Abstract
This study was aimed at characterizing the potential differences in gene expression in piglets inoculated with Porcine circovirus type 2 (PCV2), the essential causative agent of postweaning multisystemic wasting syndrome. Seven-day-old caesarean-derived, colostrum-deprived piglets were distributed into two groups: control (n = 8) and pigs inoculated with 105.2 TCID50 of the Burgos PCV2 isolate (n = 16). One control and three inoculated pigs were necropsied on days 1, 2, 5, and 8 post-infection (p.i.). The remaining pigs (four of each group) were sequentially bled on days 0, 7, 14, 21, and 29 p.i. (necropsy). Total RNA from the mediastinal lymph node (MLN) and lysed whole blood (LWB) samples were hybridized to Affymetrix Porcine GeneChip®. Forty-three probes were differentially expressed (DE) in MLN samples (FDR < 0.1, fold change > 2) and were distributed into three clusters: globally down-regulated genes, and up-regulated genes at early (first week p.i.) and late (day 29 p.i.) stages of infection. In LWB samples, maximal differences were observed at day 7 p.i., with 54 probes DE between control and inoculated pigs. Main Gene Ontology biological processes assigned to up-regulated genes were related to the immune response. Six common genes were found in both types of samples, all of which belonged to the interferon signaling antiviral effector pathway. Down-regulated genes were mainly related to cell adhesion and migration in MLN, and cellular organization and biogenesis in LWB. Microarray results were validated by quantitative real-time PCR. This study provides, for the first time, the characterization of the early and late molecular events taking place in response to a subclinical PCV2 infection.
Keywords: pig, time course, gene expression, porcine circovirus type 2, immune response
1. INTRODUCTION
Porcine circovirus type 2 (PCV2) is a small non-enveloped, single-stranded circular DNA virus that has been identified as the primary cause of postweaning multisystemic wasting syndrome (PMWS). Typical PMWS clinical signs are characterized by severe lost of weight (wasting), pallor of skin, respiratory distress, and jaundice, which mainly affect late nursery and fattening pigs [16]. The hallmark microscopic lesion of PMWS is moderate to severe lymphocyte depletion accompanied by histiocytic infiltration in lymphoid tissues and granulomatous inflammation in a variety of organs such as the lungs, liver, kidney, heart, and intestines [3, 41].
Some experimental infections have been able to reproduce the histopathological lesions observed in naturally PMWS-affected pigs; however, reproduction of disease has been limited to a few experiments [44]. In many cases, PMWS development requires a trigger such as coinfection with other pathogens (porcine parvovirus (PPV), porcine respiratory and reproductive syndrome virus (PRRSV), Mycoplasma hyopneumoniae, among others), or immune stimulation of the host [44]. Host genetics may also affect the outcome of PCV2 infection. In this sense, a genetic predisposition to suffer from PMWS has been pointed out since field observations and recent experimental studies identified certain genetic lines of pigs that tended to be more or less susceptible to PCV2 infection [25, 26, 32, 33, 36]. In addition to breed susceptibility/resistance to suffer from the disease, other individual genetic factors may also be underlying the observed differences in the ability of mounting a good adaptive immune response between susceptible and diseased pigs [10, 20, 27, 28, 39].
The recent advent of microarray technology has currently made available the determination of the gene expression level of thousands of different genes at the same time, thus allowing the profiling of the entire porcine transcriptome. This technology has been successfully applied to the study of the porcine immune response against several swine pathogens such as Salmonella [47, 50], Actinobacillus pleuropneumoniae [17], PRRSV [14, 21, 22], and pseudorabies virus [7, 8]. Previously, we performed an exploratory microarray study using lung and mesenteric lymph node samples from PCV2-inoculated Duroc pigs at 23 days p.i. (dpi) thereby identifying several genes closely related to the immune response such as cytokines, CD8, immunoglobulin, and T cell receptor (TCR) alpha molecules which were mostly up-regulated in the PCV2-inoculated group [6].
The present work is aimed at characterizing the early and late molecular mechanisms underlying the immune response of cesarean derived, colostrum deprived (CDCD) piglets subclinically infected with PCV2 using a genome-wide expression approach. Mediastinal lymph node (MLN) and peripheral blood RNA samples were collected at five different time points, and were hybridized to the Affymetrix 24K Porcine Genechip1, which is a 25-oligomer one channel chip that contains 24 123 probesets, interrogating a total of 20 201 Sus scrofa genes. This study gives new insights into the knowledge of PCV2 host-pathogen interaction and the mechanisms by which an effective immune response occurs.
2. MATERIALS AND METHODS
2.1. Experimental design
All experimental procedures and animal care were undertaken in accordance with the guidelines of the Good Experimental Practices, under the supervision of the Ethical and Animal Welfare Committee of the Universitat Autònoma de Barcelona.
Specifically, 24 seven-day-old, Landrace CDCD piglets were used. The selection of Landrace pigs was done due to the fact that this pig breed has been shown to be more susceptible to suffer from PMWS disease [32, 33]. A first group of pigs (n = 8) was kept as un-inoculated controls and the rest of the pigs (n = 16) were oronasally inoculated with 105.2 TCID50 of the Burgos isolate of PCV2 [11]. The piglets used in the present work belonged to previous studies in which the virological, clinico-pathological and immunological outcomes were evaluated [5, 12]. Briefly, all pigs remained clinically healthy during the experimental period. PCV2 subclinical infection was confirmed in all virus-inoculated pigs by quantitative real time PCR (qPCR). The PCV2 genome was detected from 7 dpi to the end of the experimental period and all pigs had seroconverted by the end of the study. Microscopic examination revealed mild PMWS-like lesions mostly in the MLN of almost all PCV2-inoculated pigs. Control piglets remained free of PCV2 infection throughout the experiment and no histological lesions were detected.
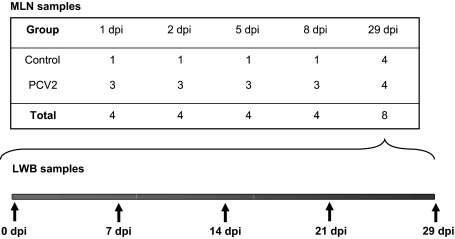
One control pig and three inoculated pigs were necropsied on days 1, 2, 5, and 8 post-inoculation (p.i.). The remaining pigs (4 of each group) were followed up throughout the experimental period, being bled at days 0, 7, 14, 21, and 29 p.i. (Fig. 1). One milliliter of whole blood samples were immediately lysed (referred to as LWB) with nucleic acid purification lysis solution (Applied Biosystems, Warrington, UK) and were immediately frozen at −96 °C. At necropsy (days 1, 2, 5, 8, and 29 p.i., Fig. 1), samples of MLN were collected by immersion in liquid nitrogen for microarray studies. All collected samples were kept at −80 °C until usage.
Figure 1.
Experimental design. MLN: mediastinal lymph node, LWB: lysed whole blood.
2.2. RNA extraction and microarray hybridization
Total RNA extraction from MLN and LWB samples was performed with the RiboPure™ kit (Ambion, Austin, USA), following the manufacturer’s instructions. RNA quality was assessed with the RNA Nano 6000 Labchip kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, USA). RNA was quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). Samples were hybridized to the Affymetrix 24K Genechip® Porcine Genome Array (Affymetrix, Santa Clara, CA, USA) following the standard Affymetrix one-cycle protocol. Reverse transcription, RNA labeling, cRNA amplification, hybridization, and scanning procedures were conducted at the Affymetrix facilities available at the Institut de Recerca Hospital Universitari Vall d’Hebron, Barcelona, Spain2. In total, 24 MLN samples (8 control and 16 PCV2-inoculated) and 39 LWB samples (corresponding to 2 groups × 4 piglets/group × 5 time points) were hybridized to microarrays. One sample of a PCV2 inoculated pig at day 21 p.i. was discarded due to low RNA quality.
2.3. Microarray data analysis
Raw data and statistical analyses were performed with Bioconductor [15] implemented in R 2.6.03. Data quality was assessed by the QC function implemented in the simpleaffy package [49]. The Robust Multichip Average (RMA, [19]) methodology was used for array normalization. The Empirical Bayes t-test statistic implemented in the limma package [40] was used to determine differential gene expression between control and inoculated pigs. For LWB, a comparison between both groups was performed for each time point. For MLN samples, where only one control pig was available at 1, 2, 5, and 8 dpi, the effect of time was included in the model as a fixed effect. The threshold of significance was set to a false discovery rate (FDR, [2]) of 0.1 and a minimum fold change of 2. Hierarchical clustering was performed with Cluster 3.0 and Java TreeView 1.1 software4, using the uncentered correlation coefficient and the average linkage method. Probes were annotated based on the chip annotation provided by Affymetrix (NetAffx), Tsai et al. [45], and the annotation of Iowa State University5. However, some of the probes were not coincident between different sources and were, therefore, validated by screening the probe nucleotide sequence available at NetAffx with the nr and EST databases available at NCBI using the Basic Local Alignment Search Tool6. These probes that could not be assigned to a known gene were not used for functional analyses. The Database for Annotation, Visualization and Integrated Discovery7 was used for assessing functional profiles of genes based on the Biological Processes (BP) category of Gene Ontology (GO). The MetaCore platform8 was used to map biological processes to canonical pathways and to construct gene interaction networks9.
2.4. Quantitative real-time PCR
Validation of porcine differentially expressed (DE) transcripts was done by qPCR for five genes (Interferon gamma, IFNG; Immunoglobulin gamma chain constant region, IgG; lectin galactoside-binding soluble 3, LGALS3; myxovirus (influenza virus) resistance 1, Mx1; and 2′,5′-oligoadenylate synthetase 1, OAS1) for the MLN samples and one gene (OAS1) for the LWB samples. The hypoxantine phosphoribosyltransferase (HPRT1) gene was used as a reference housekeeping gene in the MLN samples. The beta-actin gene (ACTB) was selected as a reference for qPCR analyses in LWB samples due to the extremely low expression of HPRT1 gene in blood. Porcine specific primers were designed with the Primer Express software (Applied Biosystems, Warrington, UK). Primer sequences are shown in Table I. cDNA synthesis was performed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using 1 μL of total RNA from MLN and LWB. Real-time qPCR was performed in triplicate in a 20 μL final volume reaction containing 4 μL of a 1:20 dilution of the cDNA, 300 nM of each primer, 0.2 μM random hexamers, and 10 μL of Power SYBR Green® PCR Master Mix on an ABI Prism H7000 (Applied Biosystems). The thermal profile consisted of a denaturalization step at 95 °C for 10 min followed by 40 cycles at 95 °C/15 s and 60 °C/1 min. PCR efficiencies between target and housekeeping genes were validated for their relative quantification following the comparative Ct method described by Livak and Schmittgen [24]. Resulting qPCR data were Log2 transformed and analyzed, on a gene-by-gene basis, with the proc GLM method of SAS software (Statistics, V 9.1; SAS Institute, Inc., Cary, NC, USA) following the models used for microarray data analysis. The significance threshold was set at α < 0.05.
Table I.
List of primers used for quantitative PCR analysis.
| Name | Sequence 5′ → 3′ | Amplicon size (bp) | GenBank accession number |
|---|---|---|---|
| ACTB-F | CGCCCAGCACGATGAAG | 63 | DQ845171 |
| ACTB-R | CCGATCCACACGGAGTACTTG | ||
| HPRT1-F | TCATTATGCCGAGGATTTGGA | 90 | DQ136030 |
| HPRT1-R | CTCTTTCATCACATCTCGAGCAA | ||
| IgG-F | CAAGAGCTACACCTGCAATGTCA | 59 | U03778-82 |
| IgG-R | CACGCTTGTCCACCTTGGT | ||
| MX1-F | CCCCTCCATAGCCGAGATCT | 55 | DQ095779 |
| MX1-R | TGCCGACCTCCTGATGGTA | ||
| OAS1-F | CTGTCGTTGGACGATGTATGCT | 63 | NM_214303 |
| OAS1-R | GCCGGGTCCAGAATCACA | ||
| LGALS3-F | AACAATTCTGGGCACAGTAAAGC | 71 | NM_001097501 |
| LGALS3-R | CAACATCATTCCCCTTCTTGAAA | ||
| IFNG-F | GAATGACTTCGAAAAGCTGATTAAAA | 61 | EU118363 |
| IFNG-R | TGGCTTTGCGCTGGATCT |
3. RESULTS
3.1. Microarray analysis
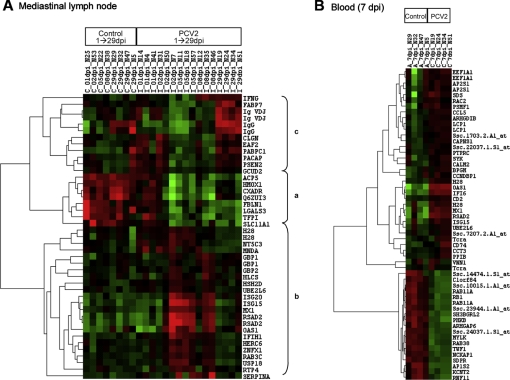
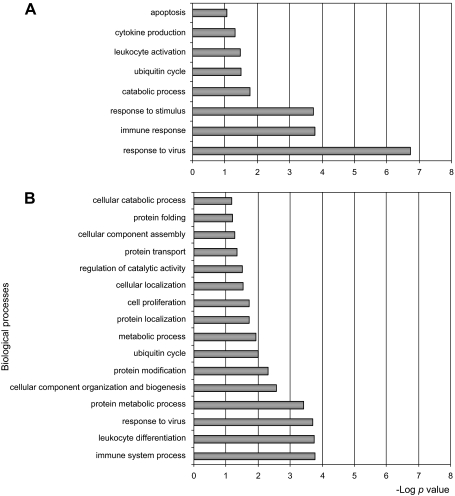
The comparison of the gene expression level between control and infected pigs in MLN samples revealed 43 DE probes (FDR < 0.1, Log2 fold change > 2, Tab. II). Gene expression differences varied with time and three differentiated clusters were identified (Fig. 2A). One cluster grouped eight probes that were globally down-regulated, from day 2 p.i. to the end of the study (cluster a). Among the up-regulated probes, two patterns were identified (clusters b and c). Cluster b grouped 23 probes that were up-regulated at early time-points after infection (5–8 dpi), while cluster c grouped 12 probes up-regulated at later stages of infection (29 dpi). Thirty-five out of 43 DE probes corresponded to well annotated genes and were, therefore, used for functional analyses. The 35 DE genes were assigned to eight biological processes (p < 0.05, Fig. 3A). The most significant biological processes over-represented in the MLN dataset were mainly related to immune system response, catabolic processes and apoptosis.
Table II.
List of differentially expressed genes between PCV2-inoculated and control pigs in mediastinal lymph node samples with the Affymetrix Porcine Genechip.
| Probe ID | Gene symbol | Gene name | Log2 FC | FDR | Biological processes |
|---|---|---|---|---|---|
| Globally down-regulated genes | |||||
| Ssc.17815.1.S1_at | LGALS3 | Lectin, galactoside-binding, soluble, 3 | −1.50 | 0.009 | Extracellular matrix organization |
| Ssc.575.1.S1_at | ACP5 | Acid phosphatase 5, tartrate resistant | −1.24 | 0.009 | Response to stimulus |
| Ssc.22441.1.A1_at | – | Annotation not clear | −1.20 | 0.024 | |
| Ssc.300.1.S1_at | SLC11A1 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | −1.08 | 0.097 | Immune response |
| Ssc.20870.1.S1_at | FBLN1 | Fibulin-1 | −1.07 | 0.007 | Cell adhesion |
| Ssc.7212.1.A1_at | TFPI | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | −1.07 | 0.051 | Blood coagulation |
| Ssc.115.1.S1_s_at | HMOX1 | Heme oxygenase (decycling) 1 | −1.05 | 0.028 | Apoptosis, cytokine production, catabolic process |
| Ssc.13115.1.A1_at | CXADR | Coxsackie virus and adenovirus receptor | −1.01 | 0.008 | Cell adhesion |
| Early up-regulated genes | |||||
| Ssc.19089.1.A1_at | Annotation not clear | 1.00 | 0.037 | ||
| Ssc.10588.1.A1_at | H28 | Histocompatibility 28 | 1.01 | 0.048 | Immune response |
| Ssc.30724.1.S1_at | HERC6 | Hect domain and RLD 6 | 1.03 | 0.069 | Ubiquitin-cycle |
| Ssc.29054.3.S1_at | GBP1 | Guanylate binding protein 1 | 1.03 | 0.044 | Immune response |
| Ssc.7116.1.A1_at | NT5C3 | 5′-nucleotidase, cytosolic III | 1.05 | 0.045 | Metabolic process |
| Ssc.26189.1.S1_a_at | RTP4 | Receptor (chemosensory) transporter protein 4 | 1.07 | 0.097 | Response to stimulus |
| Ssc.2641.1.S1_at | UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | 1.08 | 0.045 | Ubiquitin cycle |
| Ssc.883.1.S1_a_at | GBP2 | Guanylate binding protein 2 | 1.12 | 0.051 | Immune response |
| Ssc.336.1.S1_at | USP18 | Ubiquitin specific peptidase 18 | 1.13 | 0.083 | Ubiquitin cycle |
| Ssc.6433.2.S1_at | – | Annotation not clear | 1.13 | 0.084 | |
| Ssc.26009.1.S1_at | – | Annotation not clear | 1.16 | 0.082 | |
| Ssc.10593.1.S1_at | H28 | Histocompatibility 28 | 1.17 | 0.051 | Immune response |
| Ssc.9327.1.A1_at | HSH2D | Hematopoietic SH2 domain containing | 1.18 | 0.031 | Leukocyte activation |
| Ssc.7558.1.A1_at | – | Annotation not clear | 1.20 | 0.033 | |
| Ssc.29054.2.S1_at | GBP1 | Guanylate binding protein 1 | 1.30 | 0.033 | Immune response |
| Ssc.221.1.S1_at | MX1 | Myxovirus (influenza virus) resistance 1 | 1.41 | 0.070 | Response to virus, apoptosis |
| Ssc.17894.1.A1_at | – | Annotation not clear | 1.47 | 0.042 | |
| Ssc.5020.1.S1_at | SERPINA3 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 1.50 | 0.057 | Response to stimulus |
| Ssc.11557.1.A1_at | ISG15 | ISG15 ubiquitin-like modifier | 1.78 | 0.090 | Ubiquitin cycle |
| Ssc.1031.1.S1_at | OAS1 | 2′-5′-oligoadenylate synthetase 1 | 1.82 | 0.089 | Immune response |
| SscAffx.1.1.S1_at | ISG20 | Interferon stimulated exonuclease gene 20 kDa | 1.88 | 0.031 | Response to virus |
| Ssc.286.1.S1_s_at | RSAD2 | Radical S-adenosyl methionine domain containing 3 | 2.16 | 0.052 | Response to virus |
| AFFX-Ss_IRP_3_at | RSAD2 | Radical S-adenosyl methionine domain containing 2 | 2.29 | 0.064 | Response to virus |
| Late up-regulated genes | |||||
| Ssc.23658.1.S1_at | PACAP | Proapoptotic caspase adaptor protein | 1.00 | 0.019 | Apoptosis |
| Ssc.21217.1.A1_at | GCUD2 | Gastric cancer up-regulated-2 | 1.01 | 0.019 | Unknown |
| Ssc.24982.1.S1_at | FABP7 | Fatty acid-binding protein 7, brain | 1.03 | 0.041 | Cell proliferation |
| Ssc.19400.2.A1_at | - | Annotation not clear | 1.03 | 0.041 | |
| Ssc.11070.1.S1_at | IgG | Immunoglobulin G | 1.13 | 0.098 | Immune response |
| Ssc.10498.1.A1_at | EAF2 | ELL associated factor 2 | 1.14 | 0.019 | Apoptosis, regulation of transcription |
| Ssc.13778.1.S1_at | IgG | Immunoglobulin G | 1.15 | 0.024 | |
| Ssc.12505.1.A1_at | CLGN | Calmegin | 1.28 | 0.007 | Protein binding |
| Ssc.15942.2.S1_x_at | Ig VDJ | Ig heavy chain variable region (VDJ) | 1.38 | 0.046 | Immune response |
| Ssc.15942.3.S1_x_at | Ig VDJ | Ig heavy chain variable region (VDJ) | 1.47 | 0.051 | Immune response |
| Ssc.4093.1.A1_at | IFNG | Interferon gamma | 1.54 | 0.009 | Cytokine production, apoptosis |
| Ssc.23408.1.A1_s_at | – | Annotation not clear | 1.63 | 0.024 | |
LogFC: log fold change, FDR: false discovery rate.
Figure 2.
Heat maps of the differentially expressed probes between control (C) and PCV2-inoculated (I) pigs in (A) mediastinal lymph node, where a, b, and c, represent clusters for globally down-regulated probes, and early and late up-regulated probes in the mediastinal lymph node dataset, respectively; and (B) blood samples. Red represents up-regulation and green shows down-regulation for differentially expressed genes (FDR < 0.1, fold change > 2.0). (A color version of this figure is available at: www.vetres.org.)
Figure 3.
Biological process GO categorization of the significant differentially expressed genes between control and PCV2-inoculated pigs in (A) mediastinal lymph node and (B) blood samples.
In LWB samples, maximal differences in gene expression between control and PCV2-inoculated pigs were found at 7 dpi (Tab. III). Only three probes were found DE on day 21 p.i., two of them up-regulated (DEP domain containing 1B, involved in the intracellular signaling cascade, and DC2 protein, a membrane component) and one down-regulated (Exportin 7, involved in protein export from the nucleus) in PCV2-inoculated pigs. No significant differences were found at any of the remaining time-points. Among the 54 DE probes in LWB samples at 7 dpi, 35 probes were up-regulated and 19 probes were down-regulated in the PCV2-inoculated group (Fig. 2B). DE probes corresponded to 42 confirmed unique genes. Figure 3B shows the most significant BP of the LWB dataset, which were mainly related to the immune system response, protein metabolism and cellular organization and biogenesis.
Table III.
List of differentially expressed genes between PCV2-inoculated and control pigs in blood samples with the Affymetrix Porcine Genechip.
| Probe ID | Gene symbol | Gene name | log2 FC | FDR | Biological processes |
|---|---|---|---|---|---|
| Ssc.4717.1.S1_at | PHKB | Phosphorylase kinase beta | −1.59 | 0.024 | Metabolic process |
| Ssc.14474.1.S1_at | – | annotation not clear | −1.36 | 0.066 | |
| Ssc.10889.1.A1_s_at | RAB11A | RAB11A, member RAS oncogene family | −1.27 | 0.041 | Cellular component organization and biogenesis |
| Ssc.3657.1.A1_at | SH3BGRL2 | SH3 domain-binding glutamic acid-rich-like protein 2 | −1.26 | 0.05 | Unknown |
| Ssc.4572.1.S1_at | RNF11 | RING finger protein 11 | −1.24 | 0.026 | Protein metabolic process |
| Ssc.21114.1.S1_at | TWF1 | Twinfilin 1 | −1.19 | 0.034 | Protein metabolic process |
| Ssc.4135.2.A1_at | AP1S2 | Adaptor-related protein complex 1, sigma 2 subunit | −1.19 | 0.034 | Cellular component organization and biogenesis |
| Ssc.951.1.S1_at | RAB11A | RAB11A, member RAS oncogene family | −1.19 | 0.06 | Cellular component organization and biogenesis |
| Ssc.23247.1.S1_at | MYLK | Myosin light chain kinase | −1.15 | 0.097 | Protein metabolic process |
| Ssc.23944.1.A1_at | – | Annotation not clear | −1.09 | 0.09 | |
| Ssc.10015.1.A1_at | – | Annotation not clear | −1.06 | 0.071 | |
| Ssc.9586.2.S1_at | SDPR | Serum deprivation response protein | −1.05 | 0.044 | Unknown |
| Ssc.9244.1.A1_at | NCKAP1 | NCK-associated protein 1 | −1.05 | 0.055 | Protein metabolic process |
| Ssc.24037.1.S1_at | – | Annotation not clear | −1.05 | 0.096 | |
| Ssc.17063.1.A1_at | RB1 | Retinoblastoma 1 | −1.03 | 0.017 | Leukocyte differentiation |
| Ssc.494.1.S2_at | KCNT2 | Potassium channel, subfamily T, member 2 | −1.03 | 0.024 | Metabolic process |
| Ssc.20188.2.S1_at | C1orf84 | Chromosome 1 open reading frame 84 | −1.02 | 0.004 | Unknown |
| Ssc.9708.1.A1_at | ARHGAP6 | Rho-GTPase-activating protein 6 | −1.01 | 0.025 | Cellular component organization and biogenesis |
| Ssc.6940.1.A1_s_at | RAB38 | RAB38, member RAS oncogene family | −1.01 | 0.042 | Protein transport |
| Ssc.7392.1.S1_at | CALM2 | Calmodulin 2 | 1 | 0.064 | Unknown |
| Ssc.2641.1.S1_at | UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | 1.01 | 0.07 | Ubiquitin cycle |
| Ssc.902.1.S1_a_at | CCT3 | Chaperonin containing TCP1, subunit 3 (gamma) | 1.03 | 0.004 | Protein folding |
| Ssc.428.10.A1_at | Tcra | T-cell receptor alpha chain | 1.04 | 0.067 | Immune system process |
| Ssc.22037.1.S1_at | – | Annotation not clear | 1.04 | 0.088 | |
| Ssc.7207.2.A1_at | – | Annotation not clear | 1.05 | 0.037 | |
| Ssc.1703.2.A1_at | – | Annotation not clear | 1.05 | 0.094 | |
| Ssc.23248.1.S1_at | PTPRC | Protein tyrosine phosphatase, receptor type, C | 1.07 | 0.027 | Leukocyte differentiation |
| Ssc.9839.1.S1_at | PPIB | Peptidylprolyl isomerase B | 1.08 | 0.02 | Protein folding |
| Ssc.11742.2.S1_at | SYK | Spleen tyrosine kinase | 1.09 | 0.099 | Leukocyte activation differentiation |
| Ssc.17304.3.S1_at | RAC2 | Ras-related C3 botulinum toxin substrate 2 | 1.11 | 0.073 | Cellular component organization and biogenesis |
| Ssc.7158.2.A1_a_at | CAPNS1 | Calpain small subunit 1 | 1.16 | 0.019 | Cell proliferation |
| Ssc.428.6.S1_a_at | Tcra | T-cell receptor alpha chain | 1.16 | 0.076 | Immune system process |
| Ssc.15890.1.S1_at | VNN1 | Vanin 1 | 1.17 | 0.078 | Metabolic process |
| Ssc.18389.3.S1_at | AP2S1 | Adaptor-related protein complex 2, sigma 1 subunit | 1.22 | 0.081 | Cellular component organization and biogenesis |
| Ssc.23774.3.S1_at | LCP1 | Lymphocyte cytosolic protein 1 | 1.23 | 0.019 | Cellular component organization and biogenesis |
| Ssc.1230.2.S1_at | ARHGDIB | Rho GDP-dissociation inhibitor 2 | 1.27 | 0.048 | |
| Ssc.13961.2.A1_at | EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 1.28 | 0.09 | Protein metabolic process |
| Ssc.26249.2.S1_at | CCNDBP1 | Cyclin D-type binding-protein 1 | 1.32 | 0.097 | |
| Ssc.23774.2.S1_at | LCP1 | Lymphocyte cytosolic protein 1 | 1.32 | 0.098 | Cellular component organization and biogenesis |
| Ssc.22500.1.S1_at | BPGM | 2,3-bisphosphoglycerate mutase | 1.33 | 0.096 | Cellular catabolic process |
| Ssc.23793.1.S1_at | CD2 | CD2 molecule | 1.38 | 0.005 | Leukocyte differentiation |
| Ssc.10593.1.S1_at | H28 | Histocompatibility 28 | 1.39 | 0.005 | Immune system process |
| Ssc.6222.1.S1_a_at | CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | 1.42 | 0.05 | Leukocyte differentiation |
| Ssc.22030.1.S1_at | CCL5 | Chemokine (C-C motif) ligand 5 | 1.44 | 0.018 | Immune system process |
| Ssc.18389.2.S1_a_at | AP2S1 | Adaptor-related protein complex 2, sigma 1 subunit | 1.46 | 0.055 | Cellular component organization and biogenesis |
| AFFX-Ssc-ef1a-5_at | EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 1.55 | 0.096 | Protein metabolic process |
| Ssc.11557.1.A1_at | ISG15 | ISG15 ubiquitin-like modifier | 1.83 | 0.018 | Ubiquitin cycle |
| Ssc.10588.1.A1_at | H28 | Histocompatibility 28 | 1.88 | 0.004 | Immune system process |
| Ssc.6353.2.S1_at | PSMF1 | Proteasome (prosome, macropain) inhibitor subunit 1 | 1.98 | 0.055 | Protein metabolic process |
| Ssc.221.1.S1_at | MX1 | Myxovirus (influenza virus) resistance 1 | 2.31 | 0.004 | Response to virus |
| Ssc.11076.1.S1_at | SDS | Serine dehydratase | 2.42 | 0.08 | Metabolic process |
| Ssc.20101.1.S1_at | IFI6 | Interferon, alpha-inducible protein 6 | 2.99 | 0.001 | |
| Ssc.286.1.S1_s_at | RSAD2 | Radical S-adenosyl methionine domain containing 2 | 3.04 | 0.004 | Response to virus |
| Ssc.1031.1.S1_at | OAS1 | 2′-5′-oligoadenylate synthetase 1 | 3.78 | 0.004 | Response to virus |
LogFC: log fold change, FDR: false discovery rate.
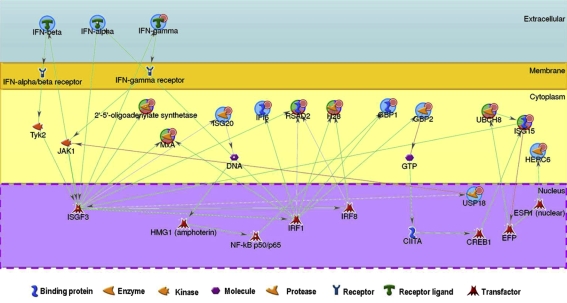
The comparison between MLN and LWB datasets revealed six common DE genes (OAS1, Mx1, ISG15, UBE2L6, RSAD2, and H28). Both datasets were jointly analyzed with the Pathway analysis option of the Metacore™ platform and revealed that the top scored map (p < 0.0001) corresponded to the antiviral action of interferons. A gene interaction network could be constructed using the common gene set as a starting point and allowing the entrance of other DE genes, unique either to the MLN or to the LWB datasets, which were proven to be involved in the same pathway (Fig. 4).
Figure 4.
Gene interaction network representing the interferon-induced antiviral effectors differentially expressed in both mediastinal lymph node and blood datasets. Small red circles indicate up-regulated genes. The Metacore software (GeneGO Inc.) was used to create the network. The six common genes to mediastinal lymph node and blood datasets, indicated by tricolor circles, were the starting nodes. Blue circles represent genes added manually. Red, green, and grey arrows indicate activation, inhibition and unspecified interactions between connected genes, respectively. (A color version of this figure is available at: www.vetres.org.)
3.2. Validation of microarray experiments by quantitative PCR
To confirm the DE genes in the microarray experiment, five (IFNG, IgG, OAS1, Mx1, LGALS3) and one (OAS1) genes were selected for real-time qPCR validation in the MLN and LWB datasets, respectively. Real-time qPCR results are shown in Table IV. All qPCR validated genes displayed significant differences in gene expression between control and PCV2-inoculated pigs and showed similar fold changes as the ones obtained by the microarray analysis (Tab. IV), thus indicating that the microarray data was highly reliable.
Table IV.
Results of the quantitative PCR validation of differentially expressed genes between PCV2-inoculated and control pigs and comparison with microarray gene expression data.
| Genes | Quantitative PCR |
Microarrays |
||||||
|---|---|---|---|---|---|---|---|---|
| FC | Log2 FC | S.E. | p value | Probes | FC | Log2 FC | p value | |
| Mediastinal lymph node | ||||||||
| IFNG | 3.63 | 1.86 | 0.38 | 0.0001 | Ssc.4093.1.A1_at | 2.91 | 1.54 | 0.0086 |
| IgG | 2.93 | 1.55 | 0.44 | 0.0026 | Ssc.11070.1.S1_at | 2.19 | 1.13 | 0.0981 |
| Ssc.13778.1.S1_at | 2.22 | 1.15 | 0.0236 | |||||
| LGALS3 | −3.70 | −1.89 | 0.53 | 0.0023 | Ssc.17815.1.S1_at | −2.86 | −1.50 | 0.0086 |
| Mx1 | 2.62 | 1.39 | 0.66 | 0.0501 | Ssc.221.1.S1_at | 2.67 | 1.41 | 0.0699 |
| OAS1 | 3.92 | 1.97 | 0.69 | 0.0103 | Ssc.1031.1.S1_at | 3.53 | 1.82 | 0.0887 |
| Blood (7 dpi) | ||||||||
| OAS1 | 11.96 | 3.58 | 1.07 | 0.0156 | Ssc.1031.1.S1_at | 13.69 | 3.78 | 0.0039 |
FC: fold change, S.E.: standard error.
4. DISCUSSION
The most important challenge for PCV2 researchers nowadays is the understanding of PMWS pathogenesis. The reason why all animals become infected but only a small percentage develops the disease is a question that remains unsolved. Several authors suggest that the complex host-virus interaction and the final ability of the pig to mount an effective immune response may be the key factors [28, 42, 46]. Here, the transcriptional profile of CDCD pigs subclinically infected with PCV2 in MLN and LWB samples was characterized to gain insight into the early and late molecular events taking place during PCV2 infection.
Overall, three patterns of gene expression were identified in MLN samples: globally down-regulated genes, and up-regulated genes at early and late stages of infection; whereas in LWB samples DE genes were mostly identified at day 7 p.i. Most down-regulated genes in MLN encoded for molecules that participate in cell adhesion and migration processes such as LGALS3 [31], FBLN1 [43], TFPI [35], HMOX1 [1], and CXADR [9]. These gene products mainly act as inhibitors of migration and cell proliferation and, therefore, the sustained reduction in their expression, found from day 2 p.i. onwards in all PCV2-inoculated pigs, may be related to the inflammatory processes (granulomatous infiltration) occurring in animals suffering PMWS.
The first days after PCV2 infection appeared to be the moment in which a higher number of genes were up-regulated both in MLN and LWB samples. The vast majority of these up-regulated genes were involved in a common pathway, the interferon-mediated antiviral effector pathway. This result agrees well with the fact that a peak of IFN-α was detected at day 5 p.i. in the PCV2-inoculated pigs from this experiment [12]. However, differences in IFNA gene expression in PCV2-inoculated pigs could not be detected, probably due to the fact that its expression took place in a different time point to those herein analyzed. Activation of interferons in response to viral infection leads to the activation of a cascade of intracellular signaling events that, ultimately, induce the expression of hundreds of genes, commonly known as interferon stimulated genes (ISG). Most of these ISG have been shown to display antiviral properties (for a review see [37]). The most prominent interferon-mediated antiviral effectors represented in the present study were OAS1, Mx1, and ISG15. The OAS1 protein catalyzes the synthesis of 2′,5′-oligomers of adenosine that bind to and activate RNase L, which degrades viral and cellular RNA, leading to the inhibition of cellular protein synthesis and impairment of viral replication [37]. Mx1 belongs to the dynamin superfamily of large GTPases and has been shown to exert their antiviral function by binding viral essential components, thereby blocking viral replication [37]. ISG15 has been recognized as an ubiquitin-like protein [18]. Protein ubiquitylation implies the post-translational labeling of a protein by covalent attachment of an ubiquitin monomer for its degradation in the proteasome. This mechanism has been shown to exert a crucial role in the regulation of immune response [34]. Several ubiquitin (UBE2L6, HERC6, and USP18) and proteasome (PSMF1) related enzymes were up-regulated in PCV2-inoculated pigs. Recently, it has been shown that the PCV2 open reading frame (ORF) 3 interferes with porcine ubiquitin E3 ligase Pirh2 [23]. The ubiquitin-proteasome system plays a key role in host-pathogen interactions and many viruses have developed different immune evasion strategies by altering this pathway [13]. The activation of several ubiquitin-proteasome related genes in PCV2 subclinically infected pigs may indicate that this pathway is crucial for the control of PCV2 pathogenesis. Other interferon-inducible genes found differentially regulated either in MLN or LWB datasets were RSAD2, H28, IFI44L, ISG20, GBP1, GBP2, and IFI6. Overall, these results indicate that an effective activation of the immune response was produced early (first week) after infection with PCV2 in lymph nodes (at least in the MLN), which is also reflected in blood samples, where a number of genes directly related to the activation of the immune system were also found up-regulated (TCRA, CCL5, CD2, CD74, and SYK).
In LWB samples at 7 dpi, several genes implicated in the organization and biogenesis of cellular components were DE between control and PCV2-inoculated pigs, such as the members of the Rab (RAB11A and RAB38) and Rho (RAC2, ARHGDIB, and ARHGP6) small GTPases, and the clathrin-associated adaptor complexes (AP1S2 and AP2S1). These genes appeared mostly down-regulated except for AP2S1 and ARHGDIB transcripts, which were up-regulated. These genes have been shown to participate in endocytosis-related processes. PCV2 internalization is produced by endocytosis, mainly through actin and Rho-GTPase mediated, dynamin-independent pathways [29, 30]. Furthermore, antigen (Ag) presentation by professional antigen presenting cells involves an active uptake of superficial Ag through macropinocytosis and/or phagocytosis processes followed by a complete arrest of this process to Ag processing and presentation to T cells in secondary lymphoid organs [4, 38, 48]. The fact that some of these genes were up-regulated while others were down-regulated might be explained by the fact that a mixture of cells at different stages can be found in LWB, and both processes (virus internalization and Ag presentation) can occur simultaneously.
In late stages of infection, a relatively low number of DE probes were found compared to the results reported by [6]. In that experiment, different pig breed, tissue samples, necropsy days, and statistical analysis were used, which may explain the differences found between both experiments. However, an increase of certain cytokines (CCL4L, CXCL9, and CXCL11 in [6], and IFNG in the present work) and IgG mRNA was detected in PCV2-inoculated pigs from both studies, thus indicating that similar immunological responses against PCV2 were obtained in Duroc [6] and Landrace (this study) subclinically infected pigs. Furthermore, the expression of IFNG and IgG genes in MLN samples correlated well with the immunological results obtained by [12] using animal material from the present experiment. In this work, a peak of IFN-γ was detected between days 14 and 21 p.i. and seroconversion took place between days 7 and 14 p.i. in all PCV2-inoculated pigs. In the current experiment, expression of IFNG gene started increasing from day 8 p.i. and was mainly over-expressed in pigs infected with PCV2 at day 29 p.i., which agrees well with its role in the early immune response against viruses.
A relatively low number of DE probes were found for both MLN and LWB datasets. This may probably be due to two main reasons. First, tissues are heterogeneous, composed by a mixture of different cell types, each with a specific transcriptional profile. Second, the low number of samples used for microarray analyses may increase data variability and reduce the statistical power to detect DE genes. This aspect might be the case for the analyses of MLN samples, since only one control pig was used for each timepoint.
Overall, this study has allowed the characterization, for the first time, of the genes that are involved in the molecular events underlying an effective immune response to counteract an infection with PCV2 and, more importantly, to control disease progression. The results from this study provide new insights into the complex host-PCV2 interaction, from a subclinical point of view. Given the difficulties in reproducing PMWS disease experimentally, further studies should be performed in healthy and naturally PMWS-affected pigs to explore the host-virus molecular interactions upon disease status.
Acknowledgments
Authors gratefully acknowledge the contribution of Dr Albert Bensaid for his critical revision of the manuscript. This study was supported by grant GEN2003-20658-C05-01/02 and project PORCIVIR (CONSOLIDER-INGENIO 2010) from the Spanish government, and research project No. 513928 of the VI Framework Programme of the European Union. Microarray hybridizations were subsidized by Fundación Genoma España10. L.T. Fernandes was funded by a CAPES grant BEX2893/04-5 from the Ministry of Education of Brazil. A. Tomas was supported by a Juan de la Cierva grant (JCI-2006-3940-3513) from the Spanish Government.
Footnotes
Tuggle C., personal communication.
DAVID 2008, http://david.abcc.ncifcrf.gov/home.jsp
GeneGo Inc., http://www.genego.com
Data from this work is available at Gene Expression Omnibus database with accession number GSE14758.
References
- 1.Alcaraz M.J., Fernández P., Guillén M.I., Anti-inflammatory actions of the heme oxygenase-1 pathway, Curr. Pharm. Des. (2003) 9:2541–2551 [DOI] [PubMed] [Google Scholar]
- 2.Benjamini Y., Hochberg Y., Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. R. Stat. Soc. Ser. B (1995) 57:289–300 [Google Scholar]
- 3.Chianini F., Majó N., Segalés J., Domínguez J., Domingo M., Immunohistochemical characterisation of PCV2 associate lesions in lymphoid and non-lymphoid tissues of pigs with natural postweaning multisystemic wasting syndrome (PMWS), Vet. Immunol. Immunopathol. (2003) 94:63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faure-André G., Vargas P., Yuseff M.I., Heuzé M., Diaz J., Lankar D., et al. , Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain, Science (2008) 322:1705–1710 [DOI] [PubMed] [Google Scholar]
- 5.Fernandes L.T., Mateu E., Sibila M., Fort M., Andaluz A., McNeilly F., et al. , Lack of in vitro and in vivo effects of lipopolysaccharide (LPS) on porcine circovirus type 2 infection, Viral Immunol. (2007) 20:541–552 [DOI] [PubMed] [Google Scholar]
- 6.Fernandes L.T., Tomás A., Bensaid A., Pérez-Enciso M., Sibila M., Sánchez A., Segalés J., Exploratory study on the transcriptional profile of pigs subclinically infected with porcine circovirus type 2, Anim. Biotechnol. (2009) 20:96–109 [DOI] [PubMed] [Google Scholar]
- 7.Flori L., Rogel-Gaillard C., Cochet M., Lemonnier G., Hugot K., Chardon P., et al. , Transcriptomic analysis of the dialogue between Pseudorabies virus and porcine epithelial cells during infection, BMC Genomics (2008) 9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flori L., Rogel-Gaillard C., Mariani V., Lemonnier G., Cochet M., Hugot K., et al. , A combined transcriptomic approach to analyse the dialogue between pseudorabies virus and porcine cells, Dev. Biol. (2008) 132:99–104 [DOI] [PubMed] [Google Scholar]
- 9.Fok P.T., Huang K.C., Holland P.C., Nalbantoglu J., The Coxsackie and adenovirus receptor binds microtubules and plays a role in cell migration, J. Biol. Chem. (2007) 282:7512–7521 [DOI] [PubMed] [Google Scholar]
- 10.Fort M., Olvera A., Sibila M., Segalés J., Mateu E., Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs, Vet. Microbiol. (2007) 125:244–255 [DOI] [PubMed] [Google Scholar]
- 11.Fort M., Sibila M., Allepuz A., Mateu E., Roerink F., Segalés J., Porcine circovirus type 2 (PCV2) vaccination of conventional pigs prevents viremia against PCV2 isolates of different genotypes and geographic origins, Vaccine (2008) 26:1063–1071 [DOI] [PubMed] [Google Scholar]
- 12.Fort M., Fernandes L.T., Nofrarias M., Díaz I., Sibila M., Pujols J., et al. , Development of cell-mediated immunity to porcine circovirus type 2 (PCV2) in caesarean-derived, colostrum-deprived piglets, Vet. Immunol. Immunopathol. (2009) 129:101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao G., Luo H., The ubiquitin-proteasome pathway in viral infections, Can. J. Physiol. Pharmacol. (2006) 84:5–14 [DOI] [PubMed] [Google Scholar]
- 14.Genini S., Delputte P.L., Malinverni R., Cecere M., Stella A., Nauwynck H.J., Giuffra E., Genome-wide transcriptional response of primary alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus, J. Gen. Virol. (2008) 89:2550–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., et al. , Bioconductor: open software development for computational biology and bioinformatics, Genome Biol. (2004) 5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding J.C.S., Clark E.G., Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS), J. Swine Health Prod. (1997) 5:201–203 [Google Scholar]
- 17.Hedegaard J., Skovgaard K., Mortensen S., Sørensen P., Jensen T.K., Hornshøj H., et al. , Molecular characterisation of the early response in pigs to experimental infection with Actinobacillus pleuropneumoniae using cDNA microarrays, Acta Vet. Scand. (2007) 49:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann J., Lerman L.O., Lerman A., Ubiquitin and ubiquitin-like proteins in protein regulation, Circ. Res. (2007) 100:1276–1291 [DOI] [PubMed] [Google Scholar]
- 19.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P., Exploration, normalization, and summaries of high density oligonucleotide array probe level data, Biostatistics (2003) 4:249–264 [DOI] [PubMed] [Google Scholar]
- 20.Ladekjaer-Mikkelsen A.S., Nielsen J., Stadejek T., Storgaard T., Krakowka S., Ellis J., et al. , Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2), Vet. Microbiol. (2002) 89:97–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C., Bachand A., Murtaugh M.P., Yoo D., Differential host cell gene expression regulated by the porcine reproductive and respiratory syndrome virus GP4 and GP5 glycoproteins, Vet. Immunol. Immunopathol. (2004) 102:189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis C.R., Ait-Ali T., Clapperton M., Archibald A.L., Bishop S., Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS), Viral Immunol. (2007) 20:343–358 [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Zhu Y., Chen I., Lau J., He F., Lau A., et al. , The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection, J. Virol. (2007) 81:9560–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods (2001) 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25.López-Soria S., Segalés J., Rose N., Viñas M.J., Blanchard P., Madec F., et al. , An exploratory study on risk factors for postweaning multisystemic wasting syndrome (PMWS) in Spain, Prev. Vet. Med. (2005) 69:97–107 [DOI] [PubMed] [Google Scholar]
- 26.Madec F., Eveno E., Morvan P., Hamon L., Blanchard P., Cariolet R., et al. , Post-weaning multisystemic wasting syndrome (PMWS) in pigs in France: clinical observations from follow-up studies on affected farms, Livest. Prod. Sci. (2000) 63:223–233 [Google Scholar]
- 27.Meerts P., Van Gucht S., Cox E., Vandebosch A., Nauwynck H.J., Correlation between type of adaptive immune response against porcine circovirus type 2 and level of virus replication, Viral Immunol. (2005) 18:333–341 [DOI] [PubMed] [Google Scholar]
- 28.Meerts P., Misinzo G., Lefebvre D., Nielsen J., Bøtner A., Kristensen C.S., Nauwynck H.J., Correlation between the presence of neutralizing antibodies against porcine circovirus 2 (PCV2) and protection against replication of the virus and development of PCV2-associated disease, BMC Vet. Res. (2006) 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misinzo G., Meerts P., Bublot M., Mast J., Weingartl H.M., Nauwynck H.J., Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31, J. Gen. Virol. (2005) 86:2057–2068 [DOI] [PubMed] [Google Scholar]
- 30.Misinzo G., Delputte P.L., Lefebvre D.J., Nauwynck H.J., Porcine circovirus 2 infection of epithelial cells is clathrin-, caveolae- and dynamin-independent, actin and Rho-GTPase-mediated, and enhanced by cholesterol depletion, Virus Res. (2009) 139:1–9 [DOI] [PubMed] [Google Scholar]
- 31.Ochieng J., Furtak V., Lukyanov P., Extracellular functions of galectin-3, Glycoconj. J. (2004) 19:527–535 [DOI] [PubMed] [Google Scholar]
- 32.Opriessnig T., Fenaux M., Thomas P., Hoogland M.J., Rothschild M.F., Meng X.J., Halbur P.G., Evidence of breed-dependent differences in susceptibility to porcine circovirus type-2-associated disease and lesions, Vet. Pathol. (2006) 43:281–293 [DOI] [PubMed] [Google Scholar]
- 33.Opriessnig T., Patterson A.R., Madson D.M., Pal N., Rothschild M., Kuhar D., et al. , Difference in severity of porcine circovirus type 2 (PCV2)-induced pathological lesions between Landrace and Pietrain pigs, J. Anim. Sci. (2009) 87:1582–1590 [DOI] [PubMed] [Google Scholar]
- 34.Petroski M.D., The ubiquitin system, disease, and drug discovery, BMC Biochem. (2008) 9:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provençal M., Michaud M., Beaulieu E., Ratel D., Rivard G.E., Gingras D., Béliveau R., Tissue factor pathway inhibitor (TFPI) interferes with endothelial cell migration by inhibition of both the Erk pathway and focal adhesion proteins, Thromb. Haemost. (2008) 99:576–585 [DOI] [PubMed] [Google Scholar]
- 36.Rose N., Abhervé-Guéguen A., Le Diguerher G., Eveno E., Jolly J.P., Blanchard P., et al. , Effet de la génétique Piétrain sur l’expression clinique de la maladie de l’amaigrissement du porcelet (MAP), étude dans 4 élevages naisseurs-engraisseurs, Journées Recherche Porcine (2004) 36:339–344 [Google Scholar]
- 37.Sadler A.J., Williams B.R., Interferon-inducible antiviral effectors, Nat. Rev. Immunol. (2008) 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallusto F., Cella M., Danieli C., Lanzavecchia A., Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products, J. Exp. Med. (1995) 182:389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez R.E. Jr, Meerts P., Nauwynck H.J., Pensaert M.B., Change of porcine circovirus 2 target cells in pigs during development from fetal to early postnatal life, Vet. Microbiol. (2003) 95:15–25 [DOI] [PubMed] [Google Scholar]
- 40.Smyth G.K., Linear models and empirical bayes methods for assessing differential expression in microarray experiments, Stat. Appl. Genet. Mol. Biol. (2004) 3:3. [DOI] [PubMed] [Google Scholar]
- 41.Sorden S.D., Update on porcine circovirus and postweaning multisystemic wasting syndrome, J. Swine Health Prod. (2000) 8:133–136 [Google Scholar]
- 42.Stevenson L.S., McCullough K., Vincent I., Gilpin D.F., Summerfield A., Nielsen J., et al. , Cytokine and C-reactive protein profiles induced by porcine circovirus type 2 experimental infection in 3-week-old piglets, Viral Immunol. (2006) 19:189–195 [DOI] [PubMed] [Google Scholar]
- 43.Timpl R., Sasaki T., Kostka G., Chu M.L., Fibulins: a versatile family of extracellular matrix proteins, Nat. Rev. Mol. Cell Biol. (2003) 4:479–489 [DOI] [PubMed] [Google Scholar]
- 44.Tomás A., Fernandes L.T., Valero O., Segalés J., A meta-analysis on experimental infections with porcine circovirus type 2 (PCV2), Vet. Microbiol. (2008) 132:260–273 [DOI] [PubMed] [Google Scholar]
- 45.Tsai S., Mir B., Martin A.C., Estrada J.L., Bischoff S.R., Hsieh W.P., et al. , Detection of transcriptional difference of porcine imprinted genes using different microarray platforms, BMC Genomics (2006) 7:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent I.E., Balmelli C., Meehan B., Allan G., Summerfield A., McCullough K.C., Silencing of natural interferon producing cell activation by porcine circovirus type 2 DNA, Immunology (2007) 120:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Couture O.P., Qu L., Uthe J.J., Bearson S.M., Kuhar D., et al. , Analysis of porcine transcriptional response to Salmonella enterica serovar choleraesuis suggests novel targets of NFkappaB are activated in the mesenteric lymph node, BMC Genomics (2008) 9:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.West M.A., Wallin R.P., Matthews S.P., Svensson H.G., Zaru R., Ljunggren H.G., et al. , Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodelling, Science (2004) 305:1153–1157 [DOI] [PubMed] [Google Scholar]
- 49.Wilson C.L., Miller C.J., Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis, Bioinformatics (2005) 21:3683–3685 [DOI] [PubMed] [Google Scholar]
- 50.Zhao S.H., Kuhar D., Lunney J.K., Dawson H., Guidry C., Uthe J.J., et al. , Gene expression profiling in Salmonella choleraesuis-infected porcine lung using a long oligonucleotide microarray, Mamm. Genome (2006) 17:777–789 [DOI] [PubMed] [Google Scholar]