Abstract
In Pseudomonas aeruginosa, conversion to the mucoid phenotype marks the onset of an irreversible state of the infection in Cystic Fibrosis (CF) patients. The main pathway for mucoid conversion is mutagenesis of the mucA gene, frequently due to −1 bp deletions in a simple sequence repeat (SSR) of 5 Gs (G5-SSR426). We have recently observed that this mucA mutation is particularly accentuated in Mismatch Repair System (MRS)-deficient cells grown in vitro. Interestingly, previous reports have shown a high prevalence of hypermutable MRS-deficient strains occurring naturally in CF chronic lung infections. Here, we used mucA as a forward mutation model to systematically evaluate the role of G5-SSR426 in conversion to mucoidy in a MRS-deficient background, with this being the first analysis combining SSR-dependent localized hypermutability and the acquisition of a particular virulence/persistence trait in P. aeruginosa. In this study, mucA alleles were engineered with different contents of G:C SSRs, and tested for their effect on the mucoid conversion frequency and mucA mutational spectra in a mutS-deficient strain of P. aeruginosa. Importantly, deletion of G5-SSR426 severely reduced the emergence frequency of mucoid variants, with no preferential site of mutagenesis within mucA. Moreover, although mutagenesis in mucA was not totally removed, this was no longer the main pathway for mucoid conversion, suggesting that G5-SSR426 biased mutations towards mucA. Mutagenesis in mucA was restored by the addition of a new SSR (C6-SSR431), and even synergistically increased when G5-SSR426 and C6-SSR431 were present simultaneously, with the mucA mutations being restricted to −1 bp deletions within any of both G:C SSRs. These results confirm a critical role for G5-SSR426 enhancing the mutagenic process of mucA in MRS-deficient cells, and shed light on another mechanism, the SSR- localized hypermutability, contributing to mucoid conversion in P. aeruginosa.
Introduction
Mutation is an essential feature of pathogenic prokaryotes, being involved in the generation of genetic variability and the acquisition of adaptive phenotypes. In fact, many virulence traits important in host colonization and sudden environmental changes (i.e., antimicrobial therapy, host immune response), are acquired by mutagenic events. Thus, factors which regulate mutagenesis may play critical roles in the pathogen's establishment and evolution within the host. Occasionally, increasing the mutation rate may facilitate the adaptation to different stimuli in bacterial populations [1]. Reports of this phenomenon involve different mechanisms, which include the presence of naturally-occurring stable mutators (i.e., DNA Repair-deficient strains), inducible or transient hypermutators (i.e., induction of error-prone DNA polymerases), and hypermutable genetic sequences (i.e., DNA simple sequence repeats [SSRs]). Regarding this last mechanism, SSRs are defined as tandem repetitions of short motives and are largely accounted for as a source of genetic variability [2], [3] through the generally accepted slipped-strand mispairing mechanism [2], [4]. In fact, there is extensive literature involving SSRs in pathogenesis of several bacterial species, frequently playing crucial roles in the antigenic- or phase-variation that occur at contingency loci (reviewed in [2]).
Pseudomonas aeruginosa is an opportunistic pathogen that chronically infects the lungs and airways of Cystic Fibrosis (CF) patients, which in order to persist in the CF lung, undergoes a genetic adaptation based on mutagenic events [5]. However, although the participation of stable hypermutators in this process has been investigated [6], [7], there are no reports about the role of SSR-localized hypermutability in the acquisition of phenotypes that allow its long-term persistence. Among these phenotypes, conversion to mucoidy (exopolysaccharide alginate-overproduction) is one of the most important virulence traits in P. aeruginosa, since it confers protection against the host immune response [8], [9], reactive oxygen intermediates [10], and pulmonary clearance [11]. In fact, the emergence of the mucoid phenotype in the CF lung marks the onset of an irreversible state of the infection and poor prognosis for the patient [12].
More than one possible pathway leading to alginate overproduction has been described [13], i.e. positive regulation by rpoN [14] or mutations in regulatory genes such as mucB and mucD [15]–[17]. However, the most frequent pathway that leads to the mucoid phenotype is the acquisition of loss-of-function mutations in a single gene, mucA, which encodes for a negative regulator of alginate production [18]. Studies of CF mucoid isolates have shown that mucA harbored loss-of-function mutations in more than 85% of isolates [11], [16], [19]. Similarly, work in our laboratory and by other researchers has shown mucA to be the main target for mutagenesis in mucoid variants obtained in vitro [20], [21]. These studies found that for several types of mutations, one of the most represented was the −1 deletion in a monomeric SSR of five Gs (G5-SSR426) located at position 426 from the mucA start codon (widely known as mucA22 allele) [11], [16], [19]–[22].
In a previous recent work, we determined in vitro that two factors involved in the regulation of the overall mutation rate, MutS (a main component of the Mismatch Repair System) and Pol IV (the error-prone DNA polymerase encoded by dinB), were essential to establish mucA as the main target for mutagenesis in mucoid conversion, with these two factors having a prominent role in the generation of the mucA22 allele [21]. Questions that still remained unsolved are: 1) why was there such a high percentage of mucoid isolates in which mutations in mucA were found? 2) what is special about mucA that makes it the main pathway to mucoid conversion (thus leaving a secondary role to other genes whose inactivation are also known to induce mucoidy, such as mucB and mucD)? 3) does mucA contain a hotspot for mutagenesis? 4) what is the role of G5-SSR426 in this phenomenon? Concerning this last question, since no study to date has evaluated the role of any SSR in P. aeruginosa, little is known about their relevance in this mutagenic adaptive processes. In an attempt to shed light on this, we designed and constructed mucA alleles with different SSR compositions by site directed mutagenesis, and then analyzed the emergence frequency of mucoid variants and the spectrum of mucA mutations in strains carrying the different mucA alleles. Assays were performed using a DNA Mismatch Repair System (MRS)-defective mutS strain for several reasons: 1) the low spontaneous rate of mucoid conversion and the low yield of mucA22 alleles in nonmutator strains do not allow an accurate analysis in this experimental system [21]; 2) MRS-deficient strains most directly reflect the mutagenesis (in frequency and nature) of the ongoing DNA synthesis [23]; 3) they provide a larger yield of mucoid variants [21]; 4) this yield is enriched in mucA22 alleles [21];. Furthermore, previous studies have reported a large proportion of P. aeruginosa hypermutator MRS-deficient strains occurring naturally in CF chronic infections [7], which has been proposed to catalyze the genetic adaptation for persistence in the CF lung environment [6]. This leads to the idea that the coexistence of SSRs and MRS deficiency might be a typical phenomenon in the CF lung.
In this work, we show that in a MRS-deficient background, G5-SSR426 was an essential hotspot biasing mutations to mucA thereby contributing, together with stable hypermutability, in the determination of mucA as the main pathway for mucoid conversion in P. aeruginosa.
Results
The Presence of SSRs in mucA Increases the Yield of Mucoid Variants in P. aeruginosa
As mentioned above, mutations in the mucA gene are known to be the major cause of mucoid conversion in P. aeruginosa [18]. Previous mucA sequence analyzes of mucoid isolates, obtained from CF patients as well as under laboratory conditions, showed that they mostly harbored the mucA22 allele (a −1 bp deletion in a homopolymeric G:C SSR here referred to as G5-SSR426) [11], [16], [19]–[22]. In order to determine the role of G5-SSR426 in mucA mutagenesis leading to P. aeruginosa mucoid conversion, we constructed strain MPA-T1 with its mucA sequence lacking G5-SSR426 (mucAT1 allele) (Figure 1). This strain was generated in a mutS deficient background in order to increase the yield of mucoid variants, and also because this background allows the direct observation of replicative errors without interference by the MRS.
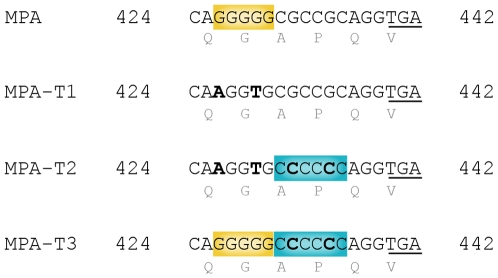
Figure 1. Site directed mutagenesis on the mucA gene.
A fragment of the P. aeruginosa mucA gene where the site directed mutagenesis was performed is shown. Base changes (bold face) were designed in order to maintain the amino acidic sequence unaltered (shown below), and codons were chosen that are commonly used by P. aeruginosa. The premature stop codon generated by a hypothetical −1 bp deletion between positions 426 and 436 of mucA is underlined. Wild type mucA allele from MPA strain with G5SSR426 is highlighted in yellow. The mucAT1 allele from strain MPA-T1 was generated by replacing G-to-A at 426 and G-to-T at 429, thus eliminating G5SSR426. The mucAT2 allele from strain MPA-T2 was generated by replacing G-to-A at 426 and G-to-T at 429 (eliminating G5SSR426), and G-to-C at 432 and 435 to generate C6SSR431 (highlighted in blue). The mucAT3 allele from strain MPA-T3 was generated by replacing G-to-C at 432 and 435 to generate C6SSR431 (highlighted in blue), and maintaining G5SSR426 (highlighted in yellow) unaltered.
Previous studies have established that mucoidy, a phenotype that is almost exclusively observed in chronic infections, could be reproducibly obtained in vitro from the effluent run-offs of continuous flow-cultured biofilms [20], [21]. Based on these antecedents, bacteria were grown in continuous cultured biofilms, with the effluents run-off being plated in order to score for mucoid variants (easily distinguishable as “mucous droplet-like” colonies). In addition, the mucoid phenotype of each variant was confirmed by the carbazole method which showed a≥3 fold increase in alginate production respect to the parental strain.
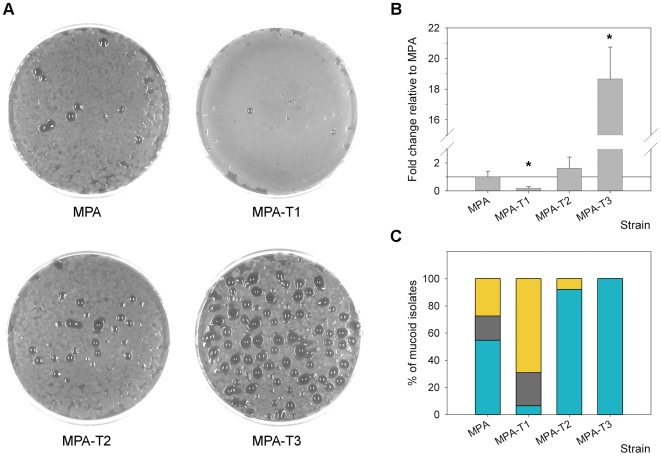
Then, the mucoid variant emergence frequencies of strains MPA-T1 and its parental MPA were compared. As shown in Figures 2A and 2B, the emergence frequency of mucoid variants suffered a significant 5.3 fold decrease (p<0.05, t-test) in strain MPA-T1 (0.09±0.05%) respect to strain MPA which carried the wild type mucA (0.48±0.20%), providing evidence that under the conditions used, G5-SSR426 was involved in the mucoid conversion process.
Figure 2. Role of SSRs in P. aeruginosa mucoid conversion.
(A) Mucoid variants, visualized as mucous droplet-like colonies emerging from MPA, MPA-T1, MPA-T2 and MPA-T3 strains plated onto MMA plates. (B) Relative emergence of mucoid variants obtained from MPA-T1, MPA-T2 and MPA-T3 strains. The values were compared to those obtained for the MPA strain. *Values for MPA-T1 and MPA-T3 were significantly different (p<0.05, as determined by the t-test) from that of MPA. (C) Spectrum of mucA mutations observed in mucoid variants obtained from MPA, MPA-T1, MPA-T2 and MPA-T3 strains. Bars indicate the observed percentage for each kind of mutation of the total number of mucoid clones analyzed in each strain (total clones analyzed: MPA, 11; MPA-T1, 13; MPA-T2, 13; and MPA-T3, 15). Bars in yellow indicate those mucoid variants that occurred in the absence of mucA alterations. The observed mutations included base substitutions (gray) and −1 bp deletions in mononucleotide G:C SSRs (blue).
To test whether the elevated frequency of mucoid conversion was an exclusive feature of G5-SSR426, and to examine whether a SSR (different from G5-SSR426) was able to restore the level of generation of mucoid variants lost in MPA-T1, we tested strain MPA-T2, which lacked G5-SSR426 in mucA, but carried a new stretch of 6 Cs (C6-SSR431) located immediately after the region previously occupied by G5-SSR426 (Figure 1). It is worth noting that the hypothetical deletion of one C in C6-SSR431 generates the same premature stop codon as the mucA22 allele. This last issue is important, in order to try to avoid as much as possible an altered emergence frequency of mucoid variants due to differential selection instead of differential mutagenesis between mucA alleles.
As shown in Figures 2A and 2B, the emergence frequency of mucoid variants in strain MPA-T2 (0.78±0.38%) showed an increase of 1.6-fold respect to MPA, which although not statistically significant (p>0.05, t-test) implied a 7.1-fold increase respect to MPA-T1. This result not only supports the previous observation that the presence of a SSR such as G5-SSR426 was determinant in the generation of mucoid variants, but also indicates that this function could be replaced by another SSR different from the wild type G5-SSR426.
In order to test the simultaneous effect of G5-SSR426 and C6-SSR431 on the emergence frequency of mucoid variants, a new strain was generated (MPA-T3) which carried both SSRs (G5C6SSR426) (mucA-T3 allele) (Figure 1). In the same way, a hypothetical −1 deletion in G5C6SSR426 generates the same premature stop codon at 440 as the mucA22 allele.
Strikingly, strain MPA-T3 showed a significant increase in the emergence frequency of mucoid variants of more than one order of magnitude (8.96±1.00%) respect to strains MPA or MPA-T2 (p<0.05, t-test) (Figure 2A and 2B). This result clearly indicates that the combination of both SSRs had a synergistic incremental effect on the yield of mucoid variants, as in the case of a larger single SSR.
To check that no variation in the global mutagenesis of the cell was acquired during strain constructions, the four strains (MPA, MPA-T1, MPA-T2 and MPA-T3) were subjected to the Rf-resistance test, which is a widely known assay to determine the overall mutagenic state of the cell [24]. The frequencies of Rf-resistant cells presented the expected values for mutS-deficient hypermutator strains, which were similar in the four strains (4.4-5×10−6 cells). Furthermore, in order to verify that the basal alginate production was not altered by the different mucA allele replacements, alginate production was measured in the four strains by the carbazole method [25]. Basal alginate production did not vary among strains MPA, MPA-T1, MPA-T2 and MPA-T3 (150–200 µg per ml of growth culture), which confirms that the modifications engineered in mucA did not produce a constitutive overproduction of alginate.
SSRs Constitute Hotspots for Mutagenesis in mucA
In a recent study [21], we showed that the overall hypermutability generated by inactivation of the MRS gene mutS produced an increase in mucA mutagenesis that was accompanied by a decrease in the variability of mutations in mucA. For this mutS strain, mutagenesis in mucA was clearly the leading pathway for mucoid conversion, and interestingly, the most represented mutation was the −1 deletion in G5-SSR426 generating the mucA22 allele [21]. In order to determine if the carriage of different mucA alleles affects the prevalence of the mucA conversion pathway, and to analyze the spectra of mutations of mucA alleles harboring different SSRs, the mucA gene of mucoid variants obtained from the four strains, MPA, MPA-T1, MPA-T2 and MPA-T3, was PCR amplified and subjected to automated direct sequence analysis.
Regarding the fraction of the mucoid isolates that harbored mutations in mucA in this study, MPA showed mutations in mucA for 73% of the mucoid isolates (Figure 2C). Analysis of mucA mutations demonstrated that 75% corresponded to the mucA22 allele, with the remaining 25% being represented by C-to-T transitions (Table 1 and Figure 2C). Both observations about the prevalence and spectrum of mucA mutations are in agreement with our previous results [21], with this being the spectrum typically observed in MRS deficient strains [26], [27].
Table 1. Mutations in the mucA gene of mucoid isolates from strains MPA, MPA-T1, MPA-T2 and MPA-T3.
| Strain and mucA mutationa | Stopb | Number of isolates |
| MPA | ||
| ΔG at 426 | TGA at 440 | 6 |
| C→T at 424 | TAG at 424 | 1 |
| C→T at 436 | TAG at 436 | 1 |
| None | none | 3 |
| MPA-T1 | ||
| ΔC at 362 | TGA at 385 | 1 |
| G→A at 249 | TGA at 247 | 1 |
| C→T at 367 | TAG at 367 | 1 |
| C→T at 424 | TAG at 424 | 1 |
| C→T at 505 | TAG at 505 | 1 |
| none | none | 9 |
| MPA-T2 | ||
| ΔC at 431 | TGA at 440 | 12 |
| none | none | 1 |
| MPA-T3 | ||
| ΔG at 426 | TGA at 440 | 5 |
| ΔC at 431 | TGA at 440 | 10 |
ΔG at 426 corresponds to a −1 bp deletion within G5SSR426; ΔC at 362 corresponds to a −1 bp deletion within a mononucleotide SSR of four Cs from 362 to 365 (C4SSR362); ΔC at 431 corresponds to a −1 bp deletion within C6SSR431. “None” refers to conversion to mucoidy occurring in the absence of mucA mutations. The nature of these non-mucA alterations leading to a mucoid phenotype was not investigated in this work.
Stop codon produced at the site of the mutation by substitutions or placed in frame by frameshift mutations.
Elimination of G5-SSR426 in MPA-T1 reduced the prevalence of mucA mutations among the mucoid isolates to 31% (Figure 2C). Notably, this result, together with the decrease in the emergence of mucoid variants in MPA-T1 (Figure 2A and 2B), suggest that after the elimination of G5-SSR426, mutagenesis of mucA was no longer the major pathway of mucoid conversion. The spectrum analysis of mucA also showed that no mutation had occurred in the region previously occupied by G5-SSR426. Instead, an increase in the variability of mutations was observed, in which four isolates (80%) harbored different C-to-T and G-to-A transitions, with one isolate (20%) having a deletion of one C in a stretch of four Cs located between 362 and 365 (C4SSR362) of the coding region of mucA, thus generating a premature stop codon at 385 (Table 1 and Figure 2C). This result demonstrates that in strain MPA-T1, mutagenesis in mucA was not biased to any particular DNA motif, indicating that there were no distinguishable hotspots for mutagenesis within the mucA-T1 allele. Furthermore, this observation suggests that in strain MPA, the emergence of mucoid variants might not be due to the effect of scattered mutagenesis along the mucA gene and further selection, but could greatly depend on the presence of G5-SSR426 biasing mutations, thereby constituting a real hotspot for mutagenesis in a MRS-deficient background.
Notably in strain MPA-T2, which carried C6-SSR431, the proportion of mucoid variants that harbored mutations in mucA increased to 93% (Figure 2C), thus restoring and even surpassing the proportion observed for the MPA wild type allele. Interestingly, this strain showed a drastic reduction in the spectrum of mucA mutations, in which 100% were represented by a −1 bp deletion within C6-SSR431 (Table 1 and Figure 2C). Therefore, it is evident that this SSR (6 bp long) has a stronger biasing capacity respect to G5-SSR426 (5 bp long), which could be logically explained due to its larger size. However, as it has been previously observed in E. coli [28], the possibility that G:C SSRs may have different levels of mutagenesis depending on their orientations respect to the replication fork should also be considered (see Discussion).
In the case of MPA-T3, which carried G5C6-SSR426, 100% of the mucoid isolates harbored mutations in mucA, with these mutations being −1 bp deletions and showing a distribution between both SSRs of 67% for C6-SSR431 and 33% for G5-SSR426 (Table 1 and Figure 2C). This distribution of mutations is also consistent with the differential capacity of each SSR to bias mutations (Figure 2C), and could explain the previous observation of the higher mucoid emergence frequency for strain MPA-T2, although this was not statistically significant compared with MPA (Figure 2B). Furthermore, spectra observed in strains MPA-T2 and MPA-T3 clearly suggest that no other kind of alterations or rearrangements are occurring in the SSRs apart from the slipped-strand mispairing mechanism.
Taken together, these results indicate that G5-SSR426 is a hotspot, which, based on its length, is able to bias but not restrict mutations towards the mucA gene. Thus, G5-SSR426 constitutes a major element, without which, mucA ceases to be the leading pathway for conversion to mucoidy in MRS-deficient P. aeruginosa.
Most of the Loss-of-Function Mutations Occur in the Periplasmic Coding Region of mucA, Independently of the SSR Content
We next performed a survey of different studies reporting mutations in the mucA gene from P. aeruginosa CF isolates, in order to analyze the way in which mucA is mutated. Four different studies [11], [16], [22], [29], along with own unpublished data, showed that the great majority of mucoid isolates obtained from CF are due to frameshifts or base substitutions along the coding sequence of mucA (Table 2). Among these mutations, different C-to-T transitions and −1 bp deletion frameshifts were the most represented, with most of these −1 bp deletions occurring in G5-SSR426, thus generating the mucA22 allele (Table 2). As we observed for the mucoid variants obtained from MPA, MPA-T1, MPA-T2 and MPA-T3 (Table 1), and also for the clinical mucoid isolates, independent of the kind of mutation (either base substitution or −1 bp frameshift), in every study the great majority of mucA mutations (64–96%) generated a premature stop codon, thus providing a truncated version of MucA.
Table 2. Classification of mucA mutations reported in different studies for P. aeruginosa CF isolates.
| Location | Analyzed isolatesa | Kinds of mutations in mucA (%) | −1 bp in G5SSR430/Total −1 bp deletions (%) | Reference | |||
| Transi-tions | Transver-sions | −1 bp deletions | Other indelsb | ||||
| USA | 41 | 34 | 9 | 32 | 25 | 67 | [11] |
| Australia | 22 | 50 | 0 | 29 | 21 | 57 | [29] |
| Germany | 14 | 41 | 0 | 53 | 6 | 88 | [22] |
| Scandinavia | 148 | 46 | 1 | 33 | 20 | 80 | [16] |
| Argentina | 24 | 29 | 8 | 63 | 0 | 67 | Unpublished |
For each study, only the isolates for which the mucA mutations were reported were considered in the analysis.
Insertion or deletion mutations different from −1 bp deletions.
It is important to point out that MucA is an anti-sigma factor located at the inner membrane of the cell, which has an N-terminal cytoplasmic region (MucA7–57), a transmembrane domain (MucA84–104) and a periplasmic C-terminal domain (MucA113–170) [30], [31]. Based on this protein domain structure, we then analyzed if the stop mutations were clustered at any particular region of mucA. With few variations among studies, stop mutations were concentrated almost exclusively in the periplasmic coding region of mucA (84–100%), in a proportion that was significantly higher than the few remaining stop mutations observed in the transmembrane (0–13%) (p<0.001, Z-test), and the cytoplasmic (0–3%) (p<0.001, Z-test) coding regions of mucA. Our own previous results with mucoid variants obtained from laboratory conditions showed a similar distribution of mutations along the mucA gene [21].
We next wondered whether the G5-SSR426 capacity of biasing mutations was a determinant factor of the region at which most mutations were found. As shown above, strains MPA, MPA-T2 and MPA-T3 (with their SSRs in the periplasmic coding region of mucA) had all the alterations affecting this domain (Table 1). In the case of strain MPA-T1, although the prevalence of mucA mutations was greatly decreased and demonstrated no defined hotspot for mutagenesis, most of the mutations found in mucA (80%) also affected the periplasmic coding region of the gene (Table 1). This domain distribution of mutations was not significantly different from that observed in clinical isolates (p = 0.989, Z-test), which indicates that the presence of G5-SSR426 does not play a major role in the selection of the functional domain of MucA in which mutations arise or are selected. Thus, other sequence properties of mucA and/or selection forces on the MucA domain that is mutated may prevail over the effect of any SSR producing a bias on mutagenesis.
In order to determine other properties in the sequence of mucA, which might lead to differential mutagenesis between domains, we further sought for all the possible positions at which premature stop codons could be generated by C-to-T transitions (which is the other large group of mutations observed in mucA). Thus, we surveyed all the CAG and CAA codons (which encode for Glutamine) and CGA (which encodes for Arginine), and their respective distributions in the mucA gene. Interestingly, MucA possesses 14 Glutamine residues, of which 12 (86%) are located within the periplasmic region, and the remaining 2 (14%) in the cytoplasmic region. No CGA codon for Arginine was observed in the whole coding sequence of mucA. If it is assumed that each of the 14 Glutamine codons possesses an equal or similar probability of changing to a stop codon, then the observation mentioned above suggests that this distributive bias of Glutamine codons towards the periplasmic coding region of mucA, might contribute to the higher prevalence of non-sense mutations within this domain. Nevertheless, since MucA is highly regulated by proteolysis in response to certain types of stress [31]–[36], this analysis cannot rule out the possibility of selection pressures favoring periplasmic over non-periplasmic truncated versions of MucA.
G5-SSR426 Is Conserved in Most of the P. aeruginosa Strains, but Not in Other Pseudomonad Related Species
Since MucA is an anti-sigma factor highly conserved among several bacterial species (also known as RseA), we next scored for G5-SSR426 or a similar SSR in the mucA gene of other Pseudomonad related species [37], some of which are also known to have the possibility of acquiring a mucoid phenotype [38], [39]. The survey of P. fluorescens Pf-5, P. putida KT2440, P. stutzeri A1501, P. syringae 1448A and DC3000, P. entomophila L48, and P. mendocina ymp, revealed that although their identity scores respect to P. aeruginosa mucA sequence are high (≥72), none of these species harbored G5-SSR426 or any other G:C SSR exceeding 4 bp in their mucA genes, which indicates that G5-SSR426 might be exclusive for P. aeruginosa. Related to this, we then analyzed if G5-SSR426 was conserved intraspecifically by comparing the mucA sequence of several P. aeruginosa strains. This survey was performed on an own collection of 38 CF strains (not shown), one environmental strain Hex1T [40], and strains PAO1, PA14, LESB58 and PA7, whose genome sequence data are available online [37]. A comparative analysis showed that the scores of the identities of the mucA gene of the different strains respect to the mucA sequence of PAO1were ≥98. Thus, it was not surprising to find that G5-SSR426 was conserved in the mucA sequences of almost every strain of P. aeruginosa. However, G5-SSR426 was not present in strain PA7. Instead, PA7 showed an intriguing feature: a new SSR, also of five Gs located at 354 (G5-SSR354) in place of the SSR of four Gs observed in every other strain of P. aeruginosa. Curiously, in strain PA7 a −1 bp deletion within G5-SSR354 generates the same premature stop codon at 440 as that produced by a −1 bp deletion within G5-SSR426 of the other P. aeruginosa strains. Thus, the presence of G5-SSR426 and/or G5-SSR354,which showed to be absent in every other Pseudomonad species here analyzed, seems to be an intraspecifically highly conserved feature, which is so far unique for P. aeruginosa.
Discussion
In P. aeruginosa, mutagenesis in the mucA gene is the main pathway for conversion to mucoidy, an alginate-overproducing phenotype most associated with chronic infections in the airways of CF patients. Mucoid conversion dramatically increases the resistance of the bacteria to pulmonary clearance mechanisms, and marks the transition to an irreversible state of the infection [12], [13]. In a previous recent work, we show that the mutagenic activity of the error-prone DNA polymerase Pol IV and the MRS loss-of-function, major factors involved in the inducible and stable hypermutability of the cell respectively, are key determinants targeting mucA for mucoid conversion in vitro [21]. Under a MutS deficiency and Pol IV proficiency background, the spectrum of mutation in mucA is dominated by frameshift mutations in a particular mononucleotide G:C SSR of five Gs (here denominated as G5SSR426), suggesting that instability of this SSR could be another main determinant which leads to mucoid conversion and makes mucA a very attractive model to study the involvement of the mutagenic mechanisms in P. aeruginosa adaptation.
Thus, in the present study, we attempted to elucidate the role of G5SSR426 in the process of conversion to mucoidy in a mutS deficient strain, which is to our knowledge the first systematic study of the mutagenic role of a SSR in P. aeruginosa. We used the mucA gene and allelic variants of mucA, engineered to contain different G:C SSR compositions (Figure 1), as a forward system for the detection of mutations that confer mucoidy. Importantly, although the system designed in this work for P. aeruginosa lacks the benefit of selection, it shares the advantage of other chromosomal forward systems used in other bacterial species for the detection of broad mutational spectra [41], [42] over reversion systems, in which the number of sites and kinds of mutations that can produce the revertant phenotype are quite limited [43]–[48].
As mentioned above, mutagenesis in mucA constitutes the main pathway for conversion to mucoidy, although other possible pathways that convergently lead to alginate overproduction have also been described [14], [15], [17]. Therefore, mucA mutations have previously been considered to be “pathoadaptive”, since their occurrence and the subsequent action of natural selection shift bacterial adaptation to the pathologic lung environment [49]. In the present study, we demonstrate in a MRS-deficient strain that mutagenesis in mucA is not scattered, but is in fact biased towards the −1 bp deletion in G5SSR426. Related to this, we show that the elimination of G5SSR426 not only significantly reduced the mutation frequency in mucA (Figures 2A and 2B), but also expanded the spectrum of mutations with no apparent hotspot (Table 1 and Figure 2C). Most importantly, in the absence of G5SSR426, mucA was no longer the major pathway for mucoid conversion, providing strong evidence that G5SSR426 makes mucA more prone to mutation. Interestingly, analysis of the coding sequences of the mucB and mucD genes (whose inactivation also lead to mucoid conversion) revealed that whereas mucD lacks mononucleotide G:C SSRs exceeding 4 bp, mucB possesses a SSR of 5 Cs. However, a recent study on a large collection of CF isolates which reported several mutations for the mucB gene showed that none of these occurred within this C-SSR [16]. This observation results intriguing since this C-SSR and G5SSR426 in mucA which could be considered comparable SSRs, are actually not. This allows the speculation that not only the preferential mucA mutagenesis may have a major selective component to it, but also that the C-SSR in mucB does not represent a hotspot for mutagenesis within the gene, as it has been observed for G5SSR426 in mucA.
Furthermore, taking into account the results on mucA mutagenesis obtained in our previous work, it seems that G5SSR426 is a hot substrate for both Pol IV-induced replication errors and correction by mismatch machinery [21]. Thus, our model of mucA mutagenesis demonstrates the combined action of the three mechanisms known to increase cellular mutability: inducible hypermutability, stable hypermutability and SSR-localized hypermutability. Also, previous studies have reported a high proportion of P. aeruginosa hypermutator MRS-deficient strains naturally occurring in CF pulmonary chronic infections [7], with SOS induction of Pol IV being recently described in P. aeruginosa [50], [51]. All these observations suggest that stressful environmental lung conditions might be propitious for mucoid conversion via MutS, Pol IV and G5SSR426-dependent mutagenesis. In this context, it is important to remember that approximately 85% of the P. aeruginosa mucoid clinical isolates harbor mutations in mucA, and that for 25–40% of these mutations, −1 bp deletions are found in G5SSR426 [11], [16], [19], [22]. In this way, given that longer SSRs (as experimentally confirmed with mucA alleles carrying C6SSR431 or G5C6SSR426) may confer a level of hypermutability that could turn mucA the exclusive genetic pathway to mucoid conversion, we conclude that not only the existence, but also the length of G5SSR426, is a major determinant of the mucA-dependent mucoid conversion process.
Regarding the different level of mutagenesis observed between G5SSR426 and C6SSR431, it should be considered that not only their length, but also their orientations respect to the replication fork may influence the mutagenesis outcome. In fact, it has been observed in E. coli, that frameshift mutagenesis on G:C SSRs displays an asymmetry during leading and lagging-strand replication [28]. Accordingly, a strand bias has also been reported for the mutagenic activity of Pol IV [52], which has been shown to be the main DNA polymerase involved in mutagenesis of G5SSR426 in vitro [21]. In this sense, further studies are necessary to elucidate the effect of strand biased mutagenesis in P. aeruginosa.
Furthermore, it results disturbing that the increase in the mutation frequency observed due to C6SSR431- and G5C6SSR426-localized mutagenesis, was produced in strains that were already stable hypermutators. As these hypermutator strains have been frequently reported for CF patients [7], our results indicate that under such a hypermutable background, genes involved in virulence which possess large G:C SSRs should deserve a special concern.
On the other hand, the survey of different studies which reported the mutations in the mucA gene observed in P. aeruginosa CF isolates, showed that the great majority of the mutations in mucA generate premature stop codons at its periplasmic domain coding region, with most of these mutations being −1 bp deletions within G5SSR426 or different C-to-T transitions (Table 2). This way of periplasmic mutagenesis has also been observed in mucoid variants emerged in vitro, from hypermutator and non-mutator strains of P. aeruginosa [20], [21]. Here we observed that elimination of G5SSR426 did not change the functional domain at which most mucA mutations were found (Table 1). This means that other sequence features of mucA and/or selective forces favoring periplasmic mutated versions of mucA might play a role in the distribution of mutations observed along the gene. As a possible explanation, we observed that the periplasmic, but not the transmembrane/cytoplasmic domains of MucA, is rich in Glutamine, with this being the substrate for the generation of premature stop codons by C-to-T transitions. Considering that the C-to-T transition is the most frequent kind of mutation, together with the −1 bp deletion in G5SSR426, this Glutamine codon distribution might contribute to the generation of stop codons at the periplasmic domain of mucA. Nevertheless, MucA is a protein which is highly and differentially regulated by proteolysis at its periplasmic, transmembrane and cytoplasmic domains [31]–[36]. As an example, it has been recently observed that in a mucoid isolate, overexpression of truncated mucA alleles at levels of saturation of the proteolytic enzymes capacities, reverts the phenotype to a non-mucoid state [31]. In this sense, mucA alleles mutated at its periplasmic region, might still retain some degree of regulatory properties on alginate production or other processes. Thus, differential selective pressures could exist favoring some truncated versions of MucA over others. Related to this, it would be interesting to observe if the addition of a new G:C SSR in the cytoplasmic or transmembrane domains could determine a biased mutagnesis towards them.
Summing up, in this work we present another mechanism, SSR-localized mutagenesis, which might act together with inducible and stable hypermutability enhancing mutagenesis in mucA. Thus, these results contribute to the understanding of the mutagenic process which leads to mucoid conversion in P. aeruginosa, one of the hallmarks of chronic infection in the airways of CF patients.
Materials and Methods
Bacterial Strains, Plasmids and Culture Media
The bacterial strains and plasmids used in this study are described in Table 3. P. aeruginosa MPA strain [53] was kindly provided by Dr Michael Jacobs from the University of Washington Genome Center (USA). Transposon insertions within the mutS gene were confirmed by PCR analysis following the provider's instructions, and the resulting hypermutable phenotype was confirmed by the rifampin resistance test (see below). To prepare inocula, bacteria were routinely cultured on LB (1% NaCl, 1% soy peptone and 0.5% yeast extract) agar plates from frozen stocks and subcultured in LB liquid medium overnight at 37°C with shaking at 250 r.p.m.
Table 3. Bacterial strains, plasmids and oligonucleotides used in this study.
| Strains, vectors and primers | Descriptiona | Source or Reference |
| Strains | ||
| P. aeruginosa | ||
| MPA | mutS::ISlacZA/hah-TcR, MPAO1 derivative | [53] |
| MPA-T1 | TcR, MPA carrying a mucAT1 allele | This study |
| MPA-T2 | TcR, MPA carrying a mucAT2 allele | This study |
| MPA-T3 | TcR, MPA carrying a mucAT3 allele | This study |
| E. coli | ||
| XL1-Blue | host strain for DNA manipulation | [60] |
| SY327 λpir | RfR; recipient to propagate pKNG-101 derivatives | [61] |
| SM10 λpir | Recipient for conjugal transfer of pKNG-101 derivatives | [61] |
| Vectors | ||
| pGEM-T Easy | ApR; PCR product cloning vector | Promega |
| pGEM-mucAT1 | ApR; P. aeruginosa mucAT1 cloned in pGEM-T Easy | This study |
| pGEM-mucAT2 | ApR; P. aeruginosa mucAT2 cloned in pGEM-T Easy | This study |
| pGEM-mucAT3 | ApR; P. aeruginosa mucAT3 cloned in pGEM-T Easy | This study |
| pKNG101 | SmR, suicide delivery plasmid containing sacB gene (SucS) | [55] |
| pKNG-mucAT1 | SmR, SucS, pKNG101 carrying mucAT1 | This study |
| pKNG-mucAT2 | SmR, SucS, pKNG101 carrying mucAT2 | This study |
| pKNG-mucAT3 | SmR, SucS, pKNG101 carrying mucAT3 | This study |
| Primers | ||
| MucPA-F | 5′-GAAGCCTGACACAGCGGCAAATGC-3′ | [21] |
| MucPA-R | 5′-CCTCAGCGGTTTTCCAGGCTGGCTGC-3′ | [21] |
| MucBamHI-F | 5′-TATGGATCCTGAAGCAATCGACAAAGCTC-3′ | This study |
| MucXbaI-R | 5′-TTATCTAGAAGCTGGGAGGGATCGAACTT-3′ | This study |
| MucT1-F | 5′-GCGAAGAGCAAGGTGCGCCGCAGG-3′ | This study |
| MucT1-R | 5′-CCTGCGGCGCACCTTGCTCTTCGC-3′ | This study |
| MucT2-F | 5′-GCAAGGTGCCCCCCAGGTGATCACCAACTCCTC-3′ | This study |
| MucT2-R | 5′-CTGGGGGGCACCTTGCTCTTCGCTGTAGCCGG-3′ | This study |
| MucT3-F | 5′-AGCAGGGGGCCCCCCAGGTGATCA-3′ | This study |
| MucT3-R | 5′-TGATCACCTGGGGGGCCCCCTGCT-3′ | This study |
Resistance markers: Tc, tetracycline; Rf, rifampin; Sm, streptomycin; Ap, ampicillin; Suc, sucrose.
AB minimal medium [20] was used to grow continuous cultured biofilms. Mucoid maintenance agar (MMA) plates were used to score for mucoid colonies [54]. Antibiotics were used at the following concentrations unless otherwise indicated: ampicillin (Ap), 50 µg ml−1; streptomycin (Sm), 200 µg ml−1; rifampin (Rf), 100 µg ml−1.
Construction of Strains MPA-T1, MPA-T2 and MPA-T3 by mucA Site Directed Mutagenesis
A set of different mucA alleles were engineered to eliminate G5-SSR426 or to contain different SSRs, by changing the specific codons for alternative codons (commonly used by P. aeruginosa) of the same amino acid and thus maintaining the primary structure of MucA intact (Figure 1).
To generate the strain MPA-T1, site directed mutagenesis in mucA was performed by replacing the endogenous mucA gene with a fragment containing a mutated version of mucA that lacked G5-SSR426 (mucAT1), with codon CAG (Gln142) changed to codon CAA, and codon GGG (Gly143) changed to codon GGT (Figure 1). The fragment was generated by PCR overlapping extension as follow: first, a PCR product was amplified using primers MucBamHI-F (containing an engineered BamHI site) and MucT1-R (containing the codon substitutions) with genomic DNA from MPA as the template. Simultaneously, a second PCR product was obtained using primers MucT1-F (which overlaps with MucT1-R) and MucXbaI-R (with an engineered XbaI site). Both resulting PCR products were gel purified (Qiagen) and ∼60 ng of these products were combined for a second PCR reaction which began with three cycles in the absence of added primers and was followed by 30 cycles with the addition of primers MucBamHI-F and MucXbaI-R. The resulting 1014 bp PCR fragment containing allele mucAT1 was cloned into pGEM-T Easy (Promega) and then subcloned into the BamHI-XbaI restriction sites of the suicide vector pKNG101 [55] in order to generate pKNG-mucAT1.
For the generation of strain MPA-T2, the endogenous mucA was replaced with the mucAT2 allele. Plasmid pKNG-mucAT2 was constructed following the same steps as pKNG-mucAT1, except that the allele mucAT2 was designed with the following codon substitutions: CAG to CAA (Gln142); GGG to GGT (Gly143); GCG to GCC (Ala144); and, CCG to CCC (Pro145) (Figure 1). The fragment containing allele mucAT2 was generated with the overlapping primers MucT2-F and MucT2-R (containing the codon substitutions) and the common primers MucBamHI-F and MucXbaI-R, as described above.
The generation of MPA-T3 was assessed following the same previous steps, except that the overlapping primers MucT3-F and MucT3-R (containing the codon substitutions) were used. In this case, codon substitutions for the generation of mucAT3 consisted in GCG to GCC (Ala144) and CCG to CCC (Pro145) (Figure 1).
The resulting pKNG101 derivatives were maintained in E. coli SY327 λpir before being transferred to E. coli SM10 λpir to perform biparental mating with P. aeruginosa MPA and to carry out allelic exchange following standard protocols [56]. Briefly, mating cells were plated on LB agar plates containing 200 µg ml−1 Sm to select a single homologous recombination event with 50 µg ml−1 Ap to counterselect the donor E. coli. Then, Sm-resistant P. aeruginosa transconjugants that showed sucrose sensitivity were grown overnight in LB medium. Finally, the second recombination event was selected by plating on LB agar supplemented with 16% sucrose and scoring for Sm-sensitive clones. Allelic exchange was confirmed by DNA sequencing analysis.
Bacterial Growth in Continuous Cultured Biofilms
Bacteria were grown in continuous-flow culture PVC tubing (USP class VI) with an inner diameter of 1.6 mm. Briefly, ∼108 cells from overnight (ON) cultures were resuspended in 1 ml of AB medium before being injected upstream of the tubing and incubated for 1 h without flow to allow bacterial attachment to the substratum. Then, the flow was resumed and biofilms were constantly irrigated with AB medium at 0.05 ml min−1 for five days. Mucoid colonies were scored by visual inspection after plating adequate aliquots of the run-off effluents on MMA agar plates, before incubating them at 37°C for 48 h and again for five days at room temperature [21].
All determinations were carried out at least in triplicate for three independent experiments.
Determination of the Mutation Frequency
Single colonies of each strain were cultured overnight in LB medium at 37°C for 24 h with shaking at 250 r.p.m. Appropriate dilutions of the cultures were plated on LB agar plates to determine the total number of viable cells, or on LB agar supplemented with 100 µg ml−1 Rf to count the number of rifampin-resistant cells, following incubation overnight at 37°C. Then, the mutation frequency was determined as the ratio of the number of rifampin–resistant cells and the number of viable cells. Determinations were carried out in duplicate for three independent experiments, and the results were expressed as means with their standard deviations.
Determination of Alginate Production
Bacteria were grown in LB medium supplemented with 2% glycerol at 37°C for 72 h with shaking at 250 r.p.m. After incubation, the cultures were centrifuged at 7000 g for 15 min and the alginate present in the supernatants was precipitated with 3 vols of ethanol at −70°C for 24 h, followed by centrifugation at 18000 g for 15 min. The pellet was resuspended in water and the alginate was quantified by the carbazole assay [25], using alginate (Sigma) and glucuronic acid as standards.
PCR Amplification and DNA Direct Sequencing of the mucA Gene
The primers MucPA-F and MucPA-R used to amplify the coding region of the mucA gene are described in Table 3. Colony PCR of the P. aeruginosa MPA, MPA-T1, MPA-T2 and MPA-T3 strains, as well as of their respective mucoid derivates, was performed as described previously [21]. The PCR products were extracted from agarose gels with a Gel purification kit (Fermentas) and subjected to automated direct DNA sequence analyzes (CRC-DNA Sequencing Facility, Univ. of Chicago, USA) by using the primers described above. To identify mutations in the mucA gene, the sequences obtained from the different mucoid variants were analyzed for homologies with the mucA (PA0763) sequence annotated in the P. aeruginosa Genome Project [57], by using the BLAST software [58].
Data Resources and Software
Sequences of the mucA (rseA) gene from P. aeruginosa strains PAO1 (PA0763),PA14 (PA14_54420), LESB58 (PALES_45801), PA7 (PSPA7_4756) and from the Pseudomonad species P. fluorescens Pf-5 (PFL_1449), P. putida KT2440 (PP_1428), P. syringae phaseolicola 1448A (PSPPH_3954), P. syringae pv. tomato str. DC3000 (PSPTO_4223), P. stutzeri A1501 (PST_1224), P. entomophila L48 (PSEEN4295), and P. mendocina ymp (Pmen_1468) were downloaded from the Pseudomonas Genome Database [37] (http://www.pseudomonas.com). Alignments were obtained using the Clustal W software [59].
Statistical Analysis
Statistical analysis of the data was performed with the two-tailed t-test using the GraphPad Instat software, and with the Z-test (normal approximation) using the Primer of Biostatistics software. Differences were considered statistically significant when p<0.05.
Acknowledgments
We are grateful to Dr Carlos Argaraña and Dr José Echenique for the valuable discussion and comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Agencia Nacional de Promoción Científica y Tecnológica (ANPCYT) Grant PICT-2007-00687 (http://www.agencia.gov.ar/), and Secretaría de Ciencia y Técnica (SECYT-UNC) Grant 05/C486 (http://www.secyt.unc.edu.ar/) supported this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blázquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 2.Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 3.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bichara M, Wagner J, Lambert IB. Mechanisms of tandem repeat instability in bacteria. Mutat Res. 2006;598:144–163. doi: 10.1016/j.mrfmmm.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 8.Mai GT, Seow WK, Pier GB, McCormack JG, Thong YH. Suppression of lymphocyte and neutrophil functions by Pseudomonas aeruginosa mucoid exopolysaccharide (alginate): reversal by physicochemical, alginase, and specific monoclonal antibody treatments. Infect Immun. 1993;61:559–564. doi: 10.1128/iai.61.2.559-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pier GB, Small GJ, Warren HB. Protection against mucoid Pseudomonas aeruginosa in rodent models of endobronchial infections. Science. 1990;249:537–540. doi: 10.1126/science.2116663. [DOI] [PubMed] [Google Scholar]
- 10.Simpson JA, Smith SE, Dean RT. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 11.Boucher JC, Yu H, Mudd MH, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 14.Boucher JC, Schurr MJ, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 15.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, et al. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, et al. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology. 2008;154:103–113. doi: 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- 17.Martin DW, Schurr MJ, Mudd MH, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, et al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon SS, Coakley R, Lau GW, Lymar SV, Gaston B, et al. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest. 2006;116:436–446. doi: 10.1172/JCI24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 21.Moyano AJ, Luján AM, Argaraña CE, Smania AM. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol. 2007;64:547–559. doi: 10.1111/j.1365-2958.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 22.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, et al. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology. 2006;152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 23.Kuban W, Jonczyk P, Gawel D, Malanowska K, Schaaper RM, et al. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J Bacteriol. 2004;186:4802–4807. doi: 10.1128/JB.186.14.4802-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 25.Knutson CA, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 26.Levy DD, Cebula TA. Fidelity of replication of repetitive DNA in mutS and repair proficient Escherichia coli. Mutat Res. 2001;474:1–14. doi: 10.1016/s0027-5107(00)00169-x. [DOI] [PubMed] [Google Scholar]
- 27.Schaaper RM, Dunn RL. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gawel D, Jonczyk P, Bialoskorska M, Schaaper RM, Fijalkowska IJ. Asymmetry of frameshift mutagenesis during leading and lagging-strand replication in Escherichia coli. Mutat Res. 2002;501:129–136. doi: 10.1016/s0027-5107(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 29.Anthony M, Rose B, Pegler MB, Elkins M, Service H, et al. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J Clin Microbiol. 2002;40:2772–2778. doi: 10.1128/JCM.40.8.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;372:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damron FH, Qiu D, Yu HD. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol. 2009;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, et al. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology. 2005;151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- 36.Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 37.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, et al. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govan JR, Fyfe JA, Jarman TR. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol. 1981;125:217–220. doi: 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- 39.Schnider-Keel U, Lejbolle KB, Baehler E, Haas D, Keel C. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl Environ Microbiol. 2001;67:5683–5693. doi: 10.1128/AEM.67.12.5683-5693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smania AM, Segura I, Pezza RJ, Becerra C, Albesa I, et al. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology. 2004;150:1327–1338. doi: 10.1099/mic.0.26751-0. [DOI] [PubMed] [Google Scholar]
- 41.Bjedov I, Dasgupta CN, Slade D, Le Blastier S, Selva M, et al. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics. 2007;176:1431–1440. doi: 10.1534/genetics.107.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. Chromosomal system for studying AmpC-mediated beta-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother. 2002;46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cupples CG, Cabrera M, Cruz C, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci U S A. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foster PL, Trimarchi JM. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 47.Tegova R, Tover A, Tarassova K, Tark M, Kivisaar M. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol. 2004;186:2735–2744. doi: 10.1128/JB.186.9.2735-2744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Lu Z, Wang A. Study of adaptive mutations in Salmonella typhimurium by using a super-repressing mutant of a trans regulatory gene purR. Mutat Res. 2001;484:95–102. doi: 10.1016/s0027-5107(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 49.Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 1999;7:191–195. doi: 10.1016/s0966-842x(99)01493-6. [DOI] [PubMed] [Google Scholar]
- 50.Blázquez J, Gómez-Gómez JM, Oliver A, Juan C, Kapur V, et al. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol. 2006;62:84–99. doi: 10.1111/j.1365-2958.2006.05366.x. [DOI] [PubMed] [Google Scholar]
- 51.Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol. 2006;188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, et al. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krieg DP, Bass JA, Mattingly SJ. Aeration selects for mucoid phenotype of Pseudomonas aeruginosa. J Clin Microbiol. 1986;24:986–990. doi: 10.1128/jcm.24.6.986-990.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 56.de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 57.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 58.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bullock WO, Fernández JM, Short JM. XL1-Blue: A high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 61.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]