Abstract
Motivation: There is a strong demand in the genomic community to develop effective algorithms to reliably identify genomic variants. Indel detection using next-gen data is difficult and identification of long structural variations is extremely challenging.
Results: We present Pindel, a pattern growth approach, to detect breakpoints of large deletions and medium-sized insertions from paired-end short reads. We use both simulated reads and real data to demonstrate the efficiency of the computer program and accuracy of the results.
Availability: The binary code and a short user manual can be freely downloaded from http://www.ebi.ac.uk/∼kye/pindel/.
Contact: k.ye@lumc.nl; zn1@sanger.ac.uk
1 INTRODUCTION
A major part of the genetic difference between individuals is encoded in the form of structural variations, such as insertions, deletions and duplications. In previous studies, hundreds of large-scale structural variants have been identified using arrayCGH (Iafrate et al., 2004; Sebat et al., 2004). Polymorphic transposon insertions (Bennett et al., 2004) and other short indel polymorphisms (Mills et al., 2006) have been discovered using capillary sequencing reads. Clone-end capillary sequencing provided a means to interrogate the intermediate size structural variations and particularly, the method is able to find relatively large insertion events (Kidd et al., 2008). Whole genome complete sequencing created a more detailed catalog of structural variations for single human individuals (Levy et al., 2007; Wheeler et al., 2008). For example, the de novo assembly of the Venter genome played an essential role in detecting long insertions, deletions and structural rearrangements.
Recent efforts on variant detection have been fueled up by next-gen high throughput sequencing. Using the Illumina platform for a complete sequencing of a human individual, Bentley et al. (2008) reported ∼4-million SNPs and ∼0.4-million short indels of size 1–16 bp. However, large indel events were not extensively studied by the authors. For the next-gen sequencing data, there are a number of ways to detect long variants, notably assembly (complete de novo or using unmapped reads only), read splitting, read coverage depth analysis, inconsistencies of insert sizes through paired-end mapping, etc. Among these, de novo assembly from short reads perhaps offers the best chance for long indels and structural rearrangements. There are a number of short read assemblers based on de Bruijn graphs. However, assembly of short read data is most successful when applied to small genoms like bacterial genomes (Chaisson and Pevzner, 2008; Zerbino and Birney, 2008). Above the eukaryotic level, it would be problematic due to repetitive genome regions. Given the current read length of 35–75 bp in the next-gen sequencing platforms, lack of high quality de novo assembly looks like to continue in the near future. Therefore, the need is eminent in the genomic community to develop read mapping related algorithms in order to reliably identify structural variants.
In this article, we present Pindel, a method that uses pattern growth algorithm to identify the break points of large deletions (1 bp–10 kb) and medium sized insertions (1–20 bp) from 36 bp paired-end short reads. We will start with introducing pattern growth for string matching. Then Pindel, the procedure of computing medium sized insertions and large deletions from paired-end short reads, will be illustrated. We test our Pindel program with simulated paired-end short reads on human chromosome X. We also report the results of Pindel using the whole human genome data of NA18507, sequenced on an Illumina platform. Finally Runtime and peak memory usage are analyzed to demonstrate the efficiency of Pindel.
2 METHODS
2.1 Pattern growth for exact string matching
In our previous study, we followed the principles of the pattern growth data structure as presented in PrefixSpan, a sequential pattern mining algorithm (Pei et al., 2004), and introduced various constraints during the data mining process to mine biological meaningful patterns from unaligned protein sequences (Ye et al., 2007). Since the data structure of pattern growth and its extensions to the analysis of protein sequences have been well documented before, here we only briefly introduce how to use pattern growth to find minimum and maximum unique substrings of a given pattern against a sequence database. In our particular application those unique substrings must start from either the leftmost or the rightmost base of pattern P.
For simplicity, we only describe the procedure of finding minimum and maximum unique substrings starting from the leftmost base of the pattern P. The inputs of the algorithm are a dataset S which consists of a series of non-empty sequences of the alphabet {A, C, G, T}, a pattern P to search for the locations of its minimum and maximum substrings. The output of the algorithm consists of all substrings (and their locations) starting from the leftmost base of P that appear exactly once in S. The algorithm works as follows. Here a is a substring that starting from the leftmost base of P and Sa is the so-called projected database that contains all sequences that contain the substring a, where the last element of each occurrence of a is marked (the so-called a-locations). The computation of Sa′ from Sa requires, for each a-location, the check whether or not the base on its right-hand side equals the newly appended item b. In case of equality this gives an a′-location in Sa′. Sequences without any a′-location are removed from the projected database. The main call is unique (Λ, SΛ), where Λ is the empty substring; note that each base in database SΛ is a Λ-location, including the position before each sequence. This call creates a projected database that marks all occurrences of the first base b of pattern P.
Let us use a simple example to explain the procedure above. Suppose we define a database S of genome sequence as ‘ATCAAGTATGCTTAGC’ and the pattern P is ‘ATGCA’. In the first step, we scan the whole database for ‘A’, the first base of pattern P. The locations of ‘A’ (red ‘A’ in ‘ATCAAGTATGCTTAGC’) are stored in a projected database of ‘A’. In the second step, we look for ‘T’ as it is the second base in pattern P at the right side of ‘A's identified previously. The projected database for ‘AT’ then only contains two locations (‘ATCAAGTATGCTTAGC’). When we search for the third base ‘G’ of pattern P at the right sides of ‘AT’, we found that ‘ATG’ appears exactly once in the database S (‘ATCAAGTATGCTTAGC’). Thus we know that ‘ATG’ is the minimum unique substring of pattern P in the database S. After we examine the fourth and fifth base of pattern P, we notice that ‘ATGC’ is also unique in the database S but ‘ATGCA’ isn't. In this case we know that ‘ATGC’ is the maximum unique substring of pattern P in this particular database S.
2.2 General procedure of Pindel
In the Pindel program, we aim to compute the precise break points as well as the fragments inserted or deleted compared to the reference genome from paired-end reads. In the preprocessing step, we first use SSAHA2 (Ning et al., 2001) to map all the reads to the reference genome. Then the mapping results are examined to keep those paired reads that only one end can be mapped. For each of those read pairs, the mapped end must be uniquely located in the genome with no mismatch bases while the other end cannot be mapped to anywhere in the genome under a given threshold alignment score (s=20 for ∼36 bp reads). For each of those pairs, our Pindel program uses the mapped end to determine the anchor point on the reference genome and the direction of the unmapped read. Knowing the anchor point, the direction to search for the unmapped read and the user defined Maximum Deletion Size (Max_D_Size) parameter, a sub-region in the reference genome can be located, where Pindel will break the unmapped read into 2 (deletion) or 3 (short insertion) fragments and map the two terminal fragment separately.
2.3 Detecting large deletions
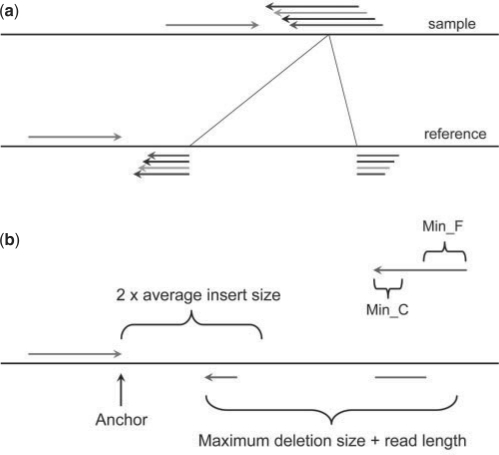
When we map paired-end reads to the reference genome, for the majority of the reads, both ends can be mapped to the reference genome. However, a small portion of them might have only one end mapped to the reference genome. One of the possibilities is that the unmapped read mate spans the break point of a deletion event in the test sample compared to the reference genome as shown in Figure 1(a). Thus, those unmapped reads actually carry the information about the precise break points of the deletion event. If we can find a proper position to split the read into two fragments, which can be mapped back to the reference separately, we will be able to compute the exact positions of the break points and thus the fragment deleted compared to the reference. If we collect multiple reads that support the same incidence, we will be more confident about the deletion event in the test sample.
Fig. 1.
Detecting deletion events. (a) When mapping paired-end reads to the reference genome, some reads may not be mapped even allowing a few mismatches because they are just across the break points of deletion events. If we can find a proper position to break the read into two fragments and map them separately, we will be able to compute the exact break points and the fragment deleted compared to the reference. If find more supporting evidences can be found, the possibility of the deletion event will be higher in the test sample. For simplicity, we only depicted one mapped read (green arrow); (b) The procedure to break the unmapped read into two parts at appropriate position and mapped them separately to the reference genome. The location and direction of the mapped read (green) define the local region to break the unmapped read into two fragments and map them separately. The 3′ end of the mapped read is defined as anchor point. Then pattern growth is used to search for minimum and maximum unique substrings from the 3′ end of unmapped reads within the range of two times of insert size starting from the anchor point. Using pattern growth again to search for minimum and maximum unique substrings from the 5′ of unmapped read within the range of read length+user defined maximum deletion size starting from the already mapped 3′ end of the unmapped read. The computed minimum and maximum substrings from both 3′ and 5′ are examined to see whether a complete unmapped read can be assembled. All possible solutions are stored in a database for sorting according to the break point coordinates. A deletion event is reported if at least two reads support it.
Although for short reads, half of a read length might be too short to be mapped uniquely in a whole genome scale as large as human's, the location and the direction of the mapped read reduces the search space dramatically. As shown in Figure 1b, when we have one read mapped (the anchor point) but the other one is not mappable, we only need to consider the local region at one side of the anchor point. The computational procedure for each unmapped read is described as following (Fig. 1b):
Read in the location and the direction of the mapped read from the mapping result obtained in the preprocessing step;
Define the 3′ end of the mapped read as anchor point;
Use pattern growth algorithm to search for minimum and maximum unique substrings from the 3′ end of the unmapped read within the range of two times of the insert size from the anchor point;
Use pattern growth to search for minimum and maximum unique substrings from the 5′ end of the unmapped read within the range of read length+Max_D_Size starting from the already mapped 3′ end of the unmapped read obtained in step 3;
Check whether a complete unmapped read can be reconstructed combining the unique substrings from 5′ and 3′ ends found in steps 3 and 4. If yes, store it in the database U. Note that exact matches and complete reconstruction of the unmapped read are required so that neither gap nor substitution is allowed.
After processing all unmapped reads, sort the database U according to the breakpoint coordinates in the reference and output every deletion event supported by at least two reads.
User has to specify the parameter Max_D_Size and also change the minimum lengths for unique substrings reported in step 3 and 4, denoted as Min_C and Min_F, respectively.
2.4 Detecting medium sized insertions
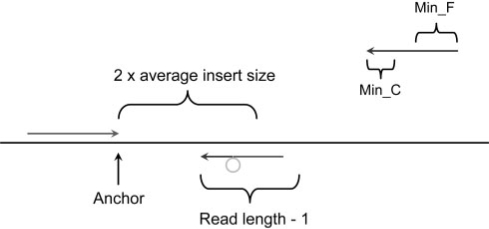
In Section 2.3, we explain how to compute the precise break points of deletion events and the deletion size could be rather large as long as we find unique matches for the two parts of the unmapped read. It is, however, difficult to infer the fragment for large insertions directly from the read sequence. In this case we aim to compute the precise break points and the fragment inserted in the medium sized range (<=20 bases for 36 bp reads). The computational procedure is very similar to that used for searching deletions. The only difference is in step 4 where the search range for the unique occurrence of minimum and maximum unique substrings from the 5′ end of the unmapped read is read length minus one. In this case, we certainly cannot reconstruct the whole read and the extra bases are an inserted fragment compared to the reference genome as shown in Fig. 2.
Fig. 2.
Detecting short insertion events. The procedure to split the unmapped read into three parts at appropriate position and mapped the terminal two separately to the reference genome. The location and direction of the mapped read (green) define the local region to split the unmapped read. The 3′ end of the mapped read is defined as anchor point. Then pattern growth is used to search for minimum and maximum unique substrings from the 3′ end of unmapped reads within the range of two times of insert size starting from the anchor point. Using pattern growth again to search for minimum and maximum unique substrings from the 5′ of unmapped read within the range of read length – 1, starting from the already mapped 3′ end of the unmapped read. The computed minimum and maximum substrings from both 3′ and 5′ are examined to see whether they are adjacent to each other. The middle fragment is the inserted fragment. All possible solutions are stored in a database for sorting according to the break point coordinates. An insertion event is reported if at least two reads support it.
2.5 Simulated data
2.5.1 Simulating paired-end reads on human X chromosome
In order to evaluate our Pindel program, we first simulated indels on human X chromosome and examined how well Pindel can detect those simulated indels in the presence of SNPs and sequencing errors. The sequence of human X chromosome was obtained at ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/Assembled_chromosomes/. We put 20 instances for each indel length we have chosen. The insertion sizes vary from 1–20 bp while the deletion sizes range from 1–10, 100, 1, 10, 100 kb to 1 Mb. SNPs are simulated at a rate of 0.001 and sequencing error rate is set to 0.005. Using the above setting, we simulated 30× coverage of 36 bp paired-end short reads with an average insert size of 200. The indels are randomly placed and their locations are recorded to facilitate comparison.
2.6 Real data
Recently the genome of a male Yoruba from Ibadan, Nigeria (YRI, sample NA18507), was sequenced (Bentley et al., 2008). The ∼4-billion paired−end reads (∼40-fold depth, 135 Gb of sequence) were obtained from the NCBI short-read archive, accession SRA000271 (ftp://ftp.ncbi.nlm.nih.gov/pub.TraceDB/ShortRead/SRA000271). The list of 378 287 short indels (1–16 bp) in their study was kindly provided by Illumina.
3 RESULTS
3.1 Implementation of Pindel
The Pindel program was implemented in C++ to identify break points of medium-sized insertion and large deletions using the principle of pattern growth. Currently no parallelization has been implemented in Pindel so that it only runs on a single CPU. The input for Pindel consists of the reference genome sequence and a file of one-end-mapped read pairs. The output contains information about each indel event at the base level. In Figure 3, for example, a deletion event (‘D’ one the start of the first line) as reported by Pindel is depicted. The indel size and its break point coordinates on the reference genome are given. There are 15 reads supporting such an event. For 9 out of 15 reads, their mapped partners are located upstream of the deletion event (+ sign) while six are located downstream (- sign). The coordinates of mapped reads are given as well. Currently we report an indel event if there are at least two reads supporting it.
Fig. 3.
An example output of Pindel. The type and size of deletion are specified first (D 321). Then the chromosome ID, coordinates of the break points and the number of reads supporting every event are given. The mapping directions of the mapped reads and their 3′ coordinates on the reference are reported for each supporting read.
3.2 Simulation on human chromosome X
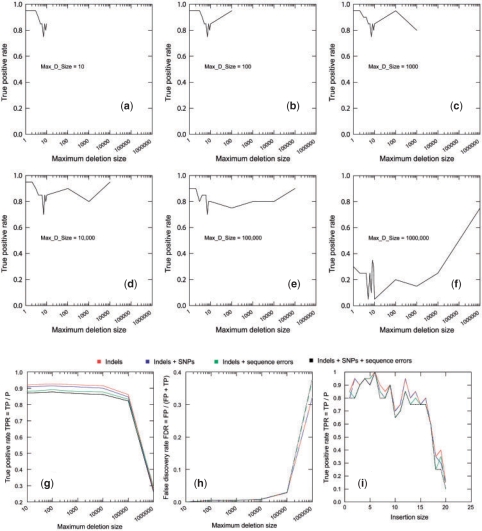
In order to evaluate performance of Pindel, we first examine how well it can retrieve randomly placed indels on human chromosome X in the presence of SNPs and sequencing errors. For deletion, user must specify the maximum size of deletion events (Max_D_size). As shown in Figure 4, when we increase Max_D_size from 10 bp to 100 kb, we are able to recover about 80% of deletions (Fig. 4a–e) with <2% false discovery rate (Fig. 4h). It should be noted that the maximum size cannot be too high as this significantly increases the false positives as well as decreases the rate of true positives (Fig. 4f and h), especially when the maximum size reaches 1 Mb. We also investigated the effect of sequencing errors and SNPs on the performance of detecting deletions by Pindel (Fig. 4g and h) and we found that data with SNPs and/or sequencing errors only slightly affects the rate of false negatives and has little effects on accuracy of variant detection due to the stringent filtering of read alignment in the preprocessing step, i.e. no mismatch is allowed. As for insertions, we can correctly detect around 80% of the insertions of 1–16 bases in the presence of SNPs and sequencing errors. For 36 bp paired-end reads, the probability for Pindel to detect insertion events longer than 16 bp is decreasing as the size of insertion events goes up (Fig. 4i).
Fig. 4.
Simulation of paired-end reads from human chromosome X. (a–f) True positive rates per each deletion size are displayed in the presence of sequencing errors and SNPs when Max_D_Size is increased from 10 to 1 000 000 bp. The impact of sequencing errors and/or SNPs on the overall true positive rates (g) and false discovery rates (h) when Max_D_Size is set to different values from 10 to 1 000 000. (i) The impact of sequencing errors and/or SNPs on the true positive rates for detecting medium sized insertions from size 1 to –20 bp.
3.3 Real data (NA18507)
The newly sequenced data (Bentley et al., 2008) of a male Yoruba from Ibadan, Nigeria (YRI, sample NA18507), provides us a chance to further examine Pindel with high depth, high quality and most importantly real paired-end read data. After preprocessing the paired-end reads with SSAHA2, we obtained 56 161 333 pairs of reads which only have one end mapped uniquely to the human reference while the other end couldn't be mapped. In order to compare the prediction of Pindel with the short indel calling (1–16 bp, indels_NA 18507) published together with the sequencing data, we first constrained our Pindel to pick up indels of length 1–16 bp. As a result, Pindel predicted 146 843 deletions and 142 908 insertions. We found that 133 974 deletions (91.2% of 146 843) and 124 559 insertions (87.2% of 142 908) are in the indel list of Bentley et al. If we take the prediction of Bentley et al. as true positive, the overall false negative rates of Pindel for deletions and insertions of length 1–16 bp will be 31.6%. Tables of comparison between predictions of Bentley et al. and Pindel are available at http://www.ebi.ac.uk/∼kye/pindel/.
Since our Pindel is able to identify deletions of size up to 10 kb from 36 bp paired-end reads, we removed the constraints on the indel size and rerun Pindel. The result was compared with the database of genomic variants [DGV, http://projects.tcag.ca/variation/, (Iafrate et al., 2004)]. For the range of 100 bp–1 kb, Pindel predicted 1399 deletions, 949 of which (67.8%) were listed in DGV. In the range of 1—10 kb, Pindel predicted 1138 deletions, 494 of which (43.4%) were listed in DGV.
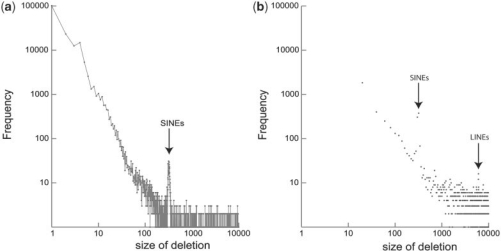
The deletion size distribution for NA18507 from 1 to 10 000 bp is shown in Figure 5a, where the frequency per each deletion size is depicted as a dot while adjacent dots are connected to illustrate the trends. There is a peak around 300 bp, which contain hundreds of putative SINEs. In Figure 5b, when we sum up the frequencies for every 20 bases, there is an additional peak at around 6 k which most likely corresponds to putative LINEs.
Fig. 5.
Plots of deletion size distribution for NA18507 from 1 to 10 000 bp. (a) The frequency per each deletion size from 1 to 10 000 bp. Adjacent dots are connected. There is a peak around 300 bp, which may contain hundreds of putative SINEs. (b) Sum of frequencies for each 20 bases is plotted. The peaks for putative SINEs and LINEs are visible.
We further characterized the 665 deletion events, whose sizes range from 300 to 350 bp. Six hundred thirty seven of 665 deletion events (95.8%) were reported as SINEs by RepeatMasker and the majority of those 637 events were flagged as potential members of the AluY family.
3.4 Runtime and memory usage for processing data of NA18507
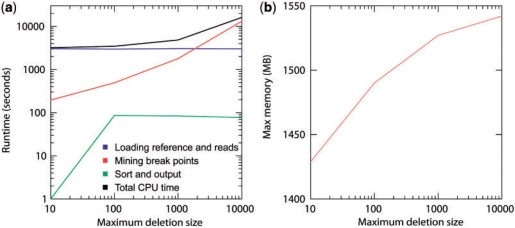
The Pindel program is efficient in time and memory. In order to reduce memory requirement, each time Pindel loads one chromosome into memory and scan the entire read alignment file, examining reads associated with the current chromosome. After Pindel processes all the reads, it outputs the results and then frees memory for the next chromosome. As shown in Figure 6a, when Max_D_Size is smaller than 1 k, the most time-consuming step is loading the reference sequence and scanning the read file. When we set Max_D_Size to 10 k, it took Pindel only 16 125 s (<4.5 h) to process the complete data of NA18507 on a single CPU.
Fig. 6.
Runtime and memory consumption for Pindel applied to the NA18507 data on a single CPU for mining indels with different Max_D_Size (10 bp, 100 bp, 1 kb and 10 kb). (a) The user runtime for Pindel is divided into three categories: (i) loading the reference genome and the reads into memory. (ii) Break unmapped reads and map them separately using pattern growth. (iii) Sort break points according to coordinates and write the results on hard disk. (b) Maximum memory consumption for Pindel to process the NA18507 data with different Max_D_Size (10 bp, 100 bp, 1 kb and 10 kb).
Because Pindel processes one chromosome at a time, the maximum memory consumption is well controlled. As shown in Figure 6b, when we increase Max_D_Size from 10 bp to 10 kb, the peak memory usage only increases slightly from 1429 to 1542 MB simply because more indel events are found and stored in memory before writing back to the hard disk.
4 DISCUSSION
4.1 Pattern growth: memory and speed
Currently major genome-wide sequence analysis programs use either hash-tables or suffix tree related data structure to index sequences to allow efficient query (Ning et al., 2001; Schulz et al., 2008). Indexing entire genomes as large as the human one requires significant amount of computer memory. Since our Pindel program only processes and breaks and unmapped reads in the defined local regions according to the position of the mapped anchor read, using pattern growth to directly search for unique minimum and maximum unique substrings is apparently more memory efficient than indexing the entire genome with hash-tables or suffix trees (Fig. 6b). In our applications, the pattern growth method is very efficient in searching for deletions up to 10 kb long, taking only 4.5 h on a single CPU to process data derived from ∼40× coverage paired-end reads of human genome. It should be noted that Pindel does need read mapping and this process is the main bulk of computational CPU time and memory usage. We are also working on the adaption of the SAM (http://samtools.sourceforge.net) format so that Pindel scans SAM or BAM files for variants regardless of the alignment tools.
4.2 Sensitivity and specificity
As one can see in the results from both simulation and real data, Pindel misses a fraction of true positives for several reasons. First of all, repeats in the reference genome may prevent unique mapping of anchor points. Pindel also requires unique exact occurrence of the terminal fragments of unmapped reads within a certain region related to the anchor point. Whenever missing anchor points or repeats occurring around the variant, Pindel will not report this event at all. The probability of finding a random substring that is identical to one of the terminal fragments increases as we enlarge the parameter Max_D_Size (Fig. 4a–g).
In the current version of Pindel, we only consider perfect matching and mismatch is not allowed. As a consequence, SNPs or sequencing base errors in the regions of anchor or indel points may lead to the miss of true positives because there might be insufficient supporting reads. We are investigating inexact matching with pattern growth to overcome the issue of SNPs and sequencing errors.
Throughout this manuscript we have used a fixed cutoff value for the number of supporting reads >=2, for the detection of deletions and insertions from paired-end short read data. Obviously this is a simplification as the actual parameters like insertion or deletion size, and the number or paired-end reads are not taken into account. For example in the simulation on chromosome X we have seen a drop in true positive rate for searching of large deletions, mostly due to the accumulated number of random matches. It was very surprising to us that the simple cutoff we used showed such promising results over a broad range of search parameters. Clearly, a fixed cutoff will be inappropriate once we introduce mismatches in the string-matching step as every mismatch may increase the number of random matches by orders of magnitudes dependent on read length. Therefore, we are currently investigating a statistical score that captures random matches due to different search parameters, inexact matching, and the total number of reads.
5 CONCLUSIONS
In this study, we present Pindel, a computational approach to detect breakpoints of large deletions and medium sized insertions from paired-end short reads. As far as we know, Pindel is one of the first programs to compute deletion events as large as 10 kb with base level precision from 36 bp paired-end short reads. Due to its high performance in sensitivity, specificity and efficiency in memory and speed, Pindel has been proved to be a promising approach to address the structural variations between individuals from next-gen high throughput sequencing.
ACKNOWLEDGEMENTS
The authors thank E. Birney, M. Fritz, M. Schuster, K. Walter and Y. Zhang for comments.
Funding: NGI/EBI fellowship 050-72-436 from Netherlands Genomics Initiative.
Conflict of Interest: none declared.
REFERENCES
- Bennett EA, et al. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, et al. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, et al. Mining sequential patterns by pattern-growth: The prefixspan approach. IEEE Trans. Knowl. Data Eng. 2004;16:1424–1440. [Google Scholar]
- Schulz MH, et al. The generalised k-truncated suffix tree for time-and space-efficient searches in multiple DNA or protein sequences. Int. J. Bioinform. Res. Appl. 2008;4:81–95. doi: 10.1504/IJBRA.2008.017165. [DOI] [PubMed] [Google Scholar]
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Smit AFA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Ye K, et al. An efficient, versatile and scalable pattern growth approach to mine frequent patterns in unaligned protein sequences. Bioinformatics. 2007;23:687–693. doi: 10.1093/bioinformatics/btl665. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]