Abstract
Motivation: Most microbial species can not be cultured in the laboratory. Metagenomic sequencing may still yield a complete genome if the sequenced community is enriched and the sequencing coverage is high. However, the complexity in a natural population may cause the enrichment culture to contain multiple related strains. This diversity can confound existing strict assembly programs and lead to a fragmented assembly, which is unnecessary if we have a related reference genome available that can function as a scaffold.
Results: Here, we map short metagenomic sequencing reads from a population of strains to a related reference genome, and compose a genome that captures the consensus of the population's sequences. We show that by iteration of the mapping and assembly procedure, the coverage increases while the similarity with the reference genome decreases. This indicates that the assembly becomes less dependent on the reference genome and approaches the consensus genome of the multi-strain population.
Contact: dutilh@cmbi.ru.nl
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
DNA sequencing is cheaper than ever before. Use of a 454 Pyrosequencer and/or Illumina Genome Analyser can produce a nearly complete bacterial genome at a cost of <€ 10 000 (for a review of next generation sequencing see Mardis, 2008). However, most microbes can not be readily obtained in pure culture, apparently because their phenotype is not compatible with growth on solid media.
A promising solution to this problem is to perform selective enrichment in continuous culture, where the conditions favorable for the species' growth can be approximated more closely. Further, in enrichment culture, interdependency on other species (e.g. the exchange of cofactors) is not problematic. Metagenomic sequencing could yield a near-complete genome if the resulting population is sufficiently enriched and if the sequencing coverage is high (degree of enrichment times sequencing coverage >20). A culture that is inoculated with a natural sample can yield a population that is highly enriched for a single species within a few months (e.g. Ettwig et al., 2008).
Currently, high sequencing coverage is achieved most cost-effectively with massive parallel sequencing methods that produce short reads [SOLiD sequencing (http://solid.appliedbiosystems.com) and Illumina/Solexa sequencing (http://www.illumina.com/pages.ilmn?ID=203; Bentley, 2006)]. Such reads are usually processed with mapping algorithms such as Eland (Bentley et al., 2008) or Maq (Li et al., 2008) if a reference genome from a closely related species is available. Truly de novo assembly directly from short reads (e.g. Velvet; Zerbino and Birney, 2008) remains difficult, although innovative techniques that use e.g. conservation at the gene level are promising (Salzberg et al., 2008).
The mapping algorithms are generally highly conservative: they permit no more than one or two mismatches per read, and do not allow the presence of gaps in the alignment. This means that any read derived from a region with a lower conservation than 30/32 ≈ 94% identity will not be mapped, and it restricts the use of a reference genome to highly similar species. Therefore, mapping reads to a reference genome has two limitations. First, it depends on an available reference genome of a closely related organism (>94% identity). Second, an enriched microbial community culture often contains multiple related strains with similar fitness (quasispecies), and the sequence diversity between such strains can be quite high (Venter et al., 2004). Such a polymorphic population can be expected to confound the highly conservative mapping and/or assembly programs leading to unnecessary fragmentation of the assembly, as well as a large fraction of the reads not being used.
Here, we set out to decipher the consensus genome of parallel populations of a quasispecies sequenced with short-read Solexa sequencing. Solexa instruments can now generate >50 nt reads, but we used an earlier version of the instrument that generates 32 nt reads. We use a related genome as a scaffold, and first map the reads to their best possible position on this reference. Then, we ask per reference position which nucleotide is the most highly represented in the population of strains. Because the resulting assembly is already a better approximation of the sequences in the strain population than the external reference, we iterate the mapping and assembly procedure to increase the coverage. The final consensus assembly captures the majority vote of the genomes in the multi-strain population.
2 METHODS
2.1 Data
We performed one single-end Illumina sequencing run of an enriched metapopulation (Ettwig et al., 2009), yielding 6 667 153 32 nt reads [details about these data and the reference genome will be published elsewhere (K.F. Ettwig et al., manuscript in preparation)].
2.2 Mapping reads to their optimal position
The list of reads were mapped against the reference genome (2 752 854 nt) with each of three programs: BlastN v2.2.20 (Altschul et al., 1997), MegaBlast v2.2.20 (Zhang et al., 2000) and Maq 0.7.1 (Li et al., 2008). For Maq, we assigned the highest sequencing quality score (∼) to every nucleotide, and then ran Maq with default parameters. For BlastN and MegaBlast, the reads were made non-redundant and given a unique identifier containing the number of instances and the sequence (e.g. 7xACGT…). We used relaxed search parameters, to make sure many reads were mapped, even if they were quite divergent. We used a short word length of 8 (other word lengths were tested as well, see Supplementary Material), turned low-complexity filtering off and used a high E-value threshold of 100, although all reads included in the assembly were mapped with much lower E-values (Fig. 1d) due to the alignment length cutoff described below. To account for the high-coding density in bacterial genomes, we performed ungapped searches (gap open and extend penalties 1000). The output for each read was immediately parsed, removing all hits with a sub-optimal score. Not only will we require none other than the highest scoring location(s) on the reference genome for each read, but this filtering step also frees disk space.
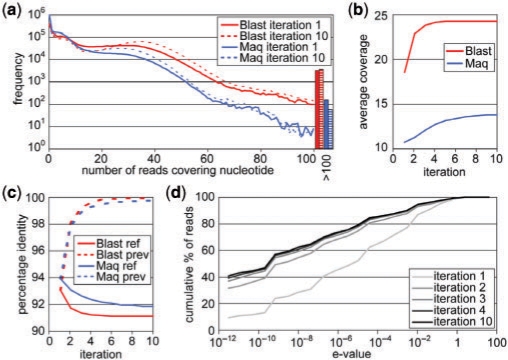
Fig. 1.
(a) Distribution of coverage scores per nucleotide in the first and 10th iterations; (b) average coverage score in each iteration; (c) percentage identity of non-zero coverage regions of the assemblies with the reference genome and with the previous assembly (i.e. the reference for that iteration); and (d) percentage of reads in the assembly for each E-value (cumulative).
2.3 Assembly
Next, we assembled the mapped reads to form a consensus genome. For Maq, we used the consensus sequence provided by the program, while for the BlastN and MegaBlast results, we wrote a custom Perl script (available from the authors on request), taking the following into account. For each position on the reference genome, we assessed which of the reads covered it with an aligned region of at least 20 nt. The nucleotide with the highest occurrence in the community was called to align to that reference position. Draws were replaced by their IUPAC nucleotide code (Cornish-Bowden, 1985). The coverage at each position equals the number of reads contributing to the consensus. Positions with no aligned reads (zero coverage) were replaced with Ns.
2.4 Iteration
After assembly, the whole procedure was iterated. Positions with zero coverage in the assembly were replaced with the nucleotide in the reference genome, and all Solexa reads were re-queried against this new reference (as above). We carried out at least 10 iterations with each read mapping algorithm.
3 RESULTS
3.1 Similarity search algorithm
Here we combine the short 32 nt Solexa sequencing reads from a metapopulation of strains to form a consensus genome describing the majority of the population. The first step in the process is to map as many of the sequencing reads as possible to their optimal position on the reference. The conservative mapping algorithm Maq (Li et al., 2008) mapped 602 120 reads, leading to an average coverage of 10.8 in the assembled regions, but 35.0% of the reference genome still has zero coverage (Supplementary Material). The large gaps remaining with this conservative mapping algorithm already shows that the reference is distant enough from the community to require a more relaxed sequence similarity search.
We used BlastN and MegaBlast as examples of less restrictive read mapping algorithms. We used very relaxed search parameters (see Section 2), allowing even quite distant reads to be mapped to their optimal position in the reference. However, this approach does require that we employ a filter for spurious short hits, so we selected only those reads that were aligned to the reference over at least 20 nt. In this preliminary search, BlastN mapped 1 598 549 reads and MegaBlast mapped 1 595 338 reads, both leading to an average coverage in the assembled regions of 18.5, while 14% of the reference nucleotides have zero coverage (Supplementary Material).
3.2 Coverage increases by iteration
Any available mapping algorithm will suffice to map highly identical reads to a reference. Our aim was also to map the more divergent reads to obtain a higher coverage of the polymorphic community on the divergent reference. The initial coverage of the Blast-based assemblies are already higher than the conservative Maq assembly, but many nucleotides still have a low coverage <10 (Fig. 1a). However, since this first assembly is composed of the metagenomic reads themselves, we expected that using it iteratively as a new mapping scaffold would yield a higher sequencing coverage. Indeed, the average coverage clearly increases after a second round of querying and assembling the reads to the consensus genome. Additional iterations gradually increase the number of reads that could be mapped for all algorithms (Fig. 1b). The statistics for the BlastN- and MegaBlast-based approaches are almost identical (Supplementary Material). These results show that more reads can be mapped as the reference is adjusted to the reads, indicating that the assembly becomes more similar to the consensus genome of the community.
3.3 Consensus genome
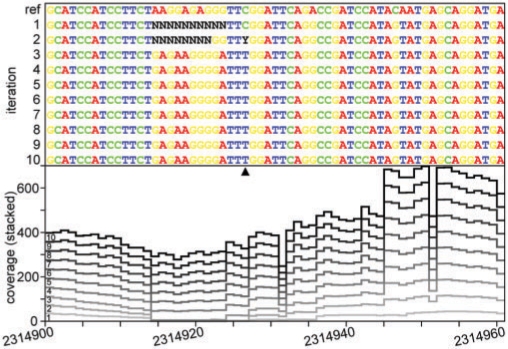
Observing this clear increase in coverage with the iterations, including a ∼15% drop of the zero-coverage nucleotides (e.g. from 387 421 to 329 788 in the BlastN-based approach), we decided to take a look in detail at how the consensus sequence changes with the iterations. Figure 2 shows a small part of the genome, illustrating some of the changes that occur as the iterations progress. For example, position 2 314 927 in the alignment (indicated with an arrow), a cytosine in the reference, changes into a Y (i.e. cytosine or thymine; Cornish-Bowden, 1985) after the second round of read mapping. Subsequently, in iteration 2, this trend is confirmed and the consensus nucleotide present in the population of reads is settled as a thymine from then on. Another example is the region with zero coverage (stretches of Ns) in the first and second iteration assemblies that are filled in the subsequent iterations. It should be noted that we map the entire list of reads against the reference or previous assembly in every iteration, and there is no source of new reads. It is possible that reads are re-mapped to a different region (e.g. to the zero-coverage region in Fig. 2) if (i) the new region has altered and gained similarity with reads that were not mapped before or that were mapped to another part of the reference; or (ii) the region where these reads were mapped before has altered and lost similarity with the reads so that they now map to this new position instead. However, as we see that the reads generally gain similarity with the evolving genome (Fig. 1c and d), explanation (i) seems the most frequent.
Fig. 2.
A region of the assembled sequence showing some of the changes that occur with the iterations. Gaps in the assembly are filled and single nucleotides are settled. The coverage per position in every iteration is shown in the bottom panel.
In general, we observe that the assembly slowly drifts away from the reference genome, as measured by the percentage identity of the mapped regions (i.e. regions with non-zero coverage) to the original reference (Fig. 1c, drawn lines). At the same time, the assembly becomes more coherent, as measured by the percentage identity of the mapped regions to the assembly from the previous iteration (Fig. 1c, dashed lines). Moreover, a larger fraction of the reads is mapped with a lower E-value (Fig. 1d). This indicates that the consensus genome of the population of strains is gradually approached. The optimum in the curves is reached around iteration four, and the reads do not obtain a better mapping than this if we include more iterations (Supplementary Material).
4 DISCUSSION
Here we show how a consensus genome can be composed by mapping metagenomic sequencing reads from a community of strains to a reference. Furthermore, this consensus genome better represents the community if we iterate the mapping and assembly at least once. This increase is independent of the read mapping algorithm. A strict mapping and assembly program such as Maq initially maps 602 120 reads, but this number is increased to 835 328 reads in iteration 10. A less strict mapping algorithm like BlastN maps 1 598 549 and 2 051 404 reads in the first and 10th iteration, respectively (Supplementary Material). Note that there is no (artificial) evolution in this method, and no optimality criteria used. The higher coverage solely results from the fact that the assembly better accommodates the reads. Thus, we profit from the best of both worlds: we use a reference to scaffold the reads, yet the iterated assembly allows the sequence to drift away from the scaffold and approach the consensus genome of the population. Iterative read mapping and assembly has previously been applied in the reconstruction of a bacterial genome from environmental sequence data (Pelletier et al., 2008), but the sequencing reads in that experiment had a much longer mean size of 633 nt, and the idea was not systematically analyzed. We show that our approach can be used with very short 32 nt reads, and the results can only be expected to improve with longer read length.
The sequence we create can be interpreted as the consensus genome of the metapopulation of strains. As always when mapping short sequencing reads, the structure of the genome is scaffolded onto the reference and therefore does not necessarily reflect the genome structure of any particular strain in the sequenced community. Thus, this approach is suited to construct the consensus genome of the most abundant lineages in the sample. Moreover, the DNA sequence at any site within the genome is not even necessarily an existing sequence, but rather the consensus of the most abundant sequences. However, we note that generally, this is also the case for the genome sequencing projects of species that can not be amplified clonally for sequencing, like animals. For example, the first human genome was composed of the combined DNA of several individuals (Lander et al., 2001). Therefore, we expect that the consensus genome we obtain using our iterated assembly method can still provide meaningful information about the encoded proteins and other genomic features. The in-depth analysis thereof will be the topic of a subsequent paper (K.F. Ettwig et al., manuscript in preparation).
Funding: Dutch Science Foundation (NWO) Horizon Project 050-71-058.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 2008;10:3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, et al. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 2009;75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina sequencing technology. Available at http://www.illumina.com/pages.ilmn?ID=203 (last accessed date June 29, 2009) [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics. Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Pelletier E, et al. ‘Candidatus Cloacamonas acidaminovorans': genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 2008;190:2572–2579. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, et al. Gene-boosted assembly of a novel bacterial genome from very short reads. PLoS Comput. Biol. 2008;4:e1000186. doi: 10.1371/journal.pcbi.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOLiD™System Sequencing. Available at http://solid.appliedbiosystems.com (last accessed date June 29, 2009) [Google Scholar]
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.