in nondemented patients with AD-related Aβ pathology. Quantitative studies, with the assistance of clinicians, on the brains of people in different Aβ stages, including nondemented people with AD-related Aβ pathology, will be helpful to elucidate if ASPDs play a role in neuronal loss in AD from the early stage of disease development.
Analyses on brains of APP-transgenic mice with or without neuronal loss would also help to elucidate the relationship between ASPDs and neuronal loss. Although the strain does not show neuronal loss, we examined Tg2576 mice, the most widely used AD-model mice carrying the human Swedish APP mutant (60), by means of immunohistochemistry and IP. ASPD-like assemblies were only minimally detected in the cerebral cortex of Tg2576 mice (supplemental Fig. S7, A and B);: they were not detected up to 14 months and only a very small amount (∼0.01 nmol/mg extracts) was detected at 23 months. As previously reported (18, 61), other Aβ assemblies such as dimers and Aβ*56 were increased in Tg2576 mice, and total Aβ reached levels comparable with those in human AD (supplemental Fig. S7C). With respect to mice with neuronal loss, in addition to certain APP transgenic mice (28, 29), there is a growing number of other AD-model mice, which have been produced by combining APP mutations with either presenilin-1 mutations (62, 63), Tau protein mutations (64), or nitric-oxide synthase knock-out (65). It should be noted that the mouse is not a perfect model of human AD, but these mice are considered to more closely resemble what occurs in the human brains. Therefore, further analysis to examine whether ASPD-like assemblies are present in these mice, which do show massive neuronal loss, will contribute to establish the relationship between neuronal loss and ASPDs.
In addition to the above, we are currently seeking to establish a direct link between native ASPDs and neuronal loss in brains from humans with AD by searching for the toxic target(s) of ASPDs on mature neurons. The identification of native ASPDs and availability of the toxicity-neutralizing antibodies should facilitate a mechanistic understanding of the cellular basis of neuronal cell loss in AD, as well as the development of therapies based on this understanding.
Supplementary Material
Acknowledgments
We thank Drs. George R. Martin, Takaomi C. Saido, Sangram S. Sisodia, R. Yu, Y. Fukazawa, D. Masui, M. Hoshino, H. Hara, and A. Sakai, for critical discussions; Dr. Charles G. Glabe for providing control blots for A11 antibody through Invitrogen; and Drs. T. Nirasawa, S. Horie, H. Kinoshita, S. Miyama, Y. Ogawa, and N. Takino for technical assistance.
This work was supported in part, by National Institutes of Health Grant NS038328 (to D. B. T.), by Health and Labour Sciences Research Grants “Research on Nanotechnical Medical” (to M. H.) and “Research on Psychiatric and Neurological Diseases and Mental Health” (to M. D.) from the Ministry of Health, Labor, and Welfare, by Special Coordination Funds for Promoting Science and Technology (to M. H.) from the Ministry of Education, Culture, Sports, Science, and Technology, and by the Program for Promotion of Fundamental Studies in Health Sciences (to M. D.) from the National Institute of Biomedical Innovation The authors declare the following conflict of interest: this work was supported in part by a grant (to M. H.) from Mitsubishi Kagaku Institute of Life Sciences, which is a nonprofit organization financially supported by Mitsubishi Chemical Corp.; this grant expires in March 2009.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables S1 and S2, Figs. S1–S7, and additional references.
A. Noguchi and M. Hoshi, unpublished data.
- AD
- Alzheimer disease
- Aβ
- amyloid β-protein
- APP
- amyloid precursor protein
- sAPPα
- human secreted form of APP
- ADDL
- Aβ-derived diffusible ligand
- AβO
- Aβ oligomer
- ASPD
- amylospheroid
- DLB
- dementia with Lewy bodies
- MALDI-TOF/MS
- matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- HFIP
- 1,1,1,3,3,3-hexafluoro-2-propanol
- PBS
- Dulbecco's phosphate-buffered saline without Ca2+ and Mg2+
- TEM
- transmission electron microscopy
- IP
- immunoprecipitation
- NCI
- noncognitively impaired
- MSCs
- bone marrow stromal cells
- NMDA-R
- N-methyl-d-aspartate-type glutamate receptor
- DIV
- days in vitro
- NCI
- noncognitively impaired.
REFERENCES
- 1.Ross C. A., Poirier M. A. (2005) Nat. Rev. Mol. Cell Biol. 6, 891–898 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J. (1991) Neuron 6, 487–498 [DOI] [PubMed] [Google Scholar]
- 3.Lansbury P. T., Lashuel H. A. (2006) Nature 443, 774–779 [DOI] [PubMed] [Google Scholar]
- 4.Iwatsubo T. (2007) Neuropathology 27, 474–478 [DOI] [PubMed] [Google Scholar]
- 5.Soto C., Estrada L. D. (2008) Arch. Neurol. 65, 184–189 [DOI] [PubMed] [Google Scholar]
- 6.Chiti F., Dobson C. M. (2009) Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 7.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 8.Tanzi R. E., Bertram L. (2005) Cell 120, 545–555 [DOI] [PubMed] [Google Scholar]
- 9.Saido T. C., Iwata N. (2006) Neurosci. Res. 54, 235–253 [DOI] [PubMed] [Google Scholar]
- 10.Klein W. L., Stine W. B., Jr., Teplow D. B. (2004) Neurobiol. Aging 25, 569–580 [DOI] [PubMed] [Google Scholar]
- 11.Walsh D. M., Selkoe D. J. (2007) J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 12.Glabe C. G. (2008) J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh D. M., Lomakin A., Benedek G. B., Condron M. M., Teplow D. B. (1997) J. Biol. Chem. 272, 22364–22372 [DOI] [PubMed] [Google Scholar]
- 15.Podlisny M. B., Ostaszewski B. L., Squazzo S. L., Koo E. H., Rydell R. E., Teplow D. B., Selkoe D. J. (1995) J. Biol. Chem. 270, 9564–9570 [DOI] [PubMed] [Google Scholar]
- 16.Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barghorn S., Nimmrich V., Striebinger A., Krantz C., Keller P., Janson B., Bahr M., Schmidt M., Bitner R. S., Harlan J., Barlow E., Ebert U., Hillen H. (2005) J. Neurochem. 95, 834–847 [DOI] [PubMed] [Google Scholar]
- 18.Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 19.Deshpande A., Mina E., Glabe C., Busciglio J. (2006) J. Neurosci. 26, 6011–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chimon S., Shaibat M. A., Jones C. R., Calero D. C., Aizezi B., Ishii Y. (2007) Nat. Struct. Mol. Biol. 14, 1157–1164 [DOI] [PubMed] [Google Scholar]
- 21.Lacor P. N., Buniel M. C., Chang L., Fernandez S. J., Gong Y., Viola K. L., Lambert M. P., Velasco P. T., Bigio E. H., Finch C. E., Krafft G. A., Klein W. L. (2004) J. Neurosci. 24, 10191–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 23.Lacor P. N., Buniel M. C., Furlow P. W., Clemente A. S., Velasco P. T., Wood M., Viola K. L., Klein W. L. (2007) J. Neurosci. 27, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankar G. M., Bloodgood B. L., Townsend M., Walsh D. M., Selkoe D. J., Sabatini B. L. (2007) J. Neurosci. 27, 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleary J. P., Walsh D. M., Hofmeister J. J., Shankar G. M., Kuskowski M. A., Selkoe D. J., Ashe K. H. (2005) Nat. Neurosci. 8, 79–84 [DOI] [PubMed] [Google Scholar]
- 26.Ashe K. H. (2001) Learn Mem. 8, 301–308 [DOI] [PubMed] [Google Scholar]
- 27.Hock B. J., Jr., Lamb B. T. (2001) Trends Genet. 17, S7–S12 [DOI] [PubMed] [Google Scholar]
- 28.Calhoun M. E., Wiederhold K. H., Abramowski D., Phinney A. L., Probst A., Sturchler-Pierrat C., Staufenbiel M., Sommer B., Jucker M. (1998) Nature 395, 755–756 [DOI] [PubMed] [Google Scholar]
- 29.Bondolfi L., Calhoun M., Ermini F., Kuhn H. G., Wiederhold K. H., Walker L., Staufenbiel M., Jucker M. (2002) J. Neurosci. 22, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo Y. M., Emmerling M. R., Vigo-Pelfrey C., Kasunic T. C., Kirkpatrick J. B., Murdoch G. H., Ball M. J., Roher A. E. (1996) J. Biol. Chem. 271, 4077–4081 [DOI] [PubMed] [Google Scholar]
- 32.Lue L. F., Kuo Y. M., Roher A. E., Brachova L., Shen Y., Sue L., Beach T., Kurth J. H., Rydel R. E., Rogers J. (1999) Am. J. Pathol. 155, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean C. A., Cherny R. A., Fraser F. W., Fuller S. J., Smith M. J., Beyreuther K., Bush A. I., Masters C. L. (1999) Ann. Neurol. 46, 860–866 [DOI] [PubMed] [Google Scholar]
- 34.Gómez-Isla T., Hollister R., West H., Mui S., Growdon J. H., Petersen R. C., Parisi J. E., Hyman B. T. (1997) Ann. Neurol. 41, 17–24 [DOI] [PubMed] [Google Scholar]
- 35.Morrison J. H., Hof P. R. (1997) Science 278, 412–419 [DOI] [PubMed] [Google Scholar]
- 36.Larrieu S., Letenneur L., Orgogozo J. M., Fabrigoule C., Amieva H., Le Carret N., Barberger-Gateau P., Dartigues J. F. (2002) Neurology 59, 1594–1599 [DOI] [PubMed] [Google Scholar]
- 37.Bouwman F. H., Schoonenboom S. N., van der Flier W. M., van Elk E. J., Kok A., Barkhof F., Blankenstein M. A., Scheltens P. (2007) Neurobiol. Aging 28, 1070–1074 [DOI] [PubMed] [Google Scholar]
- 38.Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., Sato K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6370–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lomakin A., Chung D. S., Benedek G. B., Kirschner D. A., Teplow D. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demuro A., Mina E., Kayed R., Milton S. C., Parker I., Glabe C. G. (2005) J. Biol. Chem. 280, 17294–17300 [DOI] [PubMed] [Google Scholar]
- 41.Kuwano R., Miyashita A., Arai H., Asada T., Imagawa M., Shoji M., Higuchi S., Urakami K., Kakita A., Takahashi H., Tsukie T., Toyabe S., Akazawa K., Kanazawa I., Ihara Y. (2006) Hum. Mol. Genet. 15, 2170–2182 [DOI] [PubMed] [Google Scholar]
- 42.Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirra S. S., Heyman A., McKeel D., Sumi S. M., Crain B. J., Brownlee L. M., Vogel F. S., Hughes J. P., van Belle G., Berg L. ( 1991) Neurology 41, 479– 486 [DOI] [PubMed] [Google Scholar]
- 44.Li Y. T., Woodruff-Pak D. S., Trojanowski J. Q. (1994) Neurobiol. Aging 15, 1–9 [DOI] [PubMed] [Google Scholar]
- 45.Kepe V., Huang S. C., Small G. W., Satyamurthy N., Barrio J. R. (2006) Methods Enzymol. 412, 144–160 [DOI] [PubMed] [Google Scholar]
- 46.Ince P. G., McKeith I. G. (2003) in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders (Dickson D. ed) pp. 188– 197, ISN Neuropath Press, Basel [Google Scholar]
- 47.Giasson B. I., Lee V. M.-Y., Trojanowski J. Q. (2004) in The Neuropathology of Dementia (Esiri M., Lee V. M.-Y., Trojanowski J. Q. eds) 2nd Ed., pp. 353– 375, Cambridge University Press, Cambridge [Google Scholar]
- 48.Kosaka K. (1990) J. Neurol. 237, 197–204 [DOI] [PubMed] [Google Scholar]
- 49.Dezawa M., Kanno H., Hoshino M., Cho H., Matsumoto N., Itokazu Y., Tajima N., Yamada H., Sawada H., Ishikawa H., Mimura T., Kitada M., Suzuki Y., Ide C. (2004) J. Clin. Invest. 113, 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Felice F. G., Velasco P. T., Lambert M. P., Viola K., Fernandez S. J., Ferreira S. T., Klein W. L. (2007) J. Biol. Chem. 282, 11590–11601 [DOI] [PubMed] [Google Scholar]
- 51.Zhao W. Q., De Felice F. G., Fernandez S., Chen H., Lambert M. P., Quon M. J., Krafft G. A., Klein W. L. (2008) FASEB J. 22, 246–260 [DOI] [PubMed] [Google Scholar]
- 52.Khosravani H., Zhang Y., Tsutsui S., Hameed S., Altier C., Hamid J., Chen L., Villemaire M., Ali Z., Jirik F. R., Zamponi G. W. (2008) J. Cell Biol. 181, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein W. L., Krafft G. A., Finch C. E. (2001) Trends Neurosci. 24, 219–224 [DOI] [PubMed] [Google Scholar]
- 55.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 56.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 57.Dickson D. W., Yen S. H. (1989) Neurobiol. Aging 10, 402–414 [DOI] [PubMed] [Google Scholar]
- 58.Terry R. D., Masliah E., Salmon D. P., Butters N., DeTeresa R., Hill R., Hansen L. A., Katzman R. (1991) Ann. Neurol. 30, 572–580 [DOI] [PubMed] [Google Scholar]
- 59.Thal D. R., Rüb U., Orantes M., Braak H. (2002) Neurology 58, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 60.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 61.Kawarabayashi T., Younkin L. H., Saido T. C., Shoji M., Ashe K. H., Younkin S. G. (2001) J. Neurosci. 21, 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casas C., Sergeant N., Itier J. M., Blanchard V., Wirths O., van der Kolk N., Vingtdeux V., van de Steeg E., Ret G., Canton T., Drobecq H., Clark A., Bonici B., Delacourte A., Benavides J., Schmitz C., Tremp G., Bayer T. A., Benoit P., Pradier L. (2004) Am. J. Pathol. 165, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oakley H., Cole S. L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., Berry R., Vassar R. (2006) J. Neurosci. 26, 10129–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pérez M., Ribe E., Rubio A., Lim F., Morán M. A., Ramos P. G., Ferrer I., Isla M. T., Avila J. (2005) Neuroscience 130, 339–347 [DOI] [PubMed] [Google Scholar]
- 65.Wilcock D. M., Gharkholonarehe N., Van Nostrand W. E., Davis J., Vitek M. P., Colton C. A. (2009) J. Neurosci. 29, 7957–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.