Abstract
Mutations in polycystin-1 (PC1) can cause autosomal dominant polycystic kidney disease, which is a leading cause of renal failure. The available evidence suggests that PC1 acts as a mechanosensor, receiving signals from the primary cilia, neighboring cells, and extracellular matrix. PC1 is a large membrane protein that has a long N-terminal extracellular region (about 3000 amino acids) with a multimodular structure including 16 Ig-like polycystic kidney disease (PKD) domains, which are targeted by many naturally occurring missense mutations. Nothing is known about the effects of these mutations on the biophysical properties of PKD domains. Here we investigate the effects of several naturally occurring mutations on the mechanical stability of the first PKD domain of human PC1 (HuPKDd1). We found that several missense mutations alter the mechanical unfolding pathways of HuPKDd1, resulting in distinct mechanical phenotypes. Moreover, we found that these mutations also alter the thermodynamic stability of a structurally homologous archaeal PKD domain. Based on these findings, we hypothesize that missense mutations may cause autosomal dominant polycystic kidney disease by altering the stability of the PC1 ectodomain, thereby perturbing its ability to sense mechanical signals.
Introduction
Autosomal dominant polycystic kidney disease is one of the most common life-threatening genetic diseases (with an incidence of 1 in ∼200–1000 newborns) and is a leading cause of renal failure. The majority of cases (∼85%) are caused by mutations in the PKD1 gene, which encodes for PC1. In autosomal dominant polycystic kidney disease, the sensing mechanisms for tubule size appear to be lost, and cysts develop and enlarge progressively (1–7). The function of PC1, as well as the mechanisms whereby mutations in this protein lead to the pathogenesis of the disease, remains unknown.
PC1 has a long extracellular domain (∼3000 amino acids) with a multimodular structure, containing 16 copies of a β- sandwich fold, the polycystic kidney disease (PKD)4 domain. PKD domains have a similar topology to Ig and fibronectin type III domains found in other modular proteins with structural and mechanical roles (recently reviewed in Ref. 8). PC1 is a membrane-associated protein that interacts with polycystin-2 (PC2) in the primary cilia of renal epithelial cells, which forms a mechanically sensitive ion channel complex. Bending of the cilia induces calcium flow into the cells, mediated by the PC1-PC2 complex (9–11). Mechanical signals are thus transduced into cellular responses that regulate proliferation, adhesion, and differentiation, essential for the control of renal tubules and kidney morphogenesis. It has recently been shown that the extracellular domain of PC1 has a load-bearing function where most modules are designed to resist unfolding when exposed to mechanical forces (12, 13). These data provide direct support to the hypothesis that the extracellular region is involved in the observed response of ciliated renal epithelial cells to applied forces.
To date, about 820 mutations have been identified in the PKD1 gene (available through the Autosomal Dominant Polycystic Kidney Disease: Mutation Database web site). Most are either point mutations or deletion/insertion mutations that introduce frame shifts and stop codons leading to premature termination. The most likely effect of these types of mutations is a complete loss of normal PC1 function. However, there are also about 240 missense mutations that result in non-conservative amino acid substitutions involving residues that form part of the ectodomain of PC1. Mutations may cause changes in conformation, disrupt the structure of the domains (and cause unfolding or misfolding), or affect their surface properties, as has been suggested for other Ig-like proteins (14, 15). However, very little is known about how missense mutations might alter the structure of PC1 and mechanical properties.
In this study, we used single-molecule AFM and equilibrium thermodynamics to understand the effect of missense mutations on the mechanical properties of PC1. Six missense mutations (FH26L, T36C, G43S, W38R, R57L, and V59H) were tested on the first PKD domain of PC1, HuPKDd1. We found that these mutations alter the mechanical stability of the domain, resulting in distinct mechanical PKD phenotypes. We find that point mutations can affect the free energy of mechanical unfolding and the position of the transition state. We also found that equivalent mutations in the homologous PKD domain found in Methanosarcina archaebacteria (16) affect thermodynamic stability. This indicates that pathogenic mutations can affect the normal response of the PKD domain to external mechanical forces and may help understand the molecular mechanisms underlying the physiological effects of mutations in PC1.
EXPERIMENTAL PROCEDURES
Cloning and Expression of HuPKDd1 Constructs for AFM Experiments
We cloned and expressed in bacteria a heteropolyprotein based on the first PKD domain from human PC1 (HuPKDd1, residues Val-268–Glu-354) and the titin immunoglobulin domain 27 (I27). The I27 domain has been extensively studied by force spectroscopy and hence serves as an internal fingerprint (13, 17, 18). We assembled an I27-HuPKDd1 heteropolyprotein using a multiple step cloning technique that makes use of four restriction sequences (BamHI, BglII, BstY, and KpnI) to three multiples of the I27-HuPKDd1 dimer hexamer (12, 13). The R57L mutant heteropolyprotein was obtained by mutagenesis PCR on the I27-HuPKDd1 construct.
The single-point mutations, T36C, G43S, and V59H, were produced by PCR synthesis using the QuikChange II mutagenesis kit (Stratagene). The cDNAs were subcloned into vector pAFM1–8 using the restriction sequences SacI and KpnI (19). The proteins were expressed and purified as described previously (12, 13) (see the supplemental materials for more details). The proteins were kept in phosphate-buffered saline containing 5 mm dithiothreitol at 4 °C.
Cloning, Expression, and Purification of Archaea PKD Domains (ArPKD)
The ArPKD monomer gene was cloned from a construct kindly supplied by S. Qamar and R. Sandford (University of Cambridge, UK) and ligated into a modified pRSETA vector (Invitrogen). Standard site-directed mutagenesis reactions were used to introduce mutations into individual domains. The proteins were expressed and purified as described previously (12). The two-step purification procedure involved nickel affinity chromatography followed by gel filtration.
Single-molecule Atomic Force Microscopy
The mechanical properties of single proteins were studied using a home-built single molecule AFM as described previously (20–24). The spring constant of each individual cantilever (MLCT-AUHW: silicon nitride gold-coated cantilevers; Veeco Metrology Group, Santa Barbara, CA) was calculated using the equipartition theorem (25). The cantilever spring constant varied between 30 and 50 pN/nm, and root mean square force noise (1-kHz bandwidth) was ∼15 pN. Unless noted, the pulling speed of the different force-extension curves was in the range of 0.5–0.7 nm/ms.
Single Protein Mechanics
In a typical experiment, a small aliquot of the purified proteins (∼1–50 μl, 10–100 μg/ml) was allowed to adsorb to a clean glass coverslip (for ∼10 min) and then rinsed with phosphate-buffered saline, pH 7.4. We also tested other substrates such as nickel-nitrilotriacetic acid-coated surfaces (26). We found that PKD protein constructs adsorbed well to glass, gold-coated glass, or nickel-nitrilotriacetic acid-coated coverslips. We obtained identical data with these different substrates. Proteins were picked up randomly by adsorption to the cantilever tip, which was pressed down onto the sample for 1–2 s at forces of several nanonewtons and then stretched for several hundred nanometers. The probability of picking up a protein was typically kept low (less than 1 in 50 attempts) by controlling the amount of protein used to prepare the coverslips.
Analysis of the Speed Dependence Data
The unfolding force distribution and speed dependence of the unfolding forces were fit using Monte Carlo simulation to calculate the unfolding rate constants, αo, and the position of the transition state, xu, as described previously (24, 27–29). The errors in the determination of these parameters were estimated by running the Monte Carlo simulations about 10 times.
Equilibrium Denaturation of ArPKD Domains
All experiments were carried out in phosphate-buffered saline (pH 7.4) at 25 °C. The stability of the individual ArPKD wild-type and mutant domains was determined by urea denaturation, using standard techniques (30). The protein was incubated for 3 h in varying concentrations of denaturant, and unfolding was monitored by change in intrinsic fluorescence using an Aminco Bowman fluorescence spectrometer with an excitation of 280 nm and emission monitored at the wavelength of maximum emission (320 nm for W36C and 350–360 nm for wild type and all other mutants). The raw data were fitted to a standard two-state equation, which allows [urea]50% (the concentration of denaturant where 50% of the protein is denatured) and the m-value (the dependence of ΔGD−N on the concentration of urea) to be determined (see supplemental materials). The free energy for unfolding in 0 m denaturant (ΔGD−NH2O) can be calculated from
where [urea]50% is the concentration of denaturant where 50% of the protein is denatured, and m is the dependence of ΔGD−N on the concentration of urea. The PCPMer software package was used to analyze protein sequence alignments of related proteins to detect conserved physical-chemical properties (31, 32).
Determining Mutations in an Archaea PKD Domain
The solved structures of the HuPKDd1 (Protein Data Bank (PDB) code 1B4R) and the ArPKD (PDB code 1L0Q) domain were visualized using Insight II, a commercially available program that allows structures to be manipulated and visually superimposed, and the structures were compared to determine which residues were spatially best aligned. From this alignment, it was possible to determine where to make the mutations in ArPKD that would be most nearly equivalent to those in the PC1 PKD domains.
RESULTS
Multiple Sequence Alignment of the PC1 PKD Domains and Location of Naturally Occurring Missense Mutations
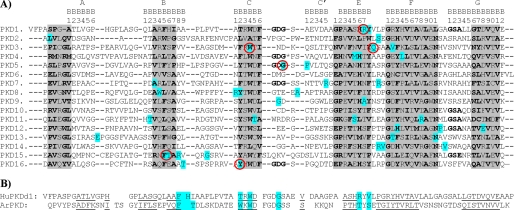
The NMR structure of the first PKD domain (HuPKDd1) shows that it has a β-sandwich structure with two sheets that pack together with a well defined hydrophobic core, centered around a conserved tryptophan located in the C strand (33). PC1 contains 16 homologous PKD domains. Fig. 1A shows a sequence alignment of the 16 domains, together with an analysis of conserved motifs. The alignment and motif detection was done by using the PCPMer program (32). This program automatically detects sequence motifs defined in terms of the conserved physical-chemical properties of residues in protein families. There are six conserved motifs in PC1 PKD domains (highlighted in gray). There are about 40 missense mutations that result in non-conservative amino acid substitutions involving residues that form part of PKD domains (highlighted in cyan). Interestingly, most of these mutations (30 out of 40) are found in conserved regions.
FIGURE 1.
A, multiple sequence alignment of the 16 human PKD domains with structural motifs highlighted. The alignment and motif detection was done by using PCPMer program (31). This program automatically detects sequence motifs defined in terms of the conserved physical-chemical properties of residues in protein families. The missense mutations are highlighted in cyan, and the selected for this work are circled in red. PKD4 has additional residues in the CC′ loop. *, EQALHQFQPPYNESFPVPD. B, structure-based sequence alignment of the first human PC1 PKD domain (HuPKDd1) and archaeal PKD domain (ArPKD) and location of pathogenic mutation positions. The best aligned residues are underlined. The equivalent positions of missense mutations in both domains are highlighted (in cyan).
Selections of Missense Mutations
Of these missense mutations, we selected six (Fig. 1A, red circles) because these have been assigned as likely to be pathogenic. These are listed in Table 1. The first mutation targets PKD domain 1 where a charged amino acid (Arg) is changed to a hydrophobic amino acid (Leu); the same position in the Fugu rubripes PKD domain 1 is occupied by a basic residue (34, 35). The second mutation targets the conserved Trp in PKD domain 3 (changed to an Arg), which results in a pathogenic phenotype (36). The third mutation also occurs in PKD domain 3 and changes a polar amino acid (Gln) to His. Interestingly, the mutated glutamine is conserved in PKD domain 3 from F. rubripes to human (34, 37). The fourth mutation is found in PKD domain 5 within the CC′ loop region. The mutation occurs within the most conserved sequence of the PKD domains, WDFGDGS (33). This sequence is conserved from archaea to humans (Fig. 1B). The glycine that is replaced (in bold) is in the C–C′ turn. Its replacement by the bulkier serine is very likely to disrupt this structure (38). The fifth mutation is a replacement of two amino acids (Phe and Thr) to Leu. This is a large change that is likely to be pathogenic (39). The last mutation is found in PKD domain 16 where a Tyr is replaced by a Cys. This mutation was found to be pathogenic (40).
TABLE 1.
List of pathogenic missense mutations in human PC1 PKD domains and equivalent residues in HuPKDd1 and ArPKD
| Residue number | PKD domain number | Mutation position in HuPKDd1 | Mutation position in ArPKD | Location strand | Surface/core |
|---|---|---|---|---|---|
| Arg-324 → Leu | 1 | R57L | T54L | E | Surface |
| Trp-967 → Arg | 3 | W38R | W38R | C | Core |
| Gln-987 → His | 3 | V59H | S56H | E–F loop | Surface |
| Gly-1166 → Ser | 5 | G43S | G43S | C–C′ loop | Short loop |
| Leu-1992, Thr-1993 → Leu | 15 | FH26L | FT26L | B | Surface, core |
| Tyr-2092 → Cys | 16 | T36C | W36C | C | Core |
Effect of Mutations on the Thermodynamic Stability of PKD Domains
We wanted to compare the effects of the selected point mutations on the thermodynamic stability of HuPKDd1. However, this domain is only marginally stable (∼1–2 kcal mol−1 (12)), so it is difficult to get accurate stability data. However, we have previously shown that it is possible to use homologous domains to model the effects of pathogenic mutations in Ig-like domains (15). The PKD domain from Methanosarcina archaebacteria (termed ArPKD) (16) has a very similar structure to HuPKDd1 (the two structures superimpose with an root mean square deviation of 2.2 Å (16)) but is thermodynamically more stable (4.4 kcal mol−1). ArPKD therefore presents itself as a good model system to study pathogenic mutations in PC1 PKD domains. To make equivalent mutations in the human PKD and ArPKD domains, the best approach was to examine the published structures (16, 33) and create an alignment of the two sequences. The results of this analysis are shown in Fig. 1B, a structure-based alignment of the ArPKD and HuPKDd1 amino acid sequences. The underlined sections of the sequence are the best aligned structurally. From this alignment, it was possible to determine where to make the mutations in ArPKD that would be most nearly equivalent to those in the PC1 PKD domains. The sites of the mutations in the aligned sequence are highlighted in cyan in Fig. 1B and are listed in Table 1.
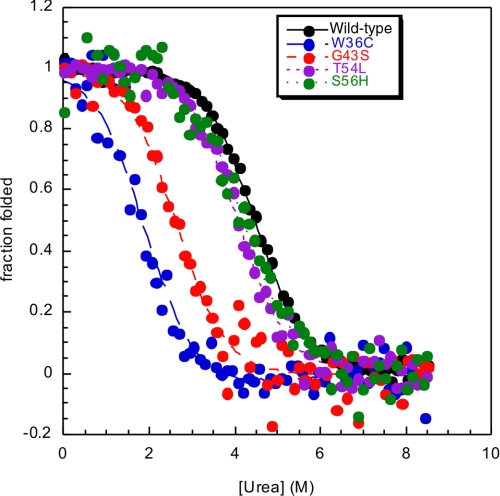
In equilibrium denaturation experiments, unfolding was monitored by following changes in intrinsic tryptophan fluorescence. The results are shown in Fig. 2, and the results are summarized in Table 2. The mutants FT26L and W38R were unfolded in buffer. All point mutations destabilize the ArPKD domain to some extent.
FIGURE 2.
Equilibrium denaturation curves for ArPKD mutants. The effects of missense mutations T54L, S56H, G43S, and W36C on ArPKD domain stability are shown. All four mutants are made to model pathogenic mutations in HuPKDd1 domains. It is clear from the changes in the midpoint that both W36C and G43S destabilize the domain significantly, whereas the mutations T54L and S56H have little effect on the stability of the PKD domain.
TABLE 2.
Thermodynamic stabilities of ArPKD mutants
| Mutation position in ArPKD | m-value | [urea]50% ArPKD | ΔGD−NH2O ArPKDa |
|---|---|---|---|
| kcal mol−1m−1 | m | kcal mol−1 | |
| Wild type | 1.0 ± 0.1 | 4.4 ± 0.1 | 4.4 ± 0.1 |
| T54L | 1.0 ± 0.1 | 4.1 ± 0.1 | 4.1 ± 0.1 |
| W38R | Unfolded | NA | NA |
| S56H | 0.9 ± 0.1 | 4.5 ± 0.1 | 4.1 ± 0.1 |
| G43S | 1.0 ± 0.1 | 2.6 ± 0.1 | 2.6 ± 0.1 |
| FT26L | Unfolded | NA | NA |
| W36C | 1.0a | 1.9 ± 0.1 | 1.9 ± 0.1 |
a Because W36C was so destabilized that there was no true folded baseline, the data were fitted with m fixed to the wild-type value of 1.0.
Effects of the Missense Mutations on the Mechanical Stability of HuPKDd1
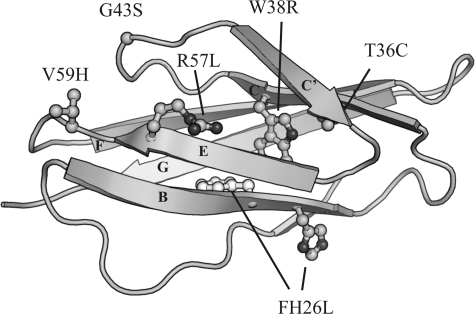
To study the effect of missense mutations on the mechanical stability of PKD domains, we used HuPKDd1 as a template because its structure is known (33) and its thermodynamic and mechanical stabilities have been characterized (12, 13). To make equivalent mutations in the HuPKDd1 domain, we used the sequence alignment shown in Fig. 1 and mutated the equivalent residues using site-directed mutagenesis. The locations of the different residues mutated in HuPKDd1 are shown in Fig. 3. For example, the change of Val-59 to His in HuPKDd1 corresponds to the natural mutation Gln-987 to His in PKD domain 3.
FIGURE 3.
NMR structure of HuPKDd1 showing the positions of mutated residues. The figure was prepared using the program PyMOL.
We used a heteropolyprotein approach to study the mechanical properties of mutant HuPKDd1 domains using single-molecule AFM techniques. In these constructs, we used the titin domain I27 as an internal mechanical fingerprint that has been extensively studied with AFM techniques (21, 41). Also, I27-based protein chimeras have been found to express well in bacteria; this strategy has proven to be useful in the analysis of several protein domains with single molecule AFM (18, 19, 23, 42, 43).
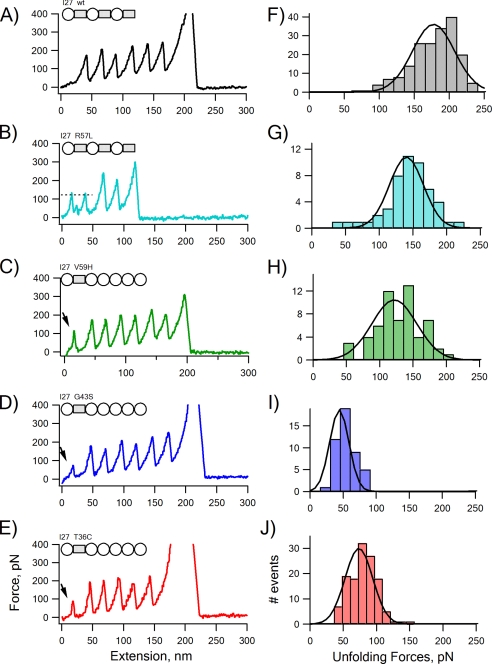
Fig. 4 shows typical examples of force-extension curves obtained for I27 heteropolyproteins harboring the wild-type and mutant HuPKDd1. As shown before, the wild-type HuPKDd1 has a mechanical stability very similar to I27 domains and unfolds at forces of about 180 pN (Fig. 4F) (12, 13). Fig. 3, B–E, show that missense mutations R57L, V59H, G43S, and T36C all result in a significant decrease in the mechanical stability. For example, the R57L mutant domain unfolds at forces of ∼140 pN (Fig. 4G), which is seen as the force peaks (marked in Fig. 4B by the dashed line) preceding the unfolding of the I27 domains (they unfold at ∼180 pN). The G43S mutation has a strong destabilizing effect of HuPKDd1. This mutant domain unfolds at ∼55 pN, which is seen as a small force peak (marked by the arrow) preceding the unfolding of the I27 domains (Fig. 4D). We found that the W38R and FT26L mutations severely destabilize the PKD domain because we were not able to express these constructs as soluble proteins.
FIGURE 4.
Force-extension relationships for wild-type (wt) and mutant HuPKDd1 measured with AFM techniques. Stretching a single molecule of each construct gave force-extension curves that followed a saw-tooth pattern with equally spaced force peaks. The schematic above each recording shows the construction of each recombinant protein chimera. The wild-type HuPKDd1 and the R57L mutant were constructed by repeating the I27-HuPKDd1 three times. The T36C, G43S, and V59H HuPKDd1 mutants were flanked by several I27 domains (to increase the solubility of expressed protein). The dashed lines in A and B present the average force used to unfold either wild-type or R57L mutant HuPKDd1. The force peaks pointed to by the solid arrow represent the unfolding of mutant domains of V59H (C), G43S (D), and T36C (E). The forces used to unfold the wild-type and mutated HuPKDd1 are shown as force histograms (F–J). The force peaks of wild-type PKD had an average force of 176 ± 32 pN (n = 189). The average forces for unfolding the HuPKDd1 mutants are: 143 ± 34 pN (n = 50) for R57L, 127 ± 33 pN (n = 59) for V59H, 54 ± 15 pN (n = 46) for G43S, and 79 ± 20 pN (n = 129) for T36C. All the experiments were carried out at the pulling speed of 0.4–0.6 nm/ms.
Kinetics of Unfolding of HuPKDd1 Mutants
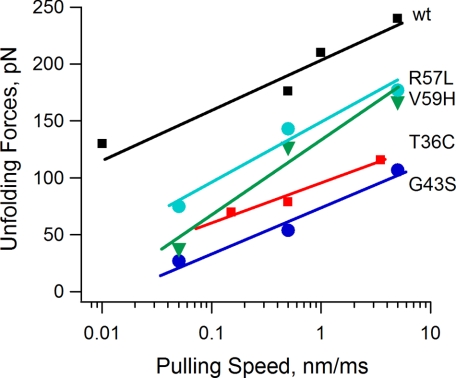
To quantify the effect of the missense mutations on the kinetics of unfolding, we analyzed the effect of pulling speed on the unfolding forces. Fig. 5 shows a plot of the average unfolding force versus the pulling rate for wild type and different mutants. The parameters used for the Monte Carlo simulation are shown in Table 3. The unfolding rate constants, αo, of the four mutants are higher than that of the wild type (9.8 × 10−4 s−1), indicating that the activation energy of unfolding was decreased by the mutations. The unfolding distances to the transition state, xu, of the G43S and T36C mutants are also larger than the wild type, indicating that there is a significant change in the unfolding pathway.
FIGURE 5.
Kinetics of unfolding of HuPKDd1 mutants. A plot of the average unfolding force versus the pulling rate for wild-type (wt, black squares), R57L (cyan circles), V59H (green triangles), T36C (red squares), and G43S (blue circles) HuPKDd1 domains is shown. The solid lines are fits of the Monte Carlo simulation to the experimental data (see “Experimental Procedures”). The parameters used for the Monte Carlo simulation are shown in Table 3.
TABLE 3.
Estimated mechanical kinetic parameters of HuPKDd1 mutants
| Fua | αo | xu | |
|---|---|---|---|
| pN | s−1 | nm | |
| Wild type | 176 ± 32 | 9.8 ± 8.1 × 10−4 | 0.25 ± 0.02 |
| R57L | 143 ± 34 | 2.6 ± 1.8 × 10−2 | 0.24 ± 0.01 |
| V59H | 127 ± 33 | 6.9 ± 5.3 × 10−2 | 0.22 ± 0.01 |
| G43S | 54 ± 15 | 2.3 ± 1.8 × 10−1 | 0.31 ± 0.02 |
| T36C | 79 ± 20 | 0.7 ± 0.5 × 10−1 | 0.32 ± 0.02 |
a At pulling speed of 0.4–0.6 nm ms−1.
DISCUSSION
We are using HuPKDd1 as a model system for assessing the effects of mutation on the mechanical stability of PKD domains in general. In general, we found that all the mutations resulted in a loss in mechanical stability. A number of studies have demonstrated that homologous proteins have the same unfolding pathways on application of force, i.e. the same mechanisms for resisting forced unfolding. Using a combination of simulations and experiments, we have recently shown that the same is true of HuPKDd1 and ArPKD.5 Hence, mutations that promote mechanical unfolding in HuPKDd1 are likely to have the same effect in other PKD domains.
All the Mutations Destabilize HuPKD and ArPKD Domains to Some Extent
Table 4 shows a summary of the effects of missense mutations on HuPKDd1 and ArPKD.
TABLE 4.
Comparison of the effect of mutations on thermodynamic and mechanical stability of PKD domains
NA, not applicable.
| Mutation position in HuPKDd1 | Mutation position in ArPKD | ΔGD−NH2O ArPKD | ΔFu HuPKDd1 |
|---|---|---|---|
| kcal mol−1 | pN | ||
| Wild type | NA | ||
| R57L | T54L | 0.3 ± 0.1 | 33 |
| W38R | W38R | >4.4 | NA |
| V59H | S56H | 0.3 ± 0.1 | 49 |
| G43S | G43S | 1.8 ± 0.1 | 122 |
| FH26L | FT26L | >4.4 | NA |
| T36C | W36C | 2.5 ± 0.1 | 97 |
Unfolded Mutants
The FH26L (or FT26L in ArPKD) and W38R mutations result in an unfolded protein domain. This is not unexpected because these target large buried residues. In particular, the Trp to Arg mutation, introducing a charged group into a buried position, causes a drastic reduction in stability. Furthermore, the tryptophan in that position is very highly conserved across the PKD domains, suggesting that it may have an importance for folding. Hence, for the W38R and FH26L, it seems very likely that the presence of the unfolded domain causes the observed disease phenotype. Perhaps the destabilized ectodomain of PC1 cannot perform its mechanosensing function, or perhaps the presence of the unfolded domain causes the protein to aggregate, to fail to be trafficked correctly, or to be degraded by the normal cell degradation machinery.
Destabilizing Mutations
Mutation T36C (or W36C in ArPKD) to cysteine replaces a large surface aromatic with a small polar side chain. This is a position with a high degree of conservation in the PKD domains. Of the other 16 PKD domains, eight have a tyrosine and four have a phenylalanine at the equivalent position, all large aromatic side chains. Rossetti et al. (44) note that this residue is conserved in the C strand of many PKD domains, including in the mouse gene, but interestingly, in F. rubripes, there is a cysteine at the equivalent position. The G43S replaces a glycine with a serine in a loop region of the protein structure. This results in a significant destabilization. This destabilization might be expected, given the conservation in this region of the structure in PKD domains of polycystin. Interestingly, this glycine residue occupies a region ϕ/ψ space of the Ramachandran plot (45), which is disallowed for residues other than glycine (positive ϕ, negative ψ) in the structures of both ArPKD and HuPKDd1.
The T36C and G43S mutants had a very significant effect on the ability of HuPKDd1 to resist forced unfolding. The unfolding force is significantly lower in these mutants, and furthermore, the distance to the transition state (xu) is significantly larger for each of these mutants, suggesting that the unfolding pathway changes significantly. Such a change in xu, associated with a change in folding mechanism, has been observed for mutants of I27 domain from human titin (29). We conclude that mutations in these positions might potentially have a significant effect on the mechanical function of PC1.
Slightly Destabilizing Mutations
The mutations that are only slightly destabilizing, R57L and V59H (T54L and S56H in ArPKD), are surface mutations. This suggests that these mutations are in regions of the protein that are not important for the mechanical stability of the PKD domains. Thomas et al. (35) reported the R57L mutation. They note the change from a basic to neutral hydrophobic residue and report that this is the only PKD domain with a basic residue in this position. Furthermore, a basic residue is found in the equivalent position in F. rubripes. Due to this conservation, they suggest that this residue may be functionally important, for example, in ligand binding or protein-protein interactions (35). Our results do not suggest that this is a position that is critical for the mechanical stability of PKD domains.
Our data suggest that the effect of the mutations G43S, T36C, and in particular, W38R and FH26L are likely to be mainly due to the large destabilization these mutations confer on the parent PKD domain (i.e. G1166S, Y2092C, W967R, and F1992L, T1993L, respectively). They will also, therefore, compromise the ability of PC1 to act as a mechanosensor.
In time, it may be possible to analyze naturally occurring variations in the genome sequence, predict the biophysical effect of the mutation, and estimate the likelihood of a given variation to be deleterious or benign. As seen here, such studies could also suggest further research into protein function, and possibly, mutation-specific therapies. There is increasing interest in human protein mutations, particularly naturally occurring mutations that may be disease-related. Although many of these proteins are experimentally intractable, the strategy presented here could make mutation studies in many human proteins possible for the first time.
Supplementary Material
Acknowledgment
We thank Jiping Wang (University of Texas Medical Branch) for helping with the expression and purification of some of the HuPKDd1 mutant proteins used in Fig. 4.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK073394 (to A. F. O.). This work was also supported by grants from the John Sealy Memorial Endowment Fund for Biomedical Research and the Polycystic Kidney Foundation (Grant 116a2r).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
J. R. Forman, J. Clarke, and E. Paci, unpublished data.
- PKD
- polycystic kidney disease domain
- ArPKD
- archaea PKD domain
- HuPKDd1
- first human PC1 PKD domain
- AFM
- atomic force microscopy
- pN
- piconewtons.
REFERENCES
- 1.Gabow P. A. (1990) Am. J. Kidney Dis. 16, 403–413 [DOI] [PubMed] [Google Scholar]
- 2.Wu G., Somlo S. (2000) Mol. Genet. Metab. 69, 1–15 [DOI] [PubMed] [Google Scholar]
- 3.Torres V. E., Harris P. C. (2006) Nat. Clin. Pract. Nephrol. 2, 40–55 [DOI] [PubMed] [Google Scholar]
- 4.Harris P. C., Torres V. E. (2009) Annu. Rev. Med. 60, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaletta T., Van der Craen M., Van Geel A., Dewulf N., Bogaert T., Branden M., King K. V., Buechner M., Barstead R., Hyink D., Li H. P., Geng L., Burrow C., Wilson P. (2003) Nephron Exp. Nephrol. 93, e9–17 [DOI] [PubMed] [Google Scholar]
- 6.Wilson P. D. (2004) Int. J. Biochem. Cell Biol. 36, 1868–1873 [DOI] [PubMed] [Google Scholar]
- 7.Sutters M., Germino G. G. (2003) J. Lab. Clin. Med. 141, 91–101 [DOI] [PubMed] [Google Scholar]
- 8.Oberhauser A. F., Carrión-Vázquez M. (2008) J. Biol. Chem. 283, 6617–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., Zhou J. (2003) Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 10.Alenghat F. J., Nauli S. M., Kolb R., Zhou J., Ingber D. E. (2004) Exp. Cell Res. 301, 23–30 [DOI] [PubMed] [Google Scholar]
- 11.Nauli S. M., Zhou J. (2004) BioEssays 26, 844–856 [DOI] [PubMed] [Google Scholar]
- 12.Forman J. R., Qamar S., Paci E., Sandford R. N., Clarke J. (2005) J. Mol. Biol. 349, 861–871 [DOI] [PubMed] [Google Scholar]
- 13.Qian F., Wei W., Germino G., Oberhauser A. (2005) J. Biol. Chem. 280, 40723–40730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman A., Jouet M., MacFarlane J., Du J. S., Kenwrick S., Chothia C. (1996) EMBO J. 15, 6050–6059 [PMC free article] [PubMed] [Google Scholar]
- 15.Randles L. G., Lappalainen I., Fowler S. B., Moore B., Hamill S. J., Clarke J. (2006) J. Biol. Chem. 281, 24216–24226 [DOI] [PubMed] [Google Scholar]
- 16.Jing H., Takagi J., Liu J. H., Lindgren S., Zhang R. G., Joachimiak A., Wang J. H., Springer T. A. (2002) Structure 10, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 17.Best R. B., Fowler S. B., Toca-Herrera J. L., Clarke J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12143–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Best R. B., Li B., Steward A., Daggett V., Clarke J. (2001) Biophys. J. 81, 2344–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steward A., Toca-Herrera J. L., Clarke J. (2002) Protein Sci. 11, 2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullard B., Linke W. A., Leonard K. (2002) J. Muscle Res. Cell Motil. 23, 435–447 [DOI] [PubMed] [Google Scholar]
- 21.Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller E., Garcia T., Hultgren S., Oberhauser A. F. (2006) Biophys. J 91, 3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberhauser A. F., Badilla-Fernandez C., Carrion-Vazquez M., Fernandez J. M. (2002) J. Mol. Biol. 319, 433–447 [DOI] [PubMed] [Google Scholar]
- 24.Oberhauser A. F., Marszalek P. E., Erickson H. P., Fernandez J. M. (1998) Nature 393, 181–185 [DOI] [PubMed] [Google Scholar]
- 25.Florin E. L., Rief M., Lehmann T., Ludvig M., Dornmair C., Moy V. T., Gaub H. E. (1995) Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 26.Klein D. C., Stroh C. M., Jensenius H., van Es M., Kamruzzahan A. S., Stamouli A., Gruber H. J., Oosterkamp T. H., Hinterdorfer P. (2003) Chemphyschem. 4, 1367–1371 [DOI] [PubMed] [Google Scholar]
- 27.Oberhauser A. F., Hansma P. K., Carrion-Vazquez M., Fernandez J. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 29.Li H., Carrion-Vazquez M., Oberhauser A. F., Marszalek P. E., Fernandez J. M. (2000) Nat. Struct. Biol. 7, 1117–1120 [DOI] [PubMed] [Google Scholar]
- 30.Pace C. N. (1986) Methods Enzymol. 131, 266–280 [DOI] [PubMed] [Google Scholar]
- 31.Mathura V. S., Schein C. H., Braun W. (2003) Bioinformatics 19, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 32.Garcia T. I., Oberhauser A. F., Braun W. (2009) Proteins 75, 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bycroft M., Bateman A., Clarke J., Hamill S. J., Sandford R., Thomas R. L., Chothia C. (1999) EMBO J. 18, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandford R., Sgotto B., Aparicio S., Brenner S., Vaudin M., Wilson R. K., Chissoe S., Pepin K., Bateman A., Chothia C., Hughes J., Harris P. (1997) Hum. Mol. Genet. 6, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 35.Thomas R., McConnell R., Whittacker J., Kirkpatrick P., Bradley J., Sandford R. (1999) Am. J. Hum. Genet. 65, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossetti S., Consugar M. B., Chapman A. B., Torres V. E., Guay-Woodford L. M., Grantham J. J., Bennett W. M., Meyers C. M., Walker D. L., Bae K., Zhang Q. J., Thompson P. A., Miller J. P., Harris P. C. (2007) J. Am. Soc. Nephrol. 18, 2143–2160 [DOI] [PubMed] [Google Scholar]
- 37.Burtey S., Lossi A. M., Bayle J., Berland Y., Fontés M. (2002) J. Med. Genet. 39, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phakdeekitcharoen B., Watnick T. J., Ahn C., Whang D. Y., Burkhart B., Germino G. G. (2000) Kidney Int. 58, 1400–1412 [DOI] [PubMed] [Google Scholar]
- 39.Rossetti S., Strmecki L., Gamble V., Burton S., Sneddon V., Peral B., Roy S., Bakkaloglu A., Komel R., Winearls C. G., Harris P. C. (2001) Am. J. Hum. Genet. 68, 46–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossetti S., Chauveau D., Kubly V., Slezak J. M., Saggar-Malik A. K., Pei Y., Ong A. C., Stewart F., Watson M. L., Bergstralh E. J., Winearls C. G., Torres V. E., Harris P. C. (2003) Lancet 361, 2196–2201 [DOI] [PubMed] [Google Scholar]
- 41.Li H., Linke W. A., Oberhauser A. F., Carrion-Vazquez M., Kerkvliet J. G., Lu H., Marszalek P. E., Fernandez J. M. (2002) Nature 418, 998–1002 [DOI] [PubMed] [Google Scholar]
- 42.Brucale M., Sandal M., Di Maio S., Rampioni A., Tessari I., Tosatto L., Bisaglia M., Bubacco L., Samorì B. (2009) Chembiochem. 10, 176–183 [DOI] [PubMed] [Google Scholar]
- 43.Li L., Huang H. H., Badilla C. L., Fernandez J. M. (2005) J. Mol. Biol. 345, 817–826 [DOI] [PubMed] [Google Scholar]
- 44.Rossetti S., Chauveau D., Walker D., Saggar-Malik A., Winearls C. G., Torres V. E., Harris P. C. (2002) Kidney Int. 61, 1588–1599 [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran G. N., Ramakrishnan C., Sasisekharan V. (1963) J. Mol. Biol. 7, 95–99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.