Abstract
Interleukin 4 (IL-4) inhibits receptor activator of NF-κB ligand (RANKL)-induced osteoclast formation and functional activity in a STAT6-dependent manner. IL-4 down-regulates expression of tartrate-resistant acid phosphatase (TRAP) in mature osteoclasts. To determine whether IL-4 regulates TRAP promoter activity, RAW264.7 cells were transfected with a TRAP promoter-luciferase reporter. Treatment with IL-4 alone modestly enhanced TRAP luciferase activity. However, IL-4 suppressed the ability of RANKL to up-regulate TRAP-luciferase activity, suggesting that IL-4 has multiple effects on TRAP transcription. IL-4 also reduced the RANKL-induced association of RNA polymerase II with the TRAP gene in osteoclasts. The TRAP promoter contains a STAT6-binding motif, and STAT6 bound to the endogenous TRAP promoter after IL-4 treatment. To determine the impact of STAT6 binding, we transfected cells with STAT6VT, a constitutively active STAT6 mutant. STAT6VT alone up-regulated TRAP-luciferase activity; this effect was abrogated by mutating the STAT6 binding site in the minimal TRAP promoter. STAT6VT did not inhibit the potent up-regulation of TRAP promoter activity caused by overexpression of NFATc1, PU.1, and microphthalmia transcription factor, downstream targets of macrophage colony-stimulating factor and RANKL. IL-4 down-regulated the expression of c-Fos and NFATc1 in mature osteoclasts. Knockdown of NFATc1 by short interfering RNA caused TRAP expression to be down-regulated, and ectopic expression of NFATc1 abrogated the IL-4-induced down-regulation of TRAP. These results suggest that STAT6 plays two distinct roles in TRAP expression. The IL-4-induced activation of STAT6 mediates suppression of the RANKL-induced TRAP promoter activity indirectly by inhibiting NFATc1 expression. However, in the absence of RANKL and osteoclast differentiation, STAT6 binds the TRAP promoter after IL-4 treatment and directly enhances TRAP expression.
Introduction
TRAP2 is a di-iron-containing metalloenzyme that is expressed in osteoclasts and in subsets of tissue macrophages and dendritic cells (1). It is also expressed at lower levels in the parenchymal cells of the liver, glomerular mesangial cells of the kidney, and pancreatic acinar cells. TRAP expression is dramatically up-regulated during osteoclast differentiation. Therefore, TRAP activity is commonly used as the identifying histochemical marker for osteoclasts (1). TRAP plays an important role in bone resorption. Mice lacking TRAP exhibited a defect in endochondral ossification, mild osteopetrosis, and disordered macrophage inflammatory responses (2–4). Conversely, transgenic mice overexpressing TRAP resulted in a decrease in trabecular bone density with characteristic mild osteoporosis (5). TRAP regulates bone resorption by mediating the degradation of endocytosed matrix products during transcytosis in activated osteoclasts (6).
Several transcription factors have been shown to bind directly to the TRAP promoter and regulate its transcriptional activity. The microphthalmia transcription factor (MITF) has been shown to bind directly to the proximal TRAP promoter and to induce promoter activity (7). MITF cooperates with PU.1, which binds to an adjacent region of the TRAP promoter, resulting in increased TRAP promoter activity (8–10). Nuclear factor of activated T-cells c1 (NFATc1) is induced and activated by receptor activator of NF-κB ligand (RANKL) treatment (11–13) and recruited to the TRAP promoter where it cooperates with PU.1 and MITF to enhance TRAP expression. PU.1-interacting protein (14), USF1/2 (15), YY1 (16), and cAMP-response element-binding protein (12) have also been shown to bind to the TRAP promoter and enhance its expression in response to RANKL treatment.
RANKL is a tumor necrosis factor family cytokine. It has been established that RANKL plays a key role in osteoclast differentiation and maintenance of their functional activity (17). RANKL induces expression of c-Fos and NFATc1, the master transcription factor of osteoclastogenesis. The autoamplification of the NFATc1 gene then results in the efficient induction of osteoclast-specific genes (18).
There have been numerous reports describing effects of IL-4 on bone biology. Previous studies, performed in vivo or by using a complex in vitro co-culture system consisting of stromal cells and bone marrow precursors, have demonstrated potent effects of IL-4 on both osteoblasts and osteoclasts, indicating that IL-4 could play a complex role in regulating bone homeostasis (19–28). Using recombinant RANKL, we previously showed that IL-4 directly prevents the RANKL-induced differentiation of myeloid precursors to osteoclasts. Furthermore, we found that IL-4 inhibited bone resorption by mature osteoclasts; these responses were STAT6-dependent (29). Several mechanisms explaining the IL-4 effect have been proposed by different groups (29–34).
In this study, we analyzed the ability of IL-4 to regulate TRAP expression in RANKL-stimulated mature osteoclasts and in macrophages. We found that IL-4 reduced TRAP expression in mature osteoclasts in a STAT6-dependent manner by suppressing the RANKL-induced TRAP transcription. Although we identified a STAT6 binding site in the TRAP promoter, binding of STAT6 to this site was not directly responsible for this transcriptional repression; in fact, IL-4-activated STAT6 binding enhanced TRAP transcription on its own. IL-4 modestly induced TRAP expression in macrophages through the activation of STAT6. We also found that IL-4 suppressed the RANKL-induced transcription of c-Fos and NFATc1 in mature osteoclasts in a STAT6-dependent manner. Furthermore, ectopic expression of NFATc1 abrogated the ability of IL-4 to down-regulate TRAP in osteoclasts. These results suggest that the IL-4-activated STAT6 can play two opposing roles in regulating TRAP expression, depending on the context. STAT6 binds the TRAP promoter after IL-4 treatment and modestly enhances TRAP mRNA transcription in macrophages, whereas the IL-4-induced activation of STAT6 indirectly mediates the suppression of the RANKL-regulated TRAP promoter activity indirectly in osteoclasts by suppressing c-Fos and NFATc1 expression.
EXPERIMENTAL PROCEDURES
Cell Culture
RAW264.7 cells or RAW264.7 cells stably expressing RANK driven by the cytomegalovirus promoter as a result of transfection (29) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with penicillin, streptomycin (BioWhittaker, Walkersville, MD), and 10% heat-inactivated fetal bovine serum (Invitrogen). Nonadherent bone marrow mononuclear cells were isolated from femurs and tibias of 4–6-week-old female wild type BALB/c (Taconic Laboratories, Germantown, NY) mice and STAT6−/− mice (obtained from Dr. William E. Paul, National Institutes of Health and bred in house). Cells were cultured overnight in α-minimum Eagle's medium (BioWhittaker) to deplete adherent stromal cells. These nonadherent bone marrow mononuclear cells were cultured for 3 days in α-minimum Eagle's medium supplemented with penicillin, streptomycin, glutamine (BioWhittaker), 10% fetal bovine serum, and 20 ng/ml recombinant mouse M-CSF (R&D Systems, Minneapolis, MN) to generate osteoclast precursors (hereafter also called BMMs). Mature osteoclasts were obtained after 6–7 days of culture in the presence of 20 ng/ml M-CSF and 150 ng/ml RANKL (provided by Dr. Mehrdad Tondravi, National Cancer Institute).
Transfection and Luciferase Assay
For the cytokine stimulation assays, 2 × 106 RAW264.7 cells were electrotransfected (Nucleofector kit V, Amaxa, Cologne, Germany) with 2 μg of TRAP promoter-firefly luciferase construct (PKB5 and STt2, obtained from Dr. S. V. Reddy, Medical University of South Carolina, Charleston, SC; Ref. 35) and 0.2 μg of PRL-TK Renilla luciferase plasmid control (Promega, Madison, WI). After transfection, cells were treated with 150 ng/ml RANKL or 10 ng/ml IL-4 (R&D Systems), respectively, for various times. Luciferase activity was measured using the Dual-Luciferase reporter assay system following the manufacturer's protocol (Promega). For the co-transfection assays, 1 μg of transcription factor cDNAs including STAT6VT (provided by Dr. Mark Kaplan, Indiana University, Indianapolis, IN; Ref. 36), NFATc1 (obtained from Dr. Deborah Galson, University of Pittsburgh School of Medicine, Pittsburgh, PA), PU.1, and MITF (both purchased from Open Biosystems) was co-transfected in various combinations with the luciferase plasmids. Luciferase activity was measured as described above after culture in the presence or absence of IL-4 for 16 h.
Electrophoretic Mobility Shift Assay
The electrophoretic mobility shift assay was performed as described previously (37) using 32P-labeled double-stranded oligos derived from the sequences of the TRAP and Cϵ promoters. For oligos on the TRAP promoter, the sequences are 5′-ATGCAGTTCTGGGGAAGTCCA-3′ (sense) and 5′-ATGCTGGACTTCCCCAGAACT-3′ (antisense). For oligos on the Cϵ promoter, the sequences are 5′-ATGCCAACTTCCCAAGAACAGA-3′ (sense) and 5′-ATGCTCTGTTCTTGGGAAGTTG-3′ (antisense). Briefly, RANK-expressing RAW264.7 cells were incubated in the presence or absence of IL-4 (10 ng/ml) and/or RANKL (150 ng/ml) for 30 min. Whole cell extracts were prepared and incubated with the labeled oligos for 30 min before running on an acrylamide gel in 1× Tris borate-EDTA buffer. In some cases, anti-STAT6 antibody (M-200, Santa Cruz Biotechnology, Santa Cruz, CA) or unlabeled oligo was added to the cell extracts for 15 min prior to incubation with the 32P-labeled oligo.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using the ChIP kit (Upstate Biotechnologies, Inc., Lake Placid, NY). Briefly, 2 × 106 cells were treated with 1% formaldehyde for 15 min at 37 °C. The cells were washed and harvested in ice-cold phosphate-buffered saline (BioWhittaker) and lysed in lysis buffer (10 mm EDTA, 1% SDS, 50 mm Tris, pH 8.1, 0.01% protease inhibitor mixture). The cells were sonicated to shear the DNA and immunoprecipitated with anti-STAT6 (M-20 or S-20, Santa Cruz Biotechnology), anti-RNA polymerase II rabbit IgG antibody (C-21, Santa Cruz Biotechnology), mouse anti-NFATc1 antibody (7A6, Santa Cruz Biotechnology), or normal rabbit IgG control at 4 °C overnight. Reverse cross-linking between protein and DNA was performed at 65 °C for 5 h. Protein was digested with proteinase K. DNA was precipitated with ethanol after phenol/chloroform extraction and resuspended in Tris-EDTA buffer. The relative quantity of precipitated DNA was measured by PCR or by real time PCR using SYBR Green Master Mix (Applied Biosystems, Foster City, CA). The results are expressed as the relative -fold enrichment of the target precipitation as compared with the normal rabbit IgG control. The following primers were used for PCR. For the STAT6 binding site on the TRAP promoter, 5′-GGGACCTACAGATGCCCAGTAC-3′ (sense) and 5′-TTCTCCGAGGATTGTCCAGAAG-3′ (antisense) (177 bp) were used. For the RNA polymerase II ChIP assay, primer pairs in the transcriptional region were used: TRAP, 5′-GCAGACCAGGGAAACTGAAGCA-3′ (sense) and 5′-CGTTGATGTCGCACAGAGGGAT-3′ (antisense), 177 bp; NFATc1, 5′-ACCCAGTCTCCATACAGTCCTC-3′ (sense) and 5′-GCCCACCCTGCTCTTTCTAC-3′ (antisense), 179 bp.
TRAP mRNA Stability Assay
RANK-RAW264.7 cells and primary BMMs were treated with 20 ng/ml M-CSF and 150 ng/ml RANKL for 4 and 7 days, respectively. Cells were treated with 10 ng/ml IL-4 for 1 additional day. Subsequently, the cells were incubated with 5 μg/ml actinomycin D (Calbiochem) for various times. RNA was extracted, and TRAP and β-actin RT-PCR was performed.
Quantitive RT-PCR Analysis
Total RNA was extracted from cultured cells using the RNeasy kit (Invitrogen). The first strand of cDNA was transcribed from 2 μg of RNA with the Superscript RT kit (Invitrogen). Quantitation of the target gene relative to that of β-actin was determined using the SYBR Green dye method with detection on an ABI7700/SDS platform (Applied Biosystems). The primers used were as follows: NFATc1, 5′-CCGTTGCTTCCAGAAAATAACA-3′ (sense) and 5′-TGTGGGATGTGAACTCGGAA-3′ (antisense), 152 bp; TRAP, 5′-CCAATGCCAAAGAGATCGCC-3′ (sense) and 5′-TCTGTGCAGAGACGTTGCCAAG-3′ (antisense), 216 bp; c-Fos, 5′-GCAGAAGGGGCAAAGTAGAG-3′ (sense) and 5′-GTGTATCTGTCAGCTCCCTC-3′ (antisense), 123 bp; cathepsin K, 5′-GACGCAGCGATGCTAACTAA-3′ (sense) and 5′-CCAGCACAGAGTCCACAACT-3′ (antisense), 146 bp; glyceraldehyde-3-phosphate dehydrogenase, 5′-GCACAGTCAAGGCCGAGAAT-3′ (sense) and 5′-GCCTTCTCCATGGTGGTGAA-3′ (antisense), 151 bp; β-actin, 5′-TGCTGTCCCTGTATGCCTCTGGTC-3′ (sense) and 5′-TCTTTGATGTCACGCACGATTTCC-3′ (antisense), 226 bp.
TRAP Staining
The cells were fixed for 10 min with 3.7% formaldehyde at room temperature. The cells were then washed once with HBS (0.9% NaCl, 10 mm HEPES, pH 7.1) and stained for 20 min at 37 °C with TRAP staining solution (0.3 mg/ml fast red violet LB, 100 μg/ml naphthol AS-MX phosphate, 50 mm sodium acetate, 30 mm sodium tartrate (Sigma), 0.1% Triton X-100, pH 5.0).
STAT6VT Stable Transfection
RANK-expressing RAW264.7 cells were electrotransfected with STAT6VT-pcDNA3.1. After transfection, cells were serially diluted in medium containing 300 μg/ml hygromycin (Invitrogen) and 800 μg/ml G418 (Invitrogen). Antibiotic-resistant clones were selected, and one clone was used in further experiments.
Mutation Analysis
The QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to mutate the putative STAT6 binding site in the TRAP-luciferase vector STt2 (−881 to +2 bp relative to the translational start site) (35, 38). The putative STAT6 site sequence TTCTGGGGAA was mutated to CCATGGGGAA. The original STt2 and mutated STt2 (muSTt2) were transfected or co-transfected with STAT6VT, NFATc1, PU.1, and MITF into RAW264.7 cells. Luciferase activity was examined after 24 h.
siRNA and NFATc1 cDNA Transfection
RANK-RAW264.7 cells were treated with RANKL for 3 days. Cells were harvested and electrotransfected with two different NFATc1 siRNAs (siRNA1, GCCAUAACUUUCUGCAAGA; siRNA2, ACGGUUACUUGGAGAAUGA; Dharmacon, Lafayette, CO) or negative control siRNA (Ambion, Austin, TX) using Nucleofector kit V (Amaxa). In another group, NFATc1 cDNA was also co-transfected with the siRNAs. After transfection, cells were maintained in the presence of RANKL (150 ng/ml) for an additional 2 days. Total RNA was extracted, and NFATc1, TRAP, and cathepsin K expression levels were examined by quantitative RT-PCR. In another experiment, RANK-RAW264.7 cells were treated with RANKL for 3 days. Cells were harvested and transfected with empty vector or NFATc1 cDNA. Cells were further treated with RANKL in the presence or absence of IL-4 for an additional 2 days. Cells were harvested, and total RNA was extracted. NFATc1 and TRAP expression levels were examined by quantitative RT-PCR.
RESULTS
IL-4 Down-regulates RANKL-induced TRAP Expression in Mature Osteoclasts
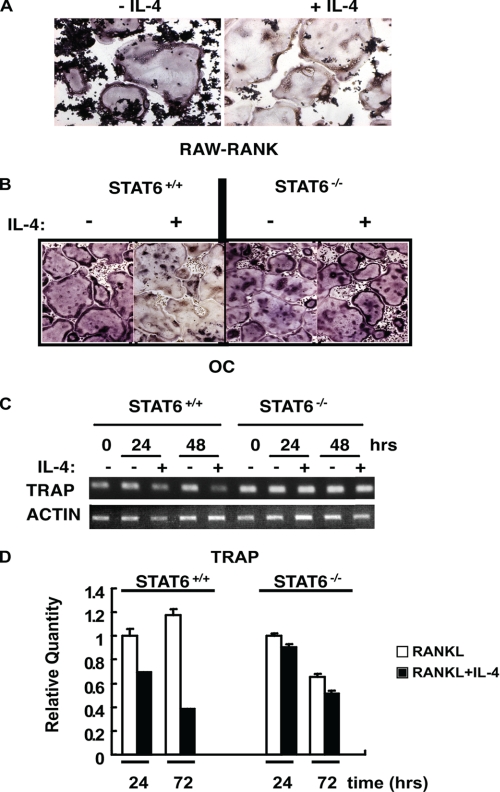
We previously showed that IL-4 suppressed the formation of osteoclasts induced by the treatment of the RAW264.7 cell line with M-CSF and RANKL (29). However, IL-4 did not inhibit the number of osteoclasts formed in RAW264.7 cells expressing RANK as a result of transfection (RANK-RAW) (29). Although IL-4 did not have an effect on osteoclast numbers, we observed less TRAP staining in the RANK-RAW cells treated with IL-4 and RANKL compared with cells treated with RANKL alone (Fig. 1A). In addition, treatment of mature STAT6+/+ osteoclasts with IL-4 for 48 h suppressed TRAP activity, whereas treatment of STAT6−/− osteoclasts with IL-4 did not affect TRAP (Fig. 1B). Furthermore, we observed the STAT6-dependent suppression of TRAP mRNA expression in primary mature osteoclasts treated with IL-4 over time (Fig. 1, C and D).
FIGURE 1.
Effect of IL-4 on TRAP expression in RANK-transfected RAW264.7 cells and primary mature osteoclasts. A, RAW264.7 cells expressing RANK as a result of transfection (RAW-RANK) were treated with RANKL (150 ng/ml) in the presence or absence of IL-4 (10 ng/ml) for 5 days. The cells were fixed and stained for TRAP as described under “Experimental Procedures.” B, BMMs were prepared from STAT6+/+ and STAT6−/− mice as indicated. These BMMs were cultured in the presence of M-CSF (20 ng/ml) and RANKL (150 ng/ml) for 5 days to generate mature multinucleated osteoclasts (OC). The cells were further cultured in the presence or absence of IL-4 for 48 h before TRAP expression was analyzed. C and D, mature osteoclasts were prepared as described in B and treated with IL-4 for various times. Total RNA was prepared and analyzed for mRNA expression by RT-PCR (C) or by quantitative PCR (D). The relative mRNA quantity of IL-4-treated osteoclasts was normalized to that of osteoclasts without IL-4 treatment on day 1. Results represent the mean ± S.D. of triplicate samples.
IL-4 Alone Enhances TRAP Expression but Inhibites RANKL-induced TRAP Expression
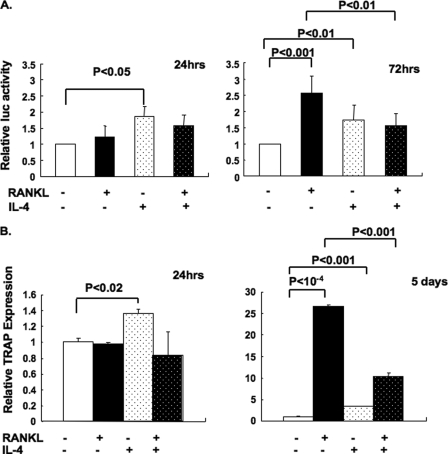
To examine the mechanism by which IL-4 regulates TRAP expression, we first transfected RAW264.7 macrophages with a construct containing the TRAP promoter (−1846 to +2 bp relative to the translational start site) fused to luciferase, PKB5 (35, 38). Transfection with a green fluorescent protein control plasmid showed a transfection efficiency of ∼90% (data not shown). Transfected cells were treated with RANKL in the presence or absence of IL-4, and luciferase activity was measured after 24 or 72 h (Fig. 2A). As published previously (15), we found that RANKL treatment up-regulated TRAP-luciferase activity ∼2.5–3-fold after 72 h of treatment with RANKL but not after 24 h. Treatment with IL-4 alone up-regulated TRAP luciferase activity by 1.7-fold after 24 or 72 h. However, IL-4 clearly inhibited the RANKL-induced TRAP luciferase activity observed after 72 h of cytokine treatment. These results suggest that IL-4 can inhibit RANKL-mediated transcription of TRAP. We previously found that IL-4 can suppress the increase in RANK expression in developing progenitors and thus limit signaling induced by RANKL (29). Therefore, to rule out an effect on RANK expression, we treated RANK-expressing RAW264.7 cells with RANKL in the presence or absence of IL-4 for 24 h or for 5 days to generate mature osteoclasts. TRAP expression level was measured by RT-PCR (Fig. 2B). Osteoclasts were fully induced after 5 days of RANKL treatment in the presence or absence of IL-4 (Fig. 1A). RANKL induced TRAP mRNA expression 27-fold over background in RANK-RAW cells after 5 days but did not up-regulate TRAP within the first 24 h. IL-4 alone slightly enhanced TRAP expression after 24 h and up-regulated TRAP expression ∼3.4-fold over background after 5 days. However, IL-4 significantly reduced the RANKL-induced TRAP expression (60% inhibition) after 5 days, whereas it did not affect osteoclast numbers (Fig. 1A and Ref. 29). These results suggest that IL-4 has opposing effects on TRAP expression that are dependent upon the presence or absence of RANKL.
FIGURE 2.
IL-4 inhibited the RANKL-induced TRAP mRNA expression and modestly enhanced TRAP mRNA expression on its own. A, the TRAP promoter fused to luciferase in the PGL2 vector (PKB5; −1846 to +2 bp relative to the translational start site) and the PRL-TK Renilla luciferase vector were co-transfected into RAW264.7 cells. The transfected cells were incubated in the presence or absence of RANKL (150 ng/ml) and IL-4 (10 ng/ml) as indicated for 24 or 72 h. Luciferase activity was measured by using the Promega Dual-Luciferase system kit. The relative luciferase activity was calculated as firefly luciferase activity divided by the Renilla luciferase activity. Luciferase activity of medium alone was normalized as 1. Data represent the mean ± S.D. of triplicate samples in 24 h. Samples in 72 h were repeated six times, and data represent the mean ± S.D. of six independent experiments. B, RANK-RAW cells were treated in the presence or absence of RANKL and IL-4 for 15 h or 5 days. Quantitative RT-PCR was performed to measure TRAP mRNA expression levels as described under “Experimental Procedures.” The abundance of TRAP mRNA was calculated relative to that of the internal reference gene, β-actin. The relative abundance of TRAP mRNA in untreated cells was normalized to 1. Data represent the mean ± the S.D. of triplicate samples.
Enhanced Expression of TRAP in Macrophages Is STAT6-dependent
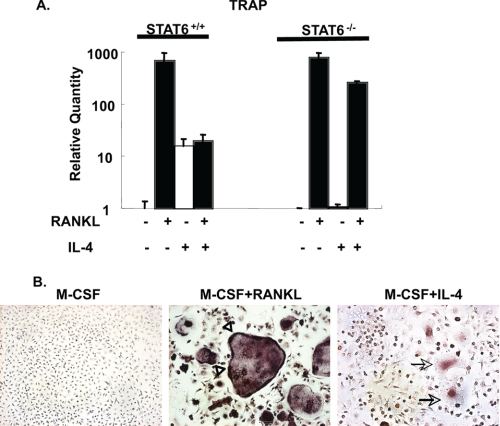
To determine whether IL-4 also enhanced TRAP in primary macrophages, STAT6+/+ BMMs and STAT6−/− BMMs were treated with IL-4 or RANKL for 5 days (Fig. 3). TRAP quantitative RT-PCR was performed to measure TRAP mRNA expression levels. RANKL alone dramatically up-regulated TRAP mRNA ∼600-fold in both STAT6+/+ and STAT6−/− mice. IL-4 alone increased TRAP mRNA by ∼15-fold in STAT6+/+ BMMs but did not up-regulate TRAP in STAT6−/− BMMs (Fig. 3A). The culture of BMMs with both RANKL and IL-4 resulted in levels of TRAP mRNA similar to those seen in the IL-4 alone group, demonstrating that IL-4 suppresses the RANKL-induced up-regulation of TRAP. These results indicate that both the IL-4-induced modest increase in TRAP mRNA and the IL-4-induced suppression of the potent RANKL-induced TRAP mRNA are STAT6-dependent. To confirm the mRNA analysis, TRAP staining was performed on wild type BMMs after cytokine treatment (Fig. 3B). BMMs cultured in M-CSF alone did not form multinucleated giant cells and were TRAP-negative (Fig. 3B). RANKL treatment induced strong TRAP staining in mature osteoclasts with TRAP concentrated along the border. IL-4 treatment induced the formation of macrophage multinucleated giant cells with a morphology distinct from that of osteoclasts (39). TRAP staining in these cells was much lighter compared with the RANKL-induced osteoclasts, and the staining was located in the center of the cell.
FIGURE 3.
IL-4 enhances TRAP expression in primary BMMs. A, STAT6+/+ and STAT6−/− BMMs were treated with M-CSF (20 ng/ml), RANKL (150 ng/ml), and/or IL-4 (10 ng/ml) as indicated for 5 days. TRAP mRNA level was measured by quantitative RT-PCR. The value of M-CSF alone was normalized to 1. B, TRAP staining was performed on wild type BMMs that were treated with the indicated cytokines for 5 days (▵, TRAP staining near the osteoclast border; →, TRAP staining in the IL-4-induced macrophage multinucleated giant cell).
IL-4 Suppresses the RANKL-induced TRAP Expression by Inhibiting Gene Transcription
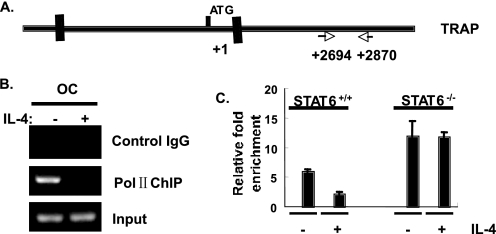
To determine whether IL-4 regulates TRAP expression at the level of transcription or mRNA stability, we performed the RNA polymerase II chromatin immunoprecipitation assay in primary mature osteoclasts as a measure of mRNA transcription (Fig. 4) (40). RANKL-stimulated osteoclasts demonstrated substantial association of RNA polymerase II with the TRAP gene that was diminished by IL-4 (Fig. 4B). As expected, untreated macrophage precursors did not show evidence of TRAP transcription as measured by RNA polymerase II ChIP (data not shown). IL-4 reduced the RANKL-induced RNA polymerase II binding to the TRAP gene in wild type mature osteoclasts but not in STAT6 knock-out osteoclasts (Fig. 4C). These results indicate that the inhibitory effect of IL-4 on TRAP expression is mediated by reducing TRAP mRNA transcription. Furthermore, we did not obtain any evidence for regulation of the half-life of TRAP mRNA by IL-4. We did not find typical AU-rich elements in the TRAP mRNA 3′-untranslated region (41), and IL-4 did not alter TRAP mRNA levels within 6 h after actinomycin D treatment (supplemental Fig. 1).
FIGURE 4.
IL-4 inhibited the RANKL-induced association of RNA polymerase II with TRAP. A, the location of the TRAP primer binding sites used in the RNA polymerase II (Pol II) chromatin immunoprecipitation assay is shown. B, mature osteoclasts (OC) were prepared from STAT6+/+ mice as described above. The osteoclasts were further cultured in the presence of RANKL (150 ng/ml) in the presence or absence of IL-4 (10 ng/ml) for 3 days. Cell lysates were prepared, and the RNA polymerase II chromatin immunoprecipitation assay was performed as described under “Experimental Procedures” using anti-RNA polymerase II antibody or normal rabbit IgG as a control. A representative TRAP ChIP PCR from wild type cells is shown. C, mature osteoclasts were prepared from STAT6+/+ or STAT6−/− mice and cultured as described above. An RNA polymerase II ChIP coupled to real time PCR was performed. The results are expressed as the relative -fold enrichment of TRAP mRNA in the anti-RNA polymerase II precipitation as compared with the normal rabbit IgG control. Results represent the mean ± S.D. of triplicate samples.
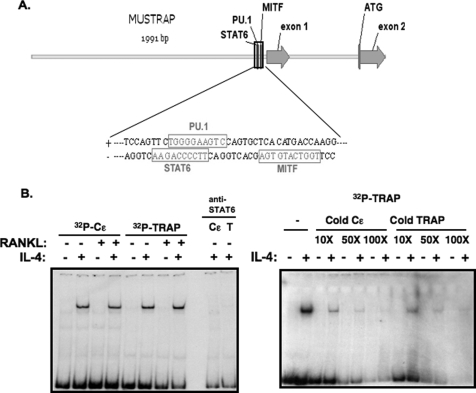
IL-4-induced STAT6 Can Directly Bind to TRAP Promoter
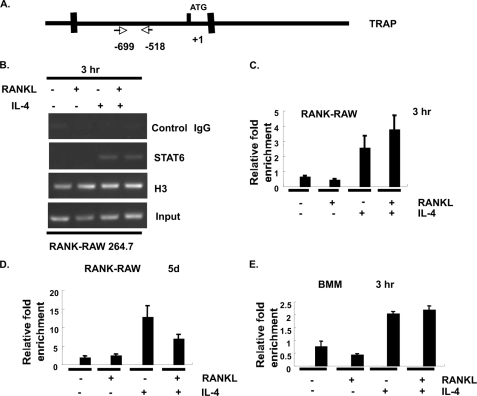
IL-4 did not suppress TRAP expression in mature osteoclasts prepared from STAT6−/− mice, indicating that the down-regulation of TRAP by IL-4 is STAT6-dependent (Fig. 1D). Interestingly, the TRAP promoter contains a STAT6 binding site that overlaps the PU.1 site shown to be a positive regulator of RANKL-induced TRAP transcription (8–10) (Fig. 5A). Based on this finding, we initially proposed that STAT6 directly regulated TRAP transcription through STAT6 binding to this site to prevent the RANKL-stimulated increase in transcription. To test whether STAT6 can bind to the TRAP promoter directly, an electrophoretic mobility shift assay was performed using a probe that contains the putative STAT6 binding site derived from the TRAP promoter using conditions optimized for STAT binding. We also used a probe containing a bona fide STAT6 site derived from the promoter of Cϵ. We found that IL-4 treatment induced a gel shift of both the TRAP- and Cϵ-derived probes; RANKL did not induce the formation of a gel shift complex and had no effect on the IL-4-induced complex (Fig. 5B). Anti-STAT6 antibody pretreatment led to a loss of this STAT6 DNA binding activity. Furthermore, the complex bound to the TRAP-derived oligo was completely abolished by cold competition using the Cϵ-derived oligo (Fig. 5B). We also performed a chromatin immunoprecipitation assay using primer pairs flanking the putative STAT6 site in the TRAP promoter to confirm STAT6 binding to the endogenous TRAP promoter in RANK-RAW cells (Fig. 6A). We found that STAT6 bound to the TRAP promoter as early as 3 h after IL-4 treatment by PCR and qPCR (Fig. 6, B and C). STAT6 was still bound to the promoter after 5 days of culture in the presence or absence of RANKL (Fig. 6D). We also could detect STAT6 binding to the TRAP promoter in primary bone marrow-derived macrophages after 3 h of treatment (Fig. 6E). RANKL treatment did not influence the detection of STAT6 bound to the TRAP promoter.
FIGURE 5.
STAT6 binds to the putative STAT6 binding site in the TRAP promoter. A, schematic of the murine TRAP promoter, highlighting the putative STAT6 binding site (TTCTGGGGAA) that overlaps the known PU.1 binding site. B, RANK-RAW264.7 cells were treated with IL-4 (10 ng/ml) or RANKL (150 ng/ml) as indicated for 30 min. Whole cell lysates were prepared and incubated with 32P-labeled double-stranded oligonucleotides derived from the sequences from the promoters of Cϵ or TRAP as described under “Experimental Procedures.” Where indicated, the lysates were preincubated for 30 min with anti-STAT6 antibody (M-200) or with excess unlabeled double-stranded oligonucleotides to inhibit the binding of the labeled probe. A representative of two independent experiments is shown.
FIGURE 6.
IL-4 induced the binding of STAT6 to the endogenous TRAP promoter as detected by ChIP assay. A, the location of the TRAP-specific PCR primers used in the STAT6 ChIP assay is shown. B, RANK-RAW264.7 cells were cultured in the presence or absence of RANKL (150 ng/ml) or IL-4 (10 ng/ml) as indicated for 3 h. Cell lysates were prepared, and the ChIP assay was performed as described under “Experimental Procedures” using anti-STAT6 antibody, anti-histone H3 as a positive control, or normal rabbit IgG as a negative control. A representative TRAP PCR for the precipitations and total input is shown. C, the samples from B were used for qPCR-coupled STAT6 ChIP. The results are expressed as the relative -fold enrichment of TRAP mRNA in the anti-STAT6 precipitation as compared with the normal rabbit IgG control. These results show the mean ± S.D. of triplicate samples of a representative experiment of three independent experiments. D, same as in C except that cytokine stimulation was for 5 days (5d). E, same as in C except that primary bone marrow-derived macrophages were treated with cytokine for 3 h.
STAT6VT Binding to TRAP Promoter Enhances TRAP Transcription
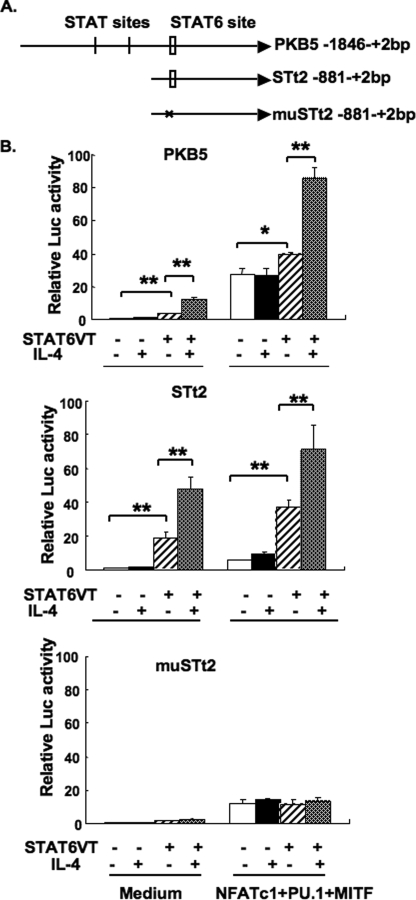
IL-4 induced the binding of STAT6 to the TRAP promoter, and it reduced RANKL-induced TRAP transcription. These results suggested that direct binding of STAT6 to the TRAP promoter may have resulted in the transcriptional repression of RANKL-induced TRAP. However, it was possible that the mechanism of suppression is through an indirect, secondary effect of the IL-4/STAT6 pathway. Indeed, IL-4 alone slightly up-regulated TRAP luciferase activity and mRNA expression (Figs. 2 and 3). To test whether STAT6 affected TRAP transcription directly, we performed transient transfection assays in RAW264.7 cells using a series of transcription factors in the absence of any RANKL. We used the three key transcription factors that are essential for osteoclastic TRAP expression, NFATc1, PU.1, and MITF (8, 11), and STAT6VT, a constitutively active form of STAT6 (42). The combination of NFATc1, PU.1, and MITF has been shown to induce TRAP transcription in the absence of RANKL treatment (8, 11); STAT6VT can bind to the STAT6-responsive element and regulate transcription without IL-4 treatment (42). Various combinations of these transcription factors were transfected into RAW264.7 cells. In all cases, the cells were co-transfected with the full-length TRAP-luciferase plasmid PKB5 (Fig. 7A). Luciferase activity was measured after 24 h. As expected, we found that the combination of NFATc1, PU.1, and MITF up-regulated TRAP-luciferase activity by 30-fold without using RANKL treatment (Fig. 7B, top panel). Interestingly, STAT6VT alone up-regulated TRAP luciferase activity ∼3-fold. Neither IL-4 nor expression of STAT6VT inhibited the NFATc1-, PU.1-, and MITF-driven luciferase activity. IL-4 significantly enhanced STAT6VT-induced luciferase activity in the absence and presence of these three transcription factors (Fig. 7B, top panel). We observed similar effects using a wild type STAT6 construct in the presence of IL-4 (supplemental Fig. 2). These results suggest that binding of STAT6 to its consensus site in the TRAP promoter does not directly suppress the NFATc1-, PU.1-, and MITF-driven transcription.
FIGURE 7.
STAT6VT directly enhanced TRAP transcription and did not inhibit TRAP transcription induced by transient transfection of NFATc1, PU. 1, and MITF. A, schematics of the TRAP-luciferase constructs. B, RAW264.7 cells were transfected with 2 μg of one of the TRAP-luciferase constructs shown above (PKB5, STt2, or mutant STt2 (muSTt2) as indicated) with or without 1 μg of cDNA encoding PU.1, NFATc1, and MITF. In some cases, the cells were also transfected with the cDNA encoding a constitutively active form of STAT6, STAT6VT. The cells were cultured overnight in the presence or absence of IL-4 (10 ng/ml) before analysis of the relative luciferase activity. The luciferase activity calculated for the luciferase vector alone group was normalized to 1. Results represent the mean ± S.D. of triplicate samples. *, p < 0.05; **, p < 0.01. This graph is representative of four independent experiments.
The TRAP promoter construct PKB5 contains two putative STAT N3 sites in addition to the STAT6 N4 binding site (Fig. 7A). Because STAT6 is capable of binding to both N3 and N4 sites, we also analyzed a truncated TRAP-luciferase plasmid, STt2 (−881 to +2 bp relative to ATG translational start site). STt2 only contains the single STAT6 N4 site and does not contain the other two STAT N3 sites (Fig. 7A). Compared with PKB5, the STAT6VT-inducing effect was more potent with the STt2 TRAP-luciferase construct (Fig. 7B, middle panel). The ability of IL-4 to enhance the STAT6VT-induced luciferase activity was also greater. Mutation of the N4 STAT6 binding site in the STt2 luciferase construct (muSTt2) abrogated the STAT6VT-induced luciferase activity, and IL-4 treatment did not enhance its activity (Fig. 7B, bottom panel). These results indicate that the STAT6VT-induced luciferase activity was mediated through the direct binding of STAT6VT to the TRAP promoter.
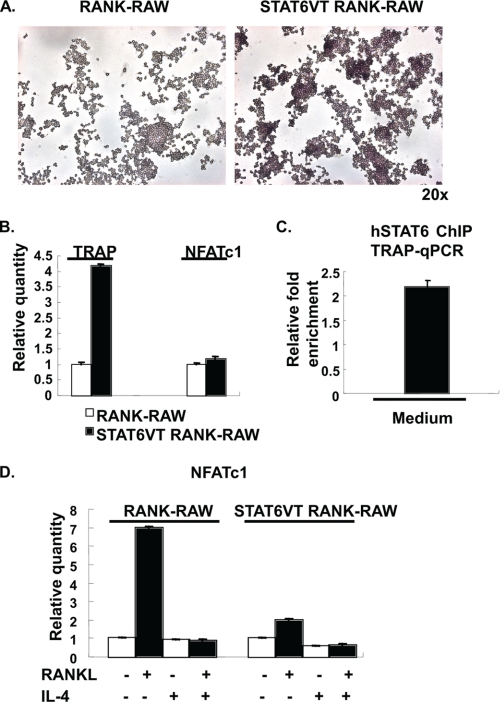
To confirm the luciferase assay results, we made STAT6VT stable transfectants using RANK-RAW cells. The untreated STAT6VT-transfected cells showed higher basal TRAP activity compared with untransfected cells (Fig. 8A). Real time PCR was performed to examine the TRAP mRNA expression levels. STAT6VT-transfected cells showed 4-fold more TRAP mRNA expression in untreated cells compared with the nontransfected cells (Fig. 8B). The up-regulation of TRAP in these cells did not correlate with expression of NFATc1; their NFATc1 expression levels were almost identical (Fig. 8B). To test whether STAT6VT directly binds to the TRAP promoter in these cells, a chromatin immunoprecipitation assay was performed by using anti-human STAT6 antibody (S-20). The ChIP assay showed that human STAT6VT can bind to the TRAP promoter directly in these transfected cells in the absence of IL-4 treatment (Fig. 8C). These results support the model that STAT6 binds directly to the TRAP promoter, resulting in relatively modest transcriptional activation.
FIGURE 8.
STAT6VT enhanced TRAP expression in stable cell lines. A, RANK-RAW cells were stably transfected with STAT6VT. Actively growing RANK-RAW cells and STAT6VT-RANK-RAW cells were stained for TRAP as described under “Experimental Procedures.” B, total RNA was isolated from RANK-RAW cells and STAT6VT-RANK-RAW cells. The expression of TRAP and NFATc1 mRNA was measured by quantitative RT-PCR. The relative abundance of TRAP and NFATc1 mRNA in untreated RANK-RAW cells was normalized to 1. C, cell lysates from STAT6VT-RANK-RAW cells were analyzed by qPCR-coupled anti-human STAT6 ChIP. The results are expressed as the relative -fold enrichment of TRAP mRNA in the anti-human STAT6 precipitation as compared with the normal rabbit IgG control. Results represent the mean ± S.D. of triplicate samples. D, RANK-RAW and STAT6VT-RANK-RAW cells were cultured in the presence or absence of RANKL (150 ng/ml) and IL-4 (10 ng/ml) as indicated for 5 days. Total RNA was prepared and analyzed for NFATc1 expression by quantitative RT-PCR. The data were normalized and are presented as in Fig. 1D.
NFATc1 is a key transcriptional regulator during osteoclast differentiation from progenitor cells. NFATc1 also can be recruited to several osteoclastic gene promoters and enhance their expression including TRAP (18). Therefore, we analyzed NFATc1 expression in parental and STAT6VT-expressing cells in the presence or absence of RANKL-induced osteoclast differentiation. We found that RANKL induced the expression of NFATc1 mRNA in parental osteoclasts by 7-fold after 5 days of treatment; this induction was completely inhibited by IL-4, consistent with previous data (Fig. 8D). The RANKL-induced expression of NFATc1 mRNA was substantially lower in the STAT6VT-expressing osteoclasts (2-fold over background), and addition of IL-4 further suppressed expression of NFATc1.
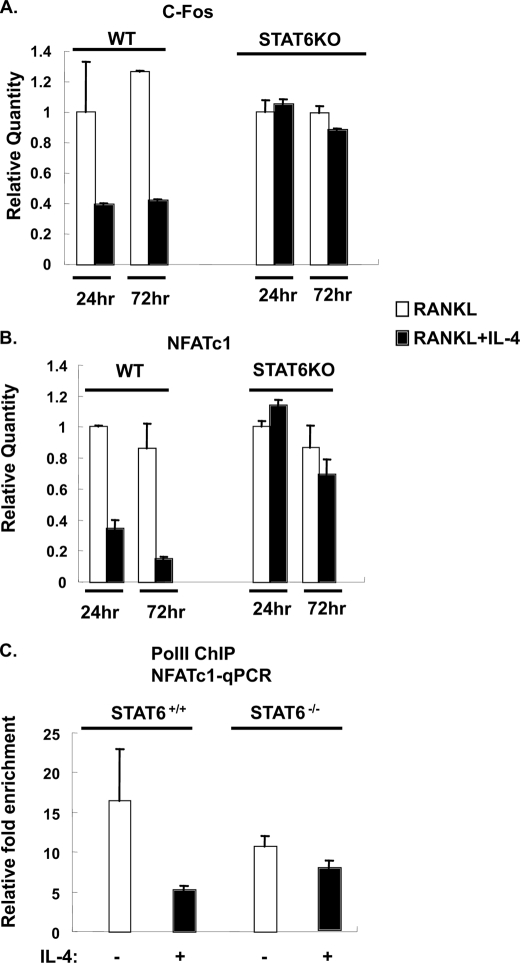
IL-4-mediated Down-regulation of RANKL-induced c-Fos and NFATc1 in Mature Osteoclasts
IL-4 inhibited TRAP expression in mature osteoclasts. Both c-Fos and NFATc1 are key transcription factors induced by RANKL during osteoclast differentiation, and they remain elevated in mature osteoclasts. NFATc1 is downstream of c-Fos induction and can be recruited to several osteoclastic gene promoters and enhance their expression, including TRAP (18). To test whether IL-4 regulated the RANKL-induced NFATc1 and c-Fos levels, we treated mature osteoclasts with IL-4 and measured the relative expression of c-Fos and NFATc1 mRNA by real time PCR. IL-4 significantly down-modulated c-Fos and NFATc1 mRNA levels in mature osteoclasts derived from STAT6+/+ mice (Fig. 9, A and B). This effect was not observed in osteoclasts derived from STAT6−/− mice. The ability of IL-4 to down-modulate NFATc1 is likely at the level of transcription. RANKL-stimulated osteoclasts demonstrated substantial association of RNA polymerase II with the NFATc1 gene that was diminished by IL-4 (Fig. 9C). IL-4 reduced the RANKL-induced RNA polymerase II binding to the NFATc1 gene in wild type mature osteoclasts but not in STAT6−/− osteoclasts (Fig. 9C).
FIGURE 9.
IL-4 down-regulated c-Fos and NFATc1 expression in STAT6+/+ osteoclasts but not in STAT6−/− osteoclasts. STAT6+/+ and STAT6−/− primary mature osteoclasts prepared as described under “Experimental Procedures” were treated with RANKL (150 ng/ml) in the presence or absence of IL-4 (10 ng/ml) for various times as indicated. A and B, total RNA was prepared and analyzed for c-Fos (A) or NFATc1 (B) expression by quantitative RT-PCR. The data were normalized and are presented as in Fig. 1D. C, cell lysates were prepared, and the ChIP assay was performed using anti-polymerase II antibody or normal rabbit IgG as a negative control. The relative abundance of NFATc1 in the sample was determined by qPCR. The results are expressed as the relative -fold enrichment of NFATc1 mRNA in the anti-polymerase II (PolII) precipitation as compared with the normal rabbit IgG control. Results represent the mean ± S.D. of triplicate samples. WT, wild type; KO, knock-out.
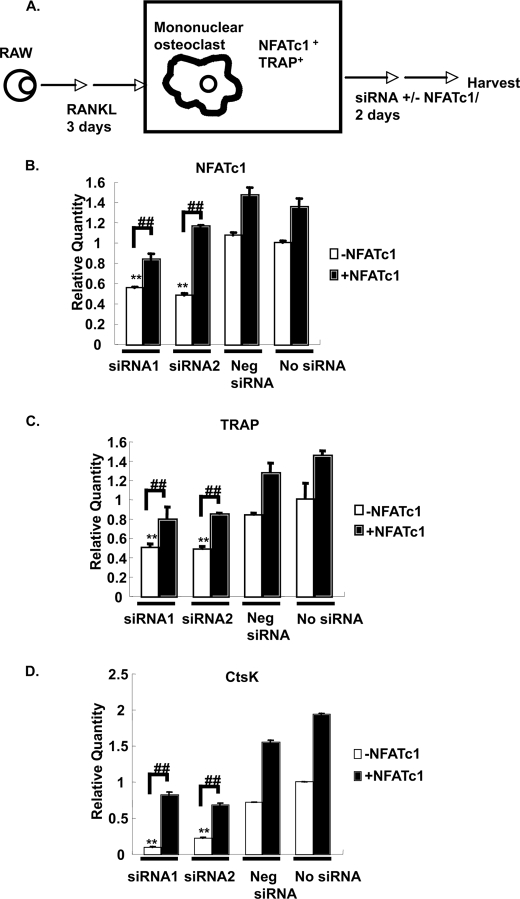
Down-regulation of NFATc1 by IL-4 Leads to Suppression of RANKL-induced TRAP Transcription
Based on these results, we hypothesized that down-regulation of NFATc1 by IL-4 may be responsible for the down-regulation of TRAP in mature osteoclasts. To test this hypothesis, RANK-RAW264.7 cells were treated with RANKL for 3 days. NFATc1 and TRAP were fully induced by this treatment (data not shown). Cells were harvested and transfected with two different NFATc1 siRNAs (Fig. 10A). After an additional 2 days in the presence of RANKL, total RNA was harvested. Quantitative PCR showed that the NFATc1 message was reduced by 50% compared with the negative control (Fig. 10B). Furthermore, TRAP and cathepsin K levels were suppressed by 50–90% (Fig. 10, C and D). To verify that the siRNA effect was specific, NFATc1 cDNA was co-transfected with the siRNA into RANK-RAW cells. NFATc1 co-transfection reversed the siRNA-induced down-regulation of NFATc1, TRAP, and cathepsin K messages (Fig. 10, B, C, and D).
FIGURE 10.
Down-regulation of NFATc1 by siRNA. A, schematic of siRNA transfection protocol. B–D, RANK-RAW cells were cultured in RANKL (150 ng/ml) for 3 days. The cells were then transfected with individual NFATc1 siRNAs (18 nm), a scrambled siRNA, or no siRNA as indicated and cultured for an additional 2 days in the presence of RANKL. In another group, cells were co-transfected with 3 μg of NFATc1 cDNA. Total RNA was harvested and analyzed by quantitative RT-PCR for NFATc1, TRAP, and cathepsin K (CtsK). For the qPCR analyses (B–D), the value of the untransfected cells was normalized to 1. Results represent the mean ± S.D. of triplicate samples. **, p < 0.01 comparing NFATc1 siRNA with the negative (Neg) scrambled siRNA transfection group; ##, p < 0.01, NFATc1 cDNA co-transfected with siRNA versus siRNA transfection only.
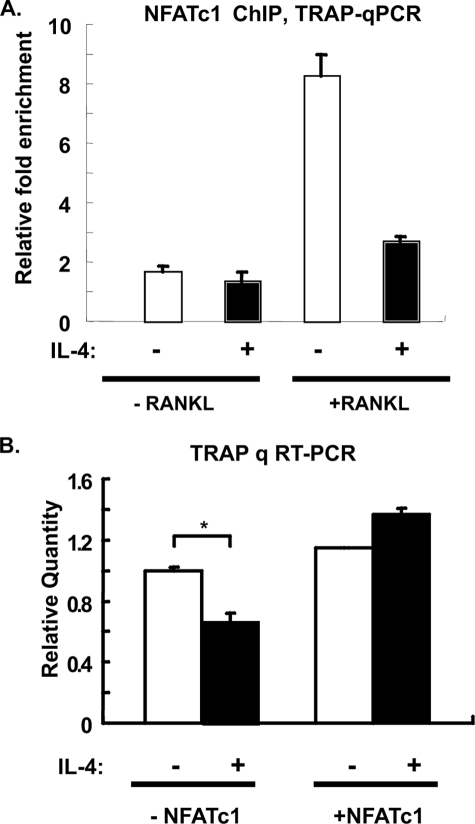
To determine whether the IL-4-induced down-regulation of NFATc1 was responsible for its inhibitory effect on TRAP, we first examined the effects of IL-4 on the binding of NFATc1 to the TRAP promoter in RANK-RAW cells by performing ChIP-coupled qPCR (Fig. 11A). ChIP assays using anti-NFATc1 antibody indicated that NFATc1 bound to the TRAP promoter after RANKL treatment but not after IL-4 treatment; the RANKL-induced binding was inhibited by IL-4 treatment. We also analyzed TRAP mRNA levels in the presence or absence of ectopic NFATc1 overexpression (Fig. 11B). Overexpression of NFATc1 abrogated the ability of IL-4 to suppress TRAP expression. Taken together, these results indicate that IL-4 inhibits the RANKL-induced TRAP transcription through the STAT6-dependent down-regulation of NFATc1 transcription.
FIGURE 11.
Ectopic expression of NFATc1 abrogates the suppressive effects of IL-4. A, RANK-RAW cells were cultured in the presence or absence of RANKL (150 ng/ml) and IL-4 (10 ng/ml) as indicated for 5 days. Cell lysates were prepared, and the ChIP assay was performed using anti-NFATc1 antibody or normal rabbit IgG as a negative control. The relative abundance of TRAP was determined by qPCR. The results are expressed as the relative -fold enrichment of TRAP mRNA in the anti-NFATc1 precipitation as compared with the normal rabbit IgG control. Results represent the mean ± S.D. of triplicate samples. B, RANK-RAW cells were treated with RANKL for 3 days as described in Fig. 9A. Cells were harvested and transfected with empty vector or NFATc1 cDNA (2 μg). After transfection, cells were cultured with RANKL in the presence of absence of IL-4 for an additional 2 days. Total RNA was harvested, and the TRAP mRNA level was examined by quantitative (q) RT-PCR. The relative quantity was normalized to the untreated control group. **, p < 0.01.
DISCUSSION
The ability of IL-4 to suppress the RANKL-induced differentiation of osteoclasts from macrophage precursors and to suppress mature osteoclast function has been reported extensively. Several mechanisms have been proposed for these effects: antagonism of the RANKL-activated NF-κB and mitogen-activate protein kinase pathways (30, 31, 34, 43), down-regulation of NFATc1 and c-Fos expression in progenitor cells (32), regulation of Ca2+ signaling (31), regulation of PPARγ1 (33), and suppression of RANK expression (29). We previously reported that the inhibitory effects of IL-4 on the bone resorbing activity of osteoclasts are STAT6-dependent (29). STAT6 is an important transcription factor activated downstream of IL-4 receptor signaling. STAT6-binding elements have been identified in the promoters of many genes including Cϵ and CD23. However, the mechanisms of STAT6-mediated gene regulation are still not completely clear. STAT6 can serve as both a positive and negative regulator of gene expression (44). STAT6 can interfere with NF-κB activity by competition for overlapping STAT6 and NF-κB DNA binding sites (45). Another potential mechanism of STAT6-mediated negative regulation of gene expression is the STAT6-mediated induction of transcriptional repressors. These repressors can directly dampen gene transcription as is the case for BCL-6 (46) or reduce the stability of target mRNA by induction of tristetraprolin (41).
In this study, we explored how IL-4 regulates the expression of TRAP, an enzyme expressed in dendritic cells and macrophages and at high levels in osteoclasts. We found distinct mechanisms of TRAP regulation in macrophages versus osteoclasts. IL-4 suppressed the RANKL-induced expression of TRAP in developing osteoclasts. IL-4 also dampened TRAP protein and mRNA expression in mature osteoclasts by regulating its transcription. Because we identified a putative STAT6 binding site in the TRAP promoter proximal to sites previously shown to be important for the RANKL-induced increase in TRAP transcription (8–11), we initially hypothesized that IL-4 suppressed TRAP expression by the direct binding of STAT6 to this site and blockade of transcription mediated by the combination of PU.1, MITF, and NFATc1. However, the TRAP promoter-luciferase experiments demonstrated a complicated pattern of effects mediated by IL-4. RANKL treatment induced significant TRAP-luciferase after 72 h of culture that was down-regulated in the presence of IL-4, whereas IL-4 alone mildly up-regulated TRAP-luciferase activity after 24 h. The relatively long culture period required to observe the RANKL-induced TRAP-luciferase activity (72 h) suggests that the expression of a cascade of essential secondary transcription factors is needed to mediate TRAP transcription. This is supported by the observation that transient transfection of the combination of NFATc1, PU.1, and MITF induced the rapid (16 h) and potent induction of TRAP-luciferase activity in the absence of RANKL signaling. These results also indicate that IL-4 may have both positive and negative effects on TRAP expression, depending on the particular cell type and stimulus.
The putative STAT6 binding site in the TRAP promoter overlaps an important PU.1 binding site and is near the MITF site. The electrophoretic mobility shift and chromatin immunoprecipitation assays demonstrated that STAT6 can bind to this site. Co-transfection with the constitutively active form of STAT6, STAT6VT, enhanced TRAP- luciferase activity and mRNA expression. IL-4 also enhanced TRAP expression in RANK+ RAW264.7 cells without STAT6VT transfection, although the magnitude was not comparable to the RANKL effect. Mutation of the putative STAT6 binding site in the TRAP promoter abrogated the STAT6VT- and IL-4-enhancing effects. Furthermore, IL-4 up-regulated TRAP expression in primary BMMs in a STAT6-dependent manner. These results indicate that STAT6 binds directly to the TRAP promoter and modestly induces transcription. However, this direct binding is likely not responsible for the suppression of the RANKL-induced increase in TRAP. Neither STAT6VT transfection nor IL-4 treatment suppressed the induction of TRAP transcription mediated by the combination of NFATc1, PU.1, and MITF.
The evidence we show here suggests that the ability of IL-4/STAT6 to suppress the RANKL-regulated TRAP transcription is most likely due to indirect mechanisms. We showed that IL-4 down-regulated c-Fos and NFATc1 mRNA induced by RANKL, and it suppressed the RANKL-induced association of NFATc1 with the TRAP promoter. Because induction of NFATc1 by RANKL is dependent on c-Fos, we focused further studies on NFATc1. We found that NFATc1 knockdown by siRNA led to down-modulation of TRAP and cathepsin K in osteoclasts, whereas overexpression of NFATc1 abrogated the ability of IL-4 to suppress TRAP transcription. These results indicate that IL-4 suppresses TRAP in mature osteoclasts by suppressing the RANKL-induced transcription of NFATc1. The mechanism by which the IL-4-activated STAT6 pathway suppresses the RANKL-induced transcription of NFATc1 is currently under investigation. We believe the regulation of NFATc1 by IL-4 is also via an indirect mechanism. The NFATc1 p1 promoter does not contain any identifiable STAT6 binding sites (47), and we have not found any evidence for direct regulation of an NFATc1 promoter-luciferase construct by IL-4/STAT6.3
Taken together, our results show that IL-4-activated STAT6 can directly bind to the TRAP promoter and up-regulate TRAP expression; this effect is distinct from the indirect mechanisms by which IL-4 suppresses the RANKL-regulated TRAP expression in osteoclasts. Both effects are mediated by STAT6. These findings illustrate the complex role that STAT6 plays in macrophage and osteoclast biology.
Our findings are in accordance with previous reports showing that IL-4 enhanced TRAP expression in both murine and human macrophages (48, 49). The significance of the ability of IL-4 to modestly induce TRAP expression in macrophages is still not clear. TRAP is highly expressed by osteoclasts and is important for bone matrix degradation (2–6). TRAP is also expressed in macrophages (50), dendritic cells, and many other tissues (51). TRAP−/− mice not only have an intrinsic defect in osteoclast resorption but also display abnormal immunomodulatory responses and cytokine secretion profiles (4). Interferon-γ-induced superoxide formation and nitrite production were enhanced in TRAP-deficient macrophages. The secretion of proinflammatory cytokines like tumor necrosis factor-α, interleukin 1β, and IL-12 was significantly greater when stimulated with lipopolysaccharide with or without addition of tumor necrosis factor-α or interferon-γ (3, 4). Production of reactive oxygen species and proinflammatory cytokines is known to be inhibited by IL-4-induced alternative macrophage activation, suggesting that TRAP may play a role in the differentiation of macrophages to the classical or alternative phenotype (52). Furthermore, dendritic cells from TRAP knock-out mice have impaired maturation and mediate defective Th1 responses (53, 54). These results suggest that TRAP may play some role in regulating type 2 inflammation by inhibiting classical macrophage activation and Th1 differentiation and/or by promoting Th2 differentiation and the differentiation of alternatively activated macrophages (52). The characterization of the specific role for the IL-4-induced TRAP in macrophages will require further investigation.
Interestingly, there are two enzyme isoforms found in serum (TRAP5a and -5b) (55). They are derived by differential, posttranslational processing of a common gene product (Acp5). TRAP5b is expressed in bone-resorbing osteoclasts and becomes elevated in diseases of increased bone resorption. TRAP5a is secreted by macrophages and dendritic cells and is increased in many patients with rheumatoid arthritis (56). An understanding of whether IL-4 regulates expression of these two isoforms would help to better understand its role in both the skeletal and immune systems.
Supplementary Material
Acknowledgments
We acknowledge Dr. Mark Kaplan, Dr. Deborah Galson, and Dr. S. V. Reddy for generously supplying plasmid cDNA for these studies; Dr. Mehrdad Tondravi for supplying recombinant RANKL and for advice during the course of these studies; Xiulan Qi for excellent technical assistance; and Dr. Nicola Heller for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI059775 and AI038985 from USPHS (to A. D. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
M. Yu, unpublished observations.
- TRAP
- tartrate-resistant acid phosphatase
- IL
- interleukin
- RANK
- receptor activator of NF-κB
- RANKL
- receptor activator of NF-κB ligand
- M-CSF
- macrophage colony-stimulating factor
- siRNA
- short interfering RNA
- MITF
- microphthalmia transcription factor
- NFATc1
- nuclear factor of activated T-cells c1
- BMM
- bone marrow-derived macrophage
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- qPCR
- quantitative PCR
- STAT
- signal transducers and activators of transcription.
REFERENCES
- 1.Walsh N. C., Cahill M., Carninci P., Kawai J., Okazaki Y., Hayashizaki Y., Hume D. A., Cassady A. I. (2003) Gene 307, 111–123 [DOI] [PubMed] [Google Scholar]
- 2.Bune A. J., Hayman A. R., Evans M. J., Cox T. M. (2001) Immunology 102, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayman A. R., Jones S. J., Boyde A., Foster D., Colledge W. H., Carlton M. B., Evans M. J., Cox T. M. (1996) Development 122, 3151–3162 [DOI] [PubMed] [Google Scholar]
- 4.Hayman A. R., Cox T. M. (2003) J. Bone Miner. Res. 18, 1905–1907 [DOI] [PubMed] [Google Scholar]
- 5.Angel N. Z., Walsh N., Forwood M. R., Ostrowski M. C., Cassady A. I., Hume D. A. (2000) J. Bone Miner. Res. 15, 103–110 [DOI] [PubMed] [Google Scholar]
- 6.Halleen J. M., Räisänen S., Salo J. J., Reddy S. V., Roodman G. D., Hentunen T. A., Lehenkari P. P., Kaija H., Vihko P., Väänänen H. K. (1999) J. Biol. Chem. 274, 22907–22910 [DOI] [PubMed] [Google Scholar]
- 7.Luchin A., Purdom G., Murphy K., Clark M. Y., Angel N., Cassady A. I., Hume D. A., Ostrowski M. C. (2000) J. Bone Miner. Res. 15, 451–460 [DOI] [PubMed] [Google Scholar]
- 8.Partington G. A., Fuller K., Chambers T. J., Pondel M. (2004) Bone 34, 237–245 [DOI] [PubMed] [Google Scholar]
- 9.Luchin A., Suchting S., Merson T., Rosol T. J., Hume D. A., Cassady A. I., Ostrowski M. C. (2001) J. Biol. Chem. 276, 36703–36710 [DOI] [PubMed] [Google Scholar]
- 10.Cassady A. I., Luchin A., Ostrowski M. C., Hume D. A. (2003) J. Bone Miner. Res. 18, 1901–1904 [DOI] [PubMed] [Google Scholar]
- 11.Sharma S. M., Bronisz A., Hu R., Patel K., Mansky K. C., Sif S., Ostrowski M. C. (2007) J. Biol. Chem. 282, 15921–15929 [DOI] [PubMed] [Google Scholar]
- 12.Sato K., Suematsu A., Nakashima T., Takemoto-Kimura S., Aoki K., Morishita Y., Asahara H., Ohya K., Yamaguchi A., Takai T., Kodama T., Chatila T. A., Bito H., Takayanagi H. (2006) Nat. Med. 12, 1410–1416 [DOI] [PubMed] [Google Scholar]
- 13.Ikeda F., Nishimura R., Matsubara T., Tanaka S., Inoue J., Reddy S. V., Hata K., Yamashita K., Hiraga T., Watanabe T., Kukita T., Yoshioka K., Rao A., Yoneda T. (2004) J. Clin. Investig. 114, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto M., Hisatake K., Nogi Y., Tsujimoto M. (2001) J. Biol. Chem. 276, 33086–33092 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Shi Z., Silveira A., Liu J., Sawadogo M., Yang H., Feng X. (2003) J. Biol. Chem. 278, 20603–20611 [DOI] [PubMed] [Google Scholar]
- 16.Shi Z., Silveira A., Patel P., Feng X. (2004) Gene 343, 117–126 [DOI] [PubMed] [Google Scholar]
- 17.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 18.Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bizzarri C., Shioi A., Teitelbaum S. L., Ohara J., Harwalkar V. A., Erdmann J. M., Lacey D. L., Civitelli R. (1994) J. Biol. Chem. 269, 13817–13824 [PubMed] [Google Scholar]
- 20.Kasono K., Sato K., Sato Y., Tsushima T., Shizume K., Demura H. (1993) Bone Miner. 21, 179–188 [DOI] [PubMed] [Google Scholar]
- 21.Lewis D. B., Liggitt H. D., Effmann E. L., Motley S. T., Teitelbaum S. L., Jepsen K. J., Goldstein S. A., Bonadio J., Carpenter J., Perlmutter R. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11618–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano Y., Watanabe K., Morimoto I., Okada Y., Ura K., Sato K., Kasono K., Nakamura T., Eto S. (1994) J. Bone Miner. Res. 9, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 23.Okada Y., Morimoto I., Ura K., Nakano Y., Tanaka Y., Nishida S., Nakamura T., Eto S. (1998) Bone 22, 361–365 [DOI] [PubMed] [Google Scholar]
- 24.Riancho J. A., Zarrabeitia M. T., Mundy G. R., Yoneda T., González-Macías J. (1993) J. Bone Miner. Res. 8, 1337–1344 [DOI] [PubMed] [Google Scholar]
- 25.Riancho J. A., Zarrabeitia M. T., Olmos J. M., Amado J. A., Gonzalez-Macias J. (1993) Bone Miner. 21, 53–61 [DOI] [PubMed] [Google Scholar]
- 26.Riancho J. A., Zarrabeitia M. T., Gonzalez-Macias J. (1993) Biochem. Biophys. Res. Commun. 196, 678–685 [DOI] [PubMed] [Google Scholar]
- 27.Roodman G. D. (1993) Calcif. Tissue Int. 53, Suppl. 1, S94– S98 [DOI] [PubMed] [Google Scholar]
- 28.Shioi A., Teitelbaum S. L., Ross F. P., Welgus H. G., Suzuki H., Ohara J., Lacey D. L. (1991) J. Cell. Biochem. 47, 272–277 [DOI] [PubMed] [Google Scholar]
- 29.Moreno J. L., Kaczmarek M., Keegan A. D., Tondravi M. (2003) Blood 102, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 30.Wei S., Wang M. W., Teitelbaum S. L., Ross F. P. (2002) J. Biol. Chem. 277, 6622–6630 [DOI] [PubMed] [Google Scholar]
- 31.Mangashetti L. S., Khapli S. M., Wani M. R. (2005) J. Immunol. 175, 917–925 [DOI] [PubMed] [Google Scholar]
- 32.Kamel Mohamed S. G., Sugiyama E., Shinoda K., Hounoki H., Taki H., Maruyama M., Miyahara T., Kobayashi M. (2005) Biochem. Biophys. Res. Commun. 329, 839–845 [DOI] [PubMed] [Google Scholar]
- 33.Bendixen A. C., Shevde N. K., Dienger K. M., Willson T. M., Funk C. D., Pike J. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2443–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abu-Amer Y. (2001) J. Clin. Investig. 107, 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcantara O., Reddy S. V., Roodman G. D., Boldt D. H. (1994) Biochem. J. 298, 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan M. H., Sehra S., Chang H. C., O'Malley J. T., Mathur A. N., Bruns H. A. (2007) Blood 110, 4367–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H. Y., Zamorano J., Keegan A. D. (1998) J. Biol. Chem. 273, 9898–9905 [DOI] [PubMed] [Google Scholar]
- 38.Reddy S. V., Scarcez T., Windle J. J., Leach R. J., Hundley J. E., Chirgwin J. M., Chou J. Y., Roodman G. D. (1993) J. Bone Miner. Res. 8, 1263–1270 [DOI] [PubMed] [Google Scholar]
- 39.Moreno J. L., Mikhailenko I., Tondravi M. M., Keegan A. D. (2007) J. Leukoc. Biol. 82, 1542–1553 [DOI] [PubMed] [Google Scholar]
- 40.Sandoval J., Rodríguez J. L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., López-Rodas G. (2004) Nucleic Acids Res. 32, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K., Nakajima H., Ikeda K., Maezawa Y., Suto A., Takatori H., Saito Y., Iwamoto I. ( 2003) J. Exp. Med. 198, 1717– 1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniel C., Salvekar A., Schindler U. (2000) J. Biol. Chem. 275, 14255–14259 [DOI] [PubMed] [Google Scholar]
- 43.Hirayama T., Dai S., Abbas S., Yamanaka Y., Abu-Amer Y. (2005) Arthritis Rheum. 52, 2719–2729 [DOI] [PubMed] [Google Scholar]
- 44.Schroder A. J., Pavlidis P., Arimura A., Capece D., Rothman P. B. (2002) J. Immunol. 168, 996–1000 [DOI] [PubMed] [Google Scholar]
- 45.Bennett B. L., Cruz R., Lacson R. G., Manning A. M. (1997) J. Biol. Chem. 272, 10212–10219 [DOI] [PubMed] [Google Scholar]
- 46.Harris M. B., Chang C. C., Berton M. T., Danial N. N., Zhang J., Kuehner D., Ye B. H., Kvatyuk M., Pandolfi P. P., Cattoretti G., Dalla-Favera R., Rothman P. B. (1999) Mol. Cell. Biol. 19, 7264–7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuvpilo S., Jankevics E., Tyrsin D., Akimzhanov A., Moroz D., Jha M. K., Schulze-Luehrmann J., Santner-Nanan B., Feoktistova E., König T., Avots A., Schmitt E., Berberich-Siebelt F., Schimpl A., Serfling E. (2002) Immunity 16, 881–895 [DOI] [PubMed] [Google Scholar]
- 48.Lacey D. L., Erdmann J. M., Tan H. L. (1994) J. Cell. Biochem. 54, 365–371 [DOI] [PubMed] [Google Scholar]
- 49.Akagawa K. S., Takasuka N., Nozaki Y., Komuro I., Azuma M., Ueda M., Naito M., Takahashi K. (1996) Blood 88, 4029–4039 [PubMed] [Google Scholar]
- 50.Janckila A. J., Slone S. P., Lear S. C., Martin A., Yam L. T. (2007) Am. J. Clin. Pathol. 127, 556–566 [DOI] [PubMed] [Google Scholar]
- 51.Hayman A. R., Macary P., Lehner P. J., Cox T. M. (2001) J. Histochem. Cytochem. 49, 675–684 [DOI] [PubMed] [Google Scholar]
- 52.Martinez F. O., Helming L., Gordon S. (2009) Annu. Rev. Immunol. 27, 451–483 [DOI] [PubMed] [Google Scholar]
- 53.Esfandiari E., Bailey M., Stokes C. R., Cox T. M., Evans M. J., Hayman A. R. (2006) J. Bone Miner. Res. 21, 1367–1376 [DOI] [PubMed] [Google Scholar]
- 54.Hayman A. R. (2008) Autoimmunity 41, 218–223 [DOI] [PubMed] [Google Scholar]
- 55.Halleen J. M., Alatalo S. L., Suominen H., Cheng S., Janckila A. J., Väänänen H. K. (2000) J. Bone Miner. Res. 15, 1337–1345 [DOI] [PubMed] [Google Scholar]
- 56.Janckila A. J., Parthasarathy R. N., Parthasarathy L. K., Seelan R. S., Hsueh Y. C., Rissanen J., Alatalo S. L., Halleen J. M., Yam L. T. (2005) J. Leukoc. Biol. 77, 209–218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.