Abstract
Cell identity is acquired in different brain structures according to a stereotyped timing schedule, by accommodating the proliferation of multipotent progenitor cells and the generation of distinct types of mature nerve cells at precise times. However, the molecular mechanisms coupling the identity of a specific neuron and its birth date are poorly understood. In the neural retina, only late progenitor cells that divide slowly can become bipolar neurons, by the activation of otx2 and vsx1 genes. In Xenopus, we found that Xotx2 and Xvsx1 translation is inhibited in early progenitor cells that divide rapidly by a set of cell cycle-related microRNAs (miRNAs). Through expression and functional screenings, we selected 4 miRNAs—mir-129, mir-155, mir-214, and mir-222—that are highly expressed at early developmental stages in the embryonic retina and bind to the 3′ UTR of Xotx2 and Xvsx1 mRNAs inhibiting their translation. The functional inactivation of these miRNAs in vivo releases the inhibition, supporting the generation of additional bipolar cells. We propose a model in which the proliferation rate and the age of a retinal progenitor are linked to each other and determine the progenitor fate through the activity of a set of miRNAs.
Keywords: cell cycle, homeodomain, translational control, neurogenesis
During brain development, different types of nerve cells are generated according to a predictable schedule, through the coordination of cell cycle progression with the sequential expression of key genes of cell fate (1, 2). The time when a neural progenitor cell stops dividing and starts differentiating—the cell birth date—is related to its fate of differentiation (3), but the underlying molecular mechanisms are largely unknown. In vertebrates, retina development is an attractive model to investigate this issue (1).
Early retinal progenitors are multipotent (4, 5). However, if a progenitor exits the cell cycle early in development it is not able to become a bipolar neuron, which is the last type of retinal neuron to be generated. The homeobox genes otx2 and vsx1 act as positive key regulators of bipolar cell generation (6–10). Why do they act late in the retinal progenitor lineage? In Xenopus, Xotx2 and Xvsx1 are transcribed from very early retina developmental stages, but their translation is repressed in early progenitors through cis-acting signals contained in their 3′ UTR (8). A crucial question concerns the mechanism able to release the translational inhibition of Xotx2 and Xvsx1 in late progenitor.
Cell cycle manipulation of retinal progenitors is sufficient to dissociate cell birth date and fate of differentiation (8, 11). The cell cycle length of vertebrate neural progenitor cells increases over time (12, 13), thus correlating with the age of a progenitor. Accordingly, in the Xenopus developing retina early progenitors have a shorter cell cycle compared with late progenitors (8). It has been shown that late retinal progenitors forced to divide as rapidly as early progenitors fail translating Xotx2 (8). Conversely, here we show that lengthening the cell cycle of early progenitors supports and anticipates Xotx2 and Xvsx1 translation. Thus, these observations suggest that the cell cycle length of a progenitor might provide a mechanism of control for the translational inhibition of Xotx2 and Xvsx1.
What is the molecular nature of the translational inhibition in early progenitors? Recent evidence implies microRNAs (miRNAs) in the control of neural development (14–16), cell cycle (17), and developmental timing (18), suggesting them as good candidates to mediate the translational inhibition of Xotx2 and Xvsx1. Here we show that 4 miRNAs bind to the 3′ UTR of Xotx2 and Xvsx1 mRNAs, inhibiting their translation and the generation of bipolar neurons.
Results
Sonic Hedgehog Inactivation Lengthens the Cell Cycle and Anticipates Xvsx1 and Xotx2 Translation in Early Retina Progenitor Cells.
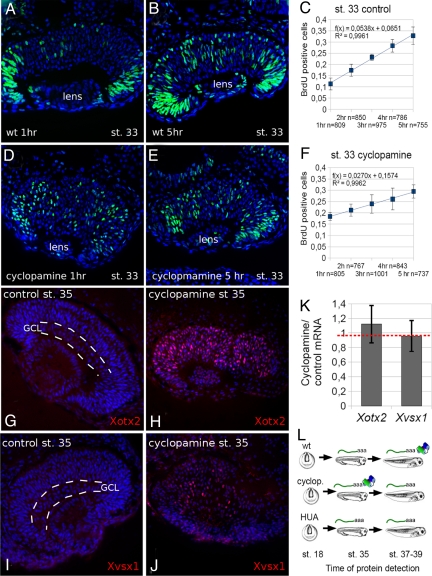
The pace of retinal progenitors proliferation is controlled by sonic hedgehog (hh) (19). Blocking hh signaling by cyclopamine lengthens G1 and G2 phases and delays cell cycle exit (20). By BrdU cumulative analysis (Fig. 1 A–F), we confirmed that stage (st.) 33 retina progenitors treated with 50 μM cyclopamine from st. 18 (after optic vesicle formation) undergo longer cell cycles compared with control progenitors, because the slope of BrdU increase over time is smaller (0.0270; Fig. 1F) compared with control (0.0538; Fig. 1C). We also confirmed that a higher proportion of progenitors remain cycling, because S index at 1 h is higher (0.18; Fig. 1F) compared with control (0.12; Fig. 1C).
Fig. 1.
hh inactivation lengthens the cell cycle and anticipates Xvsx1 and Xotx2 translation in early retina progenitor cells. (A–F) BrdU cumulative analysis of st. 33 retina. (A, B, D, and E) BrdU (green) and nuclei (Hoechst, blue) detection on retina sections. The slope of the line in C–F indicates the rate of the cell division cycles (see Materials and Methods). (n) number of scored cells. (G–J) Xotx2 and Xvsx1 immunodetection (red) and nuclear detection (Hoechst, blue) on st. 35 retina. GCL indicates ganglion cell layer, which is detectable in control retinas (G and I) but not in cyclopamine-treated retinas (H and J). (K) Cyclopamine/control relative ratio of Xvsx1 or Xotx2 mRNA as detected by qRT-PCR in st. 35 dissected retinas. Red dashed line marks ratio = 1, bars show SE. (L) Xotx2 and Xvsx1 developmental expression of mRNA (—aaa) and protein (colored item) in WT, cyclopamine-treated, and HUA-treated retinas.
In WT retina progenitors, Xvsx1 and Xotx2 are transcribed from st. 15 and st. 25, respectively (7, 10), whereas their translation is first detected at st. 37 (Xvsx1) or st. 39 (Xotx2) (8). Cyclopamine treatment from st. 18 induced a premature detection of Xotx2 (Fig. 1 G and H) and Xvsx1 (Fig. 1 I and J) proteins at st. 35, with a stronger effect on Xotx2. The cyclopamine treatment acted at the translational level, because it did not affect the mRNAs levels (Fig. 1K).
Selection of Developmentally and Cell Cycle Regulated Retina miRNAs Predicted to Bind the 3′ UTR of Xvsx1 and Xotx2.
We searched for developmentally regulated miRNAs that were predicted to bind the 3′ UTR of Xvsx1 and Xotx2. Because Xotx2 and Xvsx1 translation is de-repressed in late retina development, we reasoned that the amount of miRNAs targeting Xotx2 and Xvsx1 mRNAs ought to be high in early (st. 33) retinas and low in differentiated (st. 42) retinas. Furthermore, whereas cyclopamine removes the translational inhibition of Xvsx1 and Xotx2 (present data), we previously showed that hydroxyurea/aphidicolin (HUA) treatment maintains such inhibition (Fig. 1L) (8): therefore, the concentration of inhibitors should decrease or increase after cylopamine or HUA treatment, respectively.
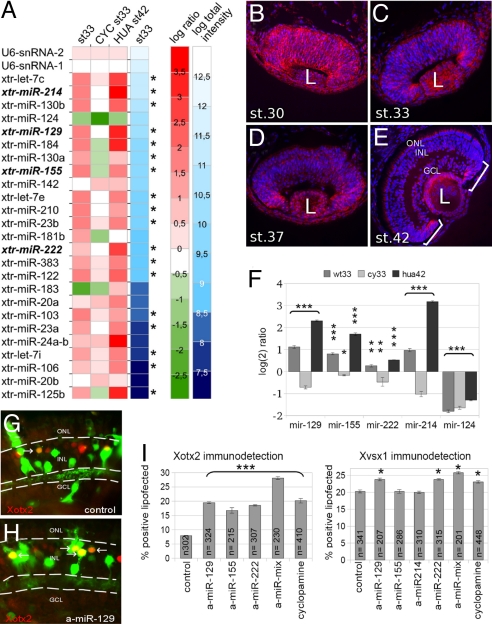
By microarray analysis, we looked for miRNAs expressed at high levels at early stages compared with late stages of retinogenesis, that were down-regulated by cyclopamine treatment and maintained high by HUA treatment. Using RNA from st. 33 dissected retinas as probe, we observed that significant hybridization signal was generated by one third of the probe-set of the Xenopus miRNAs annotated in miRbase 8.1 (21) [supporting information (SI) Table S1]. Of the 25 most expressed miRNAs, 18 had an expression pattern consistent with that expected for a putative inhibitor (asterisks in Fig. 2A). On the other hand, terminal markers of neuronal (miR-124) and photoreceptor (miR-183) differentiation showed an opposite pattern of expression.
Fig. 2.
Four developmentally regulated miRNAs inhibit the translation of Xotx2 and Xvsx1. (A) Heat map shows log ratios (red to green) of miRNA expression from WT and treated retinas with respect to WT st. 42 retina (baseline), after normalization with U6 snRNA1–2. The 25 miRNAs giving the highest hybridization signal (log total intensity, white to blue) in the st. 33 array are shown. (B–E) ISH detection (red) of mir-129; nuclei are stained by Hoechst (blue). (F) Relative quantification of miRNAs at st. 33 (wt33, WT; cy33, cyclopamine-treated) and at st. 42 after HUA treatment (hua42) as compared with their expression levels in st. 42 WT retinas (baseline). Error bars show SEM. (G and H) Xotx2 immunodetection (red) of lipofected cells (green, GFP detection). Arrows indicate double-positive cells. (I) Proportion of either Xvsx1- or Xotx2-positive lipofected cells. a-miR, antisense oligonucleotide to the indicated miRNA; a-miR-mix, equimolar mixture of antisense oligonucleotides to miR-129, miR-155, and miR-222 (Xotx2 immunodetection) or to miR-129 and miR-222 (Xvsx1 immunodetection). (n) number of cells analyzed. Error bars show SE. (L) lens; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
We then selected which of these 18 miRNAs were predicted to bind to the 3′ UTR of either Xotx2 or Xvsx1 but not to the 3′ UTR of Xotx5, a positive regulator of photoreceptor differentiation also regulated at the translational level (8, 10). According to the miRANDA algorithm (22), miR-129, miR-155, miR-214, and miR-222 (bold in Fig. 2A) possessed target sites on Xvsx1 and Xotx2 3′ UTR regions (Table S2; see Fig. 4A and Fig. S4).
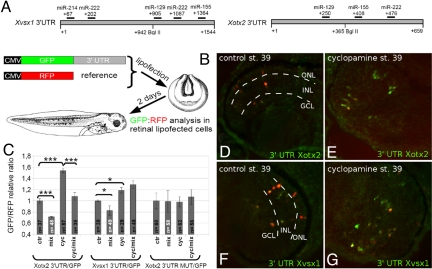
Fig. 4.
mir-129, mir-155, mir-214, and mir-222 target the 3′ UTR of Xotx2 and Xvsx1. (A) miRNA target sites as evaluated by functional assay in HEK 293 cells (Figs. S5 and S6). (B) In vivo sensor assay. (C) In vivo translational efficiency of GFP sensors as in B. Bars indicate GPF/RFP relative intensity ratio after lipofection. Cyc, cyclopamine. Mix, an equimolar mixture of antisense oligonucleotides to mir-129, miR-155, and miR-222 (for Xotx2 3′ UTR), or to mir-129, miR-155, miR-214, and miR-222 (for Xvsx1 3′ UTR). (n) number of retinal sections. Error bars show SE. Asterisks as in Fig. 2. (D–G) Examples of cells colipofected with sensors and control antisense oligonucleotide. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Layers did not form properly in cyclopamine-treated retinas (E and G), as previously described (20).
In situ hybridization (ISH) analysis (Fig. 2 B–E and Fig. S1) showed that the 4 miRNAs are expressed in retinal progenitors and that their amount decreases from an early stage (st. 30) to a late stage (st. 42) of retinogenesis. Furthermore, their amount remains high at a late stage after HUA treatment, whereas it is low after cyclopamine treatment at an early stage compared with controls (Fig. S1B). In untreated retinas at st. 42, their expression remains confined to dividing progenitors of the ciliary marginal zone and of the lens (brackets and L in Fig. 2E, respectively; see also Fig. S1A) and, to a lower extent, in non-bipolar differentiated neurons (outer nuclear layer and ganglion cell layer in Fig. 2E; see also Fig. S1A). Quantitative (q)RT-PCR results confirmed the pattern of expression observed by the microarray screening and by ISH (Fig. 2F).
In Vivo Inactivation of miR-129, miR-155, miR-214, and miR-222 Supports Xvsx1 and Xotx2 Expression.
To decoy endogenous miRNAs, we lipofected antisense oligonucleotides into the optic vesicle. Compared with control (Fig. 2 G and I and Fig. S2), the decoy of either miR-129 (Fig. 2 H and I), miR-155, or miR-222, as well as the decoy of all of the 3 miRNAs together (Fig. 2I and Fig. S2) significantly increased the proportion of Xotx2 translating cells, with a higher effect after the triple decoy. Conversely, only the decoy of miR-129 or miR-222, or their double decoy, increased, albeit to a lower extent, the proportion of Xvsx1-positive cells (Fig. 2I and Fig. S2). This result suggests that Xotx2 mRNA is more sensitive than Xvsx1 mRNA to translational repression and is consistent with the later translational onset of Xotx2 compared with Xvsx1 in retinal development (8). Notably, the high increase of Xotx2-positive cells after the knockdown of miR-129, miR-155, and miR-222 is comparable to the increase observed after cyclopamine treatment (Fig. 2I and Fig. S2), which decreases the amount of these miRNAs (Fig. 2 A and F).
In Vivo Inactivation of miR-129, miR-155, miR-214, and miR-222 Supports the Bipolar Cell Fate.
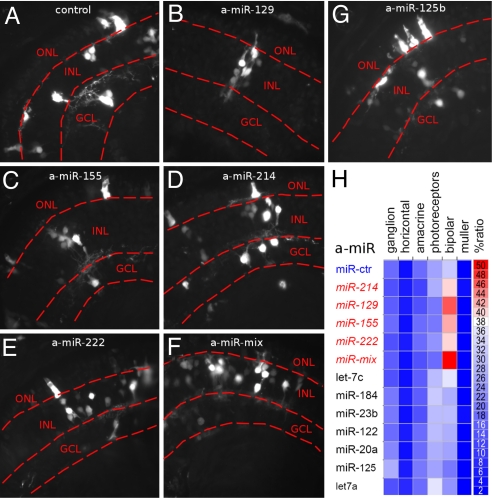
We investigated the effects of miRNAs decoy on cell fate (Fig. 3 A–H). Compared with control, the decoy of either miR-129, miR-155, miR-214, or miR-222 significantly increased the proportion of the bipolar cells in the inner nuclear layer from 32% to 44.9%, 40.9%, 39.8%, and 38.2%, respectively. Consistently, lipofecting a mixture of the 4 antisense oligonucleotides increased the proportion to 49.2%. Decoy of other miRNAs decreased the proportion of bipolar cells or had no effect (Fig. 3 and Table S3). The lipofection of antisense oligonucleotides to the 4 miRNAs did not increase the ratio of TUNEL-positive apoptotic cells compared with the lipofection of a control antisense (Fig. S3).
Fig. 3.
In vivo inactivation of miR-129, miR-155, miR-214, and miR-222 supports the bipolar cell fate. (A–G) Sections of retinas, lipofected with control (A) or antisense to miRNAs (a-miRNAs, B–G) oligonucleotides, show morphology of lipofected cells (GFP positive, white detection) in the differentiated cell layers: outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL). Bipolar cells are located in the INL. a-miR-mix is an equimolar mix of antago-miRNAs to mir-129, miR-155, miR-214, and miR-222. (H) Heat map shows the percentage ratio (red to blue) of neuronal (ganglion, horizontal, amacrine, photoreceptor, and bipolar) and glial (Müller) cells in retinas lipofected with antisense oligonucleotides to miRNAs (a-miR) as reported in Table S3.
miR-129, miR-155, miR-214, and miR-222 Target the 3′ UTR of Xvsx1 and Xotx2 in HEK 293 Cells and in Vivo.
miR-129, miR-155, and miR222 were predicted to target the 3′ UTR of Xotx2, whereas all of the 4 miRNAs were expected to target the 3′ UTR of Xvsx1 (Fig. 4Aand Fig. S4). We confirmed the activity of the identified miRNAs binding sites by measuring the translational rate of reporter constructs (sensors), carrying GFP upstream of WT or mutated 3′ UTR of Xvsx1 and Xotx2, after cotransfection with either mature miRNAs or antisense oligonucleotides to miRNA, in HEK293 cells (SI Text and Figs. S5 and S6).
The 4 miRNAs target the 3′ UTR of Xvsx1 and Xotx2 also in the developing retina. We colipofected either Xotx2 or Xvsx1 sensor with a red fluorescent protein (RFP) reporter construct into st. 18 optic vesicles and analyzed the GFP/RFP intensity ratio in lipofected retinal cells at st. 39 (Fig. 4 B and C). Xotx2 (Fig. 4 D and E) and Xvsx1 (Fig. 4 F and G) sensors translated GFP more efficiently in cyclopamine-treated retinas (Fig. 4 E and G) than in control retinas (Fig. 4 D and F; 155% and 119%, respectively), with a higher effect on the Xotx2 sensor (Fig. 4C). Thus, the sensors reproduce the translational regulation of Xotx2 and Xvsx1 (Fig. 1 G–L) in vivo. Compared with control miRNA, a mixture of the mature ds miRNAs that affected sensor translation in HEK293 cells (Fig. S5) was able to repress the translation of Xotx2 sensor by 30% (Fig. 4C). A much less significant repression of Xvsx1 sensor was also detected. The triple mutation of miR-129, miR155, and miR-222 target sites completely abolished the responsiveness of Xotx2 sensor to miRNAs. In addition, the miRNAs mixture was able to counteract the effects of cyclopamine on Xotx2 sensor, indicating that cyclopamine de-repression of Xotx2 sensor translation may work through depletion of these miRNAs.
miR-129, miR-155, miR-214, and miR-222 Act Downstream of the Cell Cycle Setting a Bipolar Cell Fate.
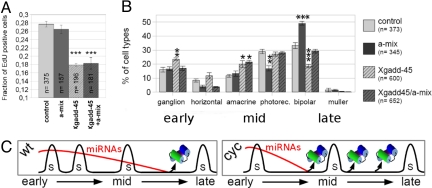
To investigate whether miRNAs affect cell fate through effects on cell cycle, we measured the proportion of lipofected cells that were postmitotic at st. 34 by 5-ethynyl-2′-deoxyuridine (EdU) birthdating (Fig. 5A and Fig. S7). The depletion of miR-129, miR-155, miR-214, and miR-222, which increases the proportion of bipolar cells by approximately 65% compared with control (Fig. 5B), did not affect the ratio of dividing progenitors significantly (26% vs. 28% of control; Fig. 5A); conversely, this proportion was decreased by lipofecting progenitors with the cell cycle inhibitor Xgadd-45-ã (23) (18%; Fig. 5A), according to previous data (8). Progenitor cells forced to withdraw early from the cell cycle by Xgadd-45-ã lipofection produced, compared with control, more of the early retinal neurons—namely ganglion cells—and fewer bipolar neurons (Fig. 5B) (8). The depletion of miR-129, miR-155, miR-214, and miR-222 counteracted both the increase of ganglion cells and the decrease of bipolar cells due to Xgadd-45-ã lipofection (Fig. 5B), with no effect on the ratio of dividing progenitors (18%; Fig. 5A). Therefore, the depletion of these miRNA in Xgadd-45-ã lipofected cells switches the fate of early progenitors from an early to a late type of retinal neuron.
Fig. 5.
miR-129, miR-155, miR-214, and miR-222 act downstream of the cell cycle setting a bipolar cell fate. (A) Fraction of EdU-positive lipofected cells. Error bars indicate SE. (n) number of cells. (B) Proportion of different lipofected retinal cell types. Error bars indicate SEM. Asterisks as in Fig. 2. (C) Model representing regulation of the timing of the translational inhibition of Xotx2 and Xvsx1 by mir-129, miR-155, miR-214, and miR-222 (miRNAs). S is the synthesis phase of the progenitor cell cycle. Early, mid, and late refer to cell birthdates in B and to retinal developmental stages in C.
Discussion
Mechanisms accounting for the generation of distinct types of neurons with a precise timing schedule have been described. ikaros, the mouse ortholog of hunchback setting early neuroblast fate in Drosophila (24), is sufficient to generate early-born neurons when misexpressed in late progenitors of the retina (25). In mouse developing cortex, the double knockdown of the 2 transcription factors COUP-TFI and -II causes sustained neurogenesis and prolonged generation of early-born neurons (26). However, how ikaros and COUP-TFI and -II are down-regulated in late progenitors remains unclear.
In the vertebrate retina, vsx1 and otx2 are both necessary and sufficient to specify the identity of the last-born neurons, the bipolar cells (6–10). In Xenopus, progenitors of st. 25–37 embryos transcribe Xvsx1 and Xotx2; upon cell cycle exit these early progenitors generate all of the retinal neurons but the late-born bipolar cells (8, 11, 27). Only after st. 37–39, when Xvsx1 and Xotx2 are translated, does differentiation of bipolar cells occur (8). Thus, translational control in frogs plays a crucial role in establishing a specific cell type at a precise developmental time.
Here we report that miR-129, miR-155, and miR-222 target the 3′ UTR of Xvsx1 and Xotx2, inhibiting their translation both in HEK 293 cells and in vivo. The in vivo decoy of each of the 4 miRNAs—miR-129, miR-155, miR-214, and miR-222—supports the generation of extra bipolar cells. However, miR-214 inactivates the translation of a reporter carrying Xvsx1 3′ UTR in HEK293 cells, but its in vivo decoy is not effective on Xvsx1 translation. We speculate that the ability of miR-214 to affect the generation of bipolar cells may be due to its action on genes other than Xvsx1 and Xotx2. Conversely, miR-129, miR-155, and miR-222 could affect the bipolar cell proportion by directly inhibiting Xvsx1 and Xotx2 translation.
The proliferation rate of a progenitor decreases over time during neural development as the progenitor cell cycle length increases (8, 12, 13). By microarray analysis, ISH, and qRT-PCR, we showed that miR-129, miR-155, miR-214, and miR-222 are highly expressed in fast cycling, early retinal progenitors of st. 30–33 embryos that do not translate Xvsx1 and Xotx2 and are down-regulated in slowly cycling, late progenitors of st. 34–37 and in st. 42 postmitotic cells. Emerging evidence indicates that the expression of these miRNAs is under the control of the cell cycle machinery. The 4 miRNAs are up-regulated in tumor cells (28–31). Moreover, the expression of mir-155 and mir-214 is directly related to the proliferation rate of primary fibroblasts (32). A functional relationship between cell cycle speed and expression of the 4 miRNAs is also supported by their decrease upon the cell cycle lengthening exerted by cyclopamine.
Interestingly, mir-222 is up-regulated as glioblastoma cells progress beyond the G1–S phase transition (33). If miR-129, miR-155, miR-214, and miR-222 were produced in a constant window of the cell cycle of retinal progenitors such as the G1–S phase transition, their amount in these cells would depend on cell cycle length because rapidly dividing cells spend a higher proportion of time in the G1–S phase transition compared with slowly dividing cells. This is consistent with the up-regulation of miR-222 (and of the other 3 miRNAs) after treatment with HUA (Fig. 2A), which blocks cells in G1–S.
We propose that miR-129, miR-155, miR-214, and miR-222 could be part of a mechanism coupling the determination of the bipolar cell identity with a low proliferation rate of retinal progenitors: they would act by inhibiting translation of the key homeobox genes Xotx2 and Xvsx1 early in development (Fig. 5E). As a consequence, these miRNAs would play a major role in matching the bipolar cell identity with a late cell birth date. A crucial question is how the expression of the 4 miRNAs may be regulated during development and, in particular, what might be the nature of the signal supporting their high level of expression in highly proliferating cells. hh signaling, which supports proliferation of embryonic retinal cells (19, 20), may be a candidate worth scrutiny.
The first isolated miRNAs were originally identified as regulators of the developmental timing in Caenorhabditis elegans (18). The Drosophila counterpart of a heterochronic miRNA gene from C. elegans, let-7, was recently shown to regulate the timing of neuromuscular tissue development, thus suggesting a widespread use of miRNAs in temporal regulation of animal development (34). Here we show that miR-129, miR-155, miR-214, and miR-222 play a similar role in the vertebrate neural development, controlling the timing of the generation of retinal bipolar cells.
Materials and Methods
BrdU cumulative analysis was performed as described previously (8). MiRNA microarrays (Exiqon miRCURY LNA Array version 8.1) were hybridized according to the manufacturer's protocol; data were analyzed using Axon Genepix and Axon Aquity software. The miRANDA algorithm (version 1.0b) was used to predict binding of the selected miRNAs. MiRNAs qRT-PCR was performed according to the miScript-System kit (Qiagen). Lipofection experiments were carried out as described previously (8, 10, 11). The Click-it kit (Invitrogen) was used to detect EdU. ISH and immunohistochemistry were performed as described previously (7, 8, 10) with minor modifications. More details are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Paolo Capelli, Viviana Guadagni, and Paola Iacopetti for assistance; and Marcella Simili, Laura Mariani, Massimiliano Andreazzoli, Simona Casarosa, Paolo Malatesta, Giuseppe Macino, Rongqiao He, and Magdalena Goetz for discussion. This work was supported by Telethon, Ministero Università e Ricerca Scientifica, Ministero Affari Esteri, and Scuola Normale Superiore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909167106/DCSupplemental.
References
- 1.Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Ohnuma S, Harris WA. Neurogenesis and the cell cycle. Neuron. 2003;40:199–208. doi: 10.1016/s0896-6273(03)00632-9. [DOI] [PubMed] [Google Scholar]
- 3.McConnell SK. Strategies for the generation of neuronal diversity in the developing central nervous system. J Neurosci. 1995;15:6987–6998. doi: 10.1523/JNEUROSCI.15-11-06987.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 5.Wong LL, Rapaport DH. Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development. 2009;136:1707–1715. doi: 10.1242/dev.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow RL, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci USA. 2004;101:1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Autilia S, et al. Cloning and developmental expression of the Xenopus homeobox gene Xvsx1. Dev Genes Evol. 2006;216:829–834. doi: 10.1007/s00427-006-0109-0. [DOI] [PubMed] [Google Scholar]
- 8.Decembrini S, Andreazzoli M, Vignali R, Barsacchi G, Cremisi F. Timing the generation of distinct retinal cells by homeobox proteins. PLoS Biol. 2006;4:e272. doi: 10.1371/journal.pbio.0040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koike C, et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- 11.Ohnuma S, Hopper S, Wang KC, Philpott A, Harris WA. Co-ordinating retinal histogenesis: Early cell cycle exit enhances early cell fate determination in the Xenopus retina. Development. 2002;129:2435–2446. doi: 10.1242/dev.129.10.2435. [DOI] [PubMed] [Google Scholar]
- 12.Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Caviness VSJ, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: A general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 14.Choi PS, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leucht C, et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 16.Visvanathan J, Lee S, Lee B, Lee JW, Lee S. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 19.Sigulinsky CL, Green ES, Clark AM, Levine EM. Vsx2/Chx10 ensures the correct timing and magnitude of Hedgehog signaling in the mouse retina. Dev Biol. 2008;317:560–575. doi: 10.1016/j.ydbio.2008.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locker M, et al. Hedgehog signaling and the retina: Insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Calle-Mustienes E, Glavic A, Modolell J, Gómez-Skarmeta JL. Xiro homeoproteins coordinate cell cycle exit and primary neuron formation by up-regulating neuronal-fate repressors and down-regulating the cell-cycle inhibitor XGadd45-gamma. Mech Dev. 2002;119:69–80. doi: 10.1016/s0925-4773(02)00296-4. [DOI] [PubMed] [Google Scholar]
- 24.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 25.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11:1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 27.Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, et al. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 29.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.le Sage C, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, et al. MicroRNA expression profiling in human ovarian cancer: MiR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 32.Brosh R, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina R, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokol NS, Xu P, Jan Y, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.