Abstract
Background
Increase of EGFR gene copy number consequent to gene amplification and/or polysomy of chromosome 7 has been significantly associated with better clinical outcome in Non Small Cell Lung Cancer (NSCLC) patients treated with Tyrosin-Kinase Inhibitors (TKIs).
The primary method to detect EGFR copy number is FISH (Fluorescence in Situ Hybridization), that in lung cancer requires a precise standardization due to the presence of intratumor heterogeneity and high frequency of chromosome 7 polysomy.
Recommendations and interpretative guidelines to discriminate NSCLC patients into FISH positive (gene amplification and high chromosome 7 polysomy) and FISH negative have been proposed by the University of Colorado Cancer Center (UCCC). However, in a subset of cases the distinction between EGFR amplification and chromosome 7 polysomy can be controversial because of a complex pattern of multiple EGFR and centromere signals.
Methods
In order to distinguish more accurately these two genetic events, 20 NSCLC FISH positive patients, showing a controversial pattern of EGFR and centromere specific signals, were further evaluated for the status of 7q31 distal region.
Results
A discrepancy between FISH results obtained with UCCC scoring system and 7q31 control was evidenced in 2 patients (10%).
Conclusion
Our data strengthen the usefulness of 7q31 region evaluation to discriminate EGFR amplification from chromosome 7 polysomy in controversial EGFR FISH positive cases. Since it has been reported a possible different contribution of amplification and polysomy to TKIs susceptibility in NSCLC, the clear distinction between these two genetic events may be important to identify a subset of patients more responsive to the therapy.
Background
The development and clinical application of Tyrosine Kinase Inhibitors (TKIs) targeting the Epidermal Growth Factor Receptor (EGFR), such as erlotinib and gefitinib, provide important insights for the treatment of non small cell lung cancer (NSCLC). However, patients derive different degrees of benefit from treatment with EGFR TKIs [1-3]. Several molecular studies showed that sensitivity to EGFR TKIs correlated very strongly with a class of somatic activating mutations of EGFR kinase domain that target mainly two exons (19 and 21), indicating that patients harbouring EGFR mutations have a higher partial and complete response rate to EGFR TKIs therapy than those without mutations [4-6]. Overall, the incidence of EGFR mutations in NSCLC among clinical responders to gefitinib or erlotinib is 77%, but this is not always associated with prolonged survival [7-11]. However, approximately 10-20% of patients who show a partial response to EGFR TKIs have not detectable EGFR mutations, suggesting other molecular mechanisms are involved in TKIs response [9].
Although a conclusive picture has yet to emerge, an association between increased EGFR gene copy number, detected by Fluorescence in Situ Hybridization (FISH), and sensitivity to TKIs therapy was observed. Cappuzzo et al. [12] reported that FISH positive patients showed a better response and an increased survival in respect to FISH negative patients (36% vs 3% and 18 months vs 7 months, respectively). Different studies [13-18] demonstrated a significant association between high EGFR copy number and response to gefitinib, time to progression and survival. In addition, it was shown that patients negative for at least two tests among mutation, FISH and immunohystochemistry had no benefit from TKI therapy [19].
According to the Molecular Assays in NSCLC Working Group, FISH represents the gold standard method to establish gene numerical status combining the gene specific probe and the control centromeric probe [20]. FISH allows to detect the real amount of gene copies in relation to the chromosome number and discriminate between gene amplification from high gene copy number due to polysomy. Normally amplification is expressed as gene copy number/chromosome control number ratio of > 2.
In NSCLC high EGFR gene copy number results mostly from chromosome 7 polysomy while less often from gene amplification. In the first case, the high number of chromosome 7 centromeres lowers the gene/chromosome ratio value to less than 2, masking possible amplification events. Moreover, EGFR gene is located at the band 7p12, very close to the centromere; as a consequence, in a subset of tumors amplification involves EGFR gene and centromeric sequences too, originating a complex pattern of multiple red and green signals difficult to classify as amplification or high polysomy.
Recently, a FISH scoring system and guidelines for EGFR FISH assay in NSCLC patients have been proposed by the University of Colorado Cancer Center (UCCC). This system stratifies results into six groups by number of copies of the EGFR gene, including disomy, low trisomy, high trisomy, low polysomy, high polysomy and gene amplification. High polysomy and gene amplification are considered FISH positive, the cut-off being assumed on a retrospective study performed in advanced NSCLC patients treated with gefitinib [21]. Although the scoring criteria of UCCC for stratification of NSCLC patients with EGFR FISH assay effectively copes with complexity of EGFR gene amplification mechanisms, it doesn't always allow to discriminate between real gene amplification and high level polysomy, particularly when almost the same number of centromere 7 and EGFR gene signals is present.
The aim of the study was to improve the discrimination between amplification and high level polysomy in those controversial cases that are characterized by multiple copies of both EGFR gene and centrome. To this purpose, we have introduced the FISH evaluation of a further chromosome 7 region (7q31), sufficiently far from EGFR locus (7p12), in 20 selected FISH positive patients showing a complex pattern of EGFR and centromere signals.
Materials and methods
Patients and samples
The study was conducted on 20 NSCLC patients selected over 90 (median age: 69; range 44-87 years) with cytologically or hystologically confirmed NSCLC in advanced stage (III-IV), according to WHO criteria: 18 adenocarcinoma, 1 mucoepidermoid carcinoma and 1 squamous cell carcinoma. The majority were females (14/20; 70%) and never smokers (16/20; 80%). Eleven patients (55%) had a history of metastasis. Mutations of EGFR gene were present in 18/20 patients (90%) who, consequently, received therapy with erlotinib. Ten patient had exon 19 deletion, 7 had mutation of exon 21 (6 cases with substitution L2573 T>G and 1 case with substitution L2558 T>C) and 1 had exon 20 deletion.
All patients had been found EGFR FISH positive for amplification or high level polisomy in a preliminary analysis. They were selected for this study because of a high number of both EGFR and centromere signals in the absence of peculiar traits of gene amplification, such as EGFR/CEP 7 ratio >2, or the presence of large or small clusters of signals. Such a picture is particularly difficult to interpret because it might reflect either an amplification involving EGFR gene together with centromeric sequences or, alternatively, a polysomy of chromosome 7.
The preliminary FISH analysis was carried out using LSI EGFR/CEP 7 (Vysis-Abbott). According to UCCC criteria, high level polysomy was defined as the presence ≥ 4 EGFR copies in > 40% of cells, and gene amplification as: a) EGFR/centromere 7 ratio ≥ 2 b) presence of small gene clusters (4-10 copies) in ≥ 10% of the tumor cells c) larger and brighter EGFR signals than expected in >10% of the tumor cells d) ≥ 15 copies of the EGFR signals in ≥ 10% of the tumor cells.
The evaluation of 7q31 region was performed by FISH on paraffin embedded specimens derived from 18 primary tumors and 2 secondary lesions (lymph node and bone). These specimens included 2 tissue sections from surgical lobectomy, 6 fine needle aspiration/core biopsies (FNAB), 3 endoscopic bronchial biopsies, 3 bronchial washings and 6 pleural effusions.
In 10 cases the median number of EGFR gene (EGFR gene/nuclei ratio) and the median number of chromosome 7 centromere (centromere 7/nuclei ratio) were very similar with a Δ < 1 unit. In the other 10 cases the median number of EGFR gene was higher than the median number of chromosome 7 centromere that, nevertheless, ranged from 6 to 23 copies.
7q31 FISH Analysis
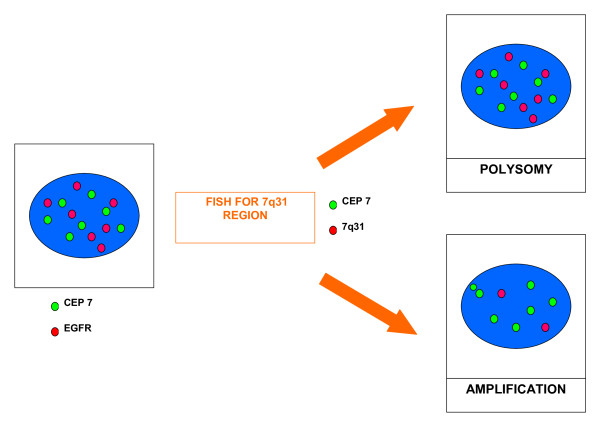
FISH analysis was performed using standard methods. In brief, 4-6 μm paraffin embedded tissue sections were deparaffinized and subsequently enzymatically digested with a commercial kit (Paraffin pretreatment reagent kit, Vysis-Abbott Molecular, Downers Grove, IL, USA). Sample DNA was denaturated at 75°C for 5 minutes. Spectrum-Orange-labeled LSI D7S522 probe, that hybridises to band 7q31, was used together with Spectrum-Green-labelled centromere 7 reference probe (LSI D7S522/CEP 7, Vysis-Abbott). The probe mix (10 μl), previously denaturated for 5' at 75°C, was applied on each slide. The slides, covered with a glass coverslip, were incubated for 5' at 79° for codenaturation and placed in a humidified chamber at 37°C overnight to allow hybridisation to occur. After post-hybridization washes, carried out according to manifacturer's protocols, tissue sections were counterstained with DAPI I (Vysis-Abbott) and examined with a fluorescence microscope (Olympus, Tokyo, Japan). An average of 40 non-overlapping nuclei with intact morphology were analysed using H&E-stained sections as hysto-topographic reference. Copy number of 7q31 specific signals was estimated for each tumor sample and the median number of 7q31 band signals (7q31 number/nuclei) was calculated for each patient. Particular attention was focused on those nuclei exhibiting high number of chromosome 7 centromeres. If the copy number of 7q31 region was not increased similarly to chromosome 7 centromere, polysomy could be excluded; conversely, increased 7q31 signals paralleling the increased centromere signals was considered as a signature of polysomy (Fig. 1).
Figure 1.
EGFR/CEP7 co-amplification or Polisomy of chromosome 7?. In case of polysomy, the copy number of 7q31 region increases similarly to centromere signals. EGFR high copy number in the presence of an eusomic status of 7q31 region is indicative of gene amplification.
Results
According to UCCC scoring system, 13 out of 20 patients (65%) showed high level polysomy. Seven patients (35%) were classified as amplified cases, because of the presence of EGFR/CEP7 ratio >2 (cases n° 4 and 20), tight gene clusters and larger and brighter EGFR signals in > 10% of cells (n° 5 and 19) and of >15 EGFR signals in ≥ 10% of cells (n° 2, 7 and 13).
Using 7q31 band FISH evaluation, almost the same mean number of 7q31 band and chromosome 7 centromeres was observed in 10 cases (n° 3, 5, 6, 9, 14, 15, 16, 17, 18, 19), confirming a polysomic status. In cases n° 5 and 19, however, a cell population with amplification was co-present. In 4 cases (cases n° 4, 7, 10, 13) 7q31 mean number was lower than centromere mean number as expected in case of gene amplification. Patients n° 2 and 11 who had very similar value for EGFR gene and 7q31 band, even higher than centromere number, were considered polysomic. Patients n° 1, 8, 12, because of their relatively low EGFR copy number, were supposed to be polysomic even if 7q31 mean number was lower than centromere mean number. In these cases the presence of complex chromosomal rearrangements, resulting in chromosome 7 long arm losses and short arm gains, was more conceivable.
The results obtained applying UCCC criteria alone and UCCC criteria together with 7q31 FISH control were in agreement in 18/20 cases (90%). Case n° 2, classified as amplified having 10% of cells with >15 EGFR copies, had to be defined polysomic because of the presence of about the same number of 7q31 band and EGFR gene. In case n° 10, according to UCCC protocol, the presence of nuclei with EGFR gene high number (>15) confined just in 2,7% of cells suggested chromosome 7 polysomy rather than amplification. The eusomic status of 7q31 region (7q31/nuclei ratio = 2.2), much lower than centromere mean number, might be the consequence of chromosome 7 long arm deletion, resulting in 7q31 loss.
However, the presence of EGFR increased copy number with a percentage of cells harbouring >15 EGFR signals (even <10% of neoplastic population), together with a high number of chromosome 7 centromeres and a very low number of 7q31 region, suggested a gene amplification rather than chromosome 7 polysomy associated with deletion 7q31 (Fig. 2).
Figure 2.

FISH analysis of 7q31 region. Case n° 10: a) FISH in bronchial biopsy section showing gain of both EGFR gene and chromosome 7 centromere; b) the patient is classified as amplified on the basis of eusomic status of 7q31 region. Case n° 2: c) FISH in pleural effusion sample with EGFR and centromere balanced aneusomy; d) increased copy number of 7q31 region is a signature of polysomy.
Results obtained with UCCC criteria alone and UCCC criteria with 7q31 FISH control are compared in Table 1.
Table 1.
FISH results according to UCCC criteria and 7q31 control
| Case N° |
EGFR/CEP7 Ratio | EGFR/NUCLEI Ratio (Range) | CEP7/NUCLEI Ratio(Range) | % of cells with >4 EGFR copies | NOTES | RESULTS (UCCC criteria) | 7q31/NUCLEI Ratio (Range) | CEP7/NUCLEI Ratio (Range) | RESULTS (7q31 control) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.13 | 5.35 (3-8) | 4.7 (2-8) | 85% | P | 2.02 (1-4) | 4 (2-8) | P | |
| 2 | 1.50 | 8.4 (4-17) | 5.55 (2-10) | 100% | 10% of cells with >15 EGFR copies | A | 8.58 (4-14) | 7.2 (2-13) | P |
| 3 | 1.09 | 4.48 (3-9) | 4.1 (2-9) | 89% | P | 3.97 (2-5) | 4.8 (2-7) | P | |
| 4 | 2.80 | 12.42 (5-23) | 4 (2-7) | 100% | EGFR/CEP7 Ratio >2 | A | 2(2) | 5.3 (4-8) | A |
| 5 | 1.50 | 7.4 (3-16) | 4.9 (2-8) | 100% | 22,5% of cells with tight gene clusters | A | 4.48 (2-9) | 4 (2-7) | A |
| 6 | 1.45 | 6.2 (3-12) | 4.2 (2-8) | 84% | P | 4.44 (3-7) | 4.72 (2-7) | P | |
| 7 | 1.0 | 15.2 (10-40) | 14.5 (8-35) | 100% | 80% of cells with >15 EGFR copies | A | 1.70 | 18 (12-45) | A |
| 8 | 1.16 | 5.58 (3-10) | 4.78 (3-9) | 97% | P | 2.37 (1-4) | 5.2 (4-10) | P | |
| 9 | 1.05 | 4.92 (2-11) | 4.64 (2-9) | 82% | P | 5.07 (2-9) | 6.7 (2-11) | P | |
| 10 | 1.69 | 8.52 (4-22) | 5.04 (2-9) | 97% | 2.4% of cells with >15 EGFR copies | P | 2.2 (1-4) | 5 (2-12) | A |
| 11 | 1.24 | 4.89 (2-10) | 3.93 (2-8) | 75% | P | 6.07 (3-12) | 6 (2-10) | P | |
| 12 | 1.32 | 7.96 (3-15) | 6.01 (3-9) | 97% | 2,7% of cells with >15 EGFR copies | P | 3.2 (1-6) | 6 (3-10) | P |
| 13 | 1.25 | 10.5 (4-26) | 8.36 (4-23) | 100% | 16% of cells with >15 EGFR copies | A | 2.6 (2-5) | 7.7 (4-18) | A |
| 14 | 1.06 | 5.95 (2-13) | 5.6 (2-12) | 100% | P | 4.1 (2-5) | 6.3 (2-11) | P | |
| 15 | 1.18 | 4.39 (2-13) | 3.72 (1-13) | 72% | P | 3.36(2-5) | 3.9 (2-10) | P | |
| 16 | 1.22 | 4.64 (2-9) | 3.78 (2-6) | 71% | P | 2.35 (1-3) | 4.2 (2-7) | P | |
| 17 | 1.70 | 3.9 (1-8) | 2.2 (1-5) | 55% | P | 2.24 (1-4) | 3 (2-6) | P | |
| 18 | 1.10 | 4.9 (2-8) | 4.4 (2-7) | 90% | P | 6.02 (4-9) | 7.4 (2-10) | P | |
| 19 | 1.40 | 6.4 (3-23) | 4.7 (1-10) | 93% | Tight gene clusters in 11,5% of cells | A | 4.15 (2-7) | 5.6 (2-10) | A |
| 20 | 2.90 | 8.8 (2-13) | 3.1 (2-6) | 88% | EGFR/CEP7 Ratio >2 | A | 1.6 (1-3) | 2.1 (2-5) | A |
A = Amplification; P = Polysomy
Discussion
The development of small molecule inhibitors of EGFR resulted in new therapeutics options for patients with advanced NSCLC. Clinical trials provide sufficient evidence that identification of molecular markers for predicting the effects of TKIs may become an integral part of the future standard of care for patients with NSCLC [22-27]. As a consequence, standardized assay procedures must be developed.
Although the significance is still controversial [28-30], EGFR copy number may be predictive of response to EGFR TKIs therapy and, presently, FISH is the gold standard method to detect EGFR gene numerical status. Normally, the gene specific probe is used coupled with a centromeric probe as control to assist in distinguishing gene amplification from chromosomal polysomy. Increased gene copy number, in association with an increase in the number of the chromosome the gene maps on, is interpreted as polysomy. However, FISH analysis in NSCLC is complicated by multiple factors. Lung cancers are frequently heterogeneous with significant variability of EGFR copy number within the same tumor. Moreover, in NSCLC high EGFR gene copy number results more often from chromosome 7 polysomy rather than gene amplification. This depends on the high level of chromosomal instability, as demonstrated by the extremely complex karyotypes. The modal chromosome numbers usually cluster in the near-triploid to near-tetraploid range and the most frequent numerical alteration is polysomy 7 or 7p, with the number of 7/7p exceeding the number of copies expected based on the ploidy of the tumor. Even among the structural rearrangements, the most frequently affected band is 7p11-12, where EGFR gene maps [31-33]. Furthermore, EGFR gene is located at the band 7p12 very close to centromere and EGFR amplicon may include centromeric sequences too, originating a coamplification-type gene amplification, with a complex pattern of signals. Finally, polysomy may coexist with gene amplification, giving rise to a high number of both centromere and EGFR signals.
A useful FISH scoring system for EGFR FISH evaluation in NSCLC patients has been introduced by the University of Colorado Cancer Center (UCCC) [21]. This system stratifies results into six groups (disomy, low trisomy, high trisomy, low polysomy, high polysomy and gene amplification) according to the number of copies of the EGFR gene, chromosome 7 centromere and their frequency in tumor cells of the sample. High polysomy and gene amplification are considered FISH positive.
The guidelines suggested by the Molecular Assays in NSCLC Working Group recommend a two color FISH with centromeric probe as control and encourage the scoring system described by the Colorado group to be used for FISH assay interpretation. Nevertheless authors invite other investigators to propose algorithms for EGFR FISH evaluation [20].
In our study, we introduced a further FISH control, analysing chromosome 7 distal region (7q31) in a series of 20 FISH positive patients with advanced NSCLC and a complex pattern of signals. Fair concordance was seen in 18/20 (90%) cases. However in 2 cases (10%) we found discrepancy between FISH results obtained with UCCC scoring system and 7q31 FISH control. According to UCCC criteria, patient n° 2, because of the presence of 10% of cells with >15 EGFR gene copies, was considered amplified. On the contrary, FISH evaluation of 7q31 region revealed almost the same number of both 7q31 and centromere signals (8,4 vs. 8,58), showing that the EGFR gene increased copy number is consequent to chromosome 7 polysomy. Patient n° 10, having <10% of cells with >15 EGFR gene copies, was classified as polysomic. However, the eusomic status of 7q31 band suggested the presence of EGFR gene amplification. Our results show that in controversial cases, characterized by multiple EGFR and centromere specific signals, 7q31 FISH detection increases accuracy in discriminate EGFR amplification from chromosome 7 polysomy.
From the molecular point of view, amplification and polysomy are different genetic events. Amplification is an alteration by which a cell acquires multiple copies of a DNA sequence, the size ranging from a few hundred kilobases to megabases and is one of the mechanisms by which protooncogenes may be activated in tumor cells. In that case chromosomes may display homogeneously staining regions or small acentric circular autonomously replicating double minutes. Polisomy, reflecting genetic instability, may arise from mitotic non-disjunction followed by secondary endoreduplication events, and deregulate global gene transcription in cancer cells.
In contrast to breast cancer, where HER2 amplification, but not chromosome 17 polysomy seems to confer sensitivity to monoclonal antibody trastuzumab [34,35], it has been reported that patients with NSCLC can derive a clinical benefit from TKIs therapy when EGFR increased copy number (ICN) is found, without distinction between amplification and/or high level of chromosome 7p polisomy. However, recent in vitro and clinical studies, focused on the relationship between mutations and ICN of EGFR gene, have shown that mutations are preferentially associated with amplification of the gene. The observation that four of six human NSCLC cell lines harbouring EGFR mutations were positive for EGFR amplification, whereas none of the ten cell lines negative for EGFR mutations manifested EGFR amplification, supported a close association between these two genetic alterations [36]. As far as clinical studies, Nomura et al (2007) observed a close relationship between three polymorphisms of EGFR gene, activating mutations, and selective amplification of the mutant allele, suggesting that just mutation promotes and drives mechanisms of amplification [37]. Similar evidence was reported in glioblastomas, where the variant form of EGFR gene frequently demonstrates allele-specific amplification [38,39]. Recent studies have reported that EGFR mutations occur early during multistage pathogenesis of NSCLC and precede amplification, but not necessarily high polysomy [37,40,41]. In addition, Morinaga et al (2008) observed a specific association between mutations and amplification, underlying that EGFR amplification, but not high polysomy, correlates with mutations [42].
As a consequence, a higher sensitivity and specificity of the EGFR FISH assay can turn out in an improved identification of those subsets of patients who can benefit more from TKI therapy.
We are currently investigating a wider cohort to characterize furtherly the relationship among ICN, mutations and TKI response. Preliminary data, regarding 16 EGFR mutated patients, indicate that the median overall survival is 16 months in presence of an EGFR amplification vs 9.5 months in cases associated with 7p polysomy (unpublished data).
In conclusion, as it is conceivable that amplification and polysomy differently affects EGFR expression, activation, and sensitivity to TKIs, we underlie the importance of discriminating these two genetic events with accuracy.
Conclusion
Results from this study strengthen the applicability and usefulness of the scoring criteria developed at UCCC for the assessment of EGFR gene patterns, however the further control of a different and distal region of chromosome 7, specifically 7q31, adds more accuracy in interpreting controversial FISH results, when multiple centromeric and gene specific signals are present. Distinction between EGFR gene amplification and high chromosome 7 polysomy is required, till the role of increased EGFR copy number in NSCLC patients in contributing to lung cancer oncogenesis and susceptibility to TKIs will be definitely established.
Abbreviations
(FISH): Fluorescence in Situ Hybridization; (NSCLC): Non Small Cell Lung Carcinoma; (EGFR): Epidermal Growth Factor Receptor; (TKIs): Tyrosine Kinase Inhibitors.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LC: FISH analysis, conception, and design of the study; MC and PF: technical execution; TV: critical revision of the data and the manuscript; MR: revision of the manuscript.
All authors read and approved the final manuscript.
Contributor Information
Laura Casorzo, Email: lcasorzo@mauriziano.it.
Mara Corigliano, Email: corimar@tiscali.it.
Paolo Ferrero, Email: ferreropaolo69@tiscali.it.
Tiziana Venesio, Email: tiziana.venesio@ircc.it.
Mauro Risio, Email: mrisio@mauriziano.it.
Acknowledgements
This work was supported by the Piedmont Region "Ricerca finalizzata 2007".
References
- Barlési F, Tchouhadjian C, Doddoli C, Villani P, Greillier L, Kleisbauer JP, Thomas P, Astoul P. Gefitinib (ZD Iressa) in non-small-cell lung cancer: a review of clinical trials from a daily practice perspective. Fundam Clin Pharmacol. 1839;19:385–93. doi: 10.1111/j.1472-8206.2005.00323.x. [DOI] [PubMed] [Google Scholar]
- Giaccone G. Epidermal growth factor receptor inhibitors in the treatment of non-small-cell lung cancer. J Clin Oncol. 2005;23:3235–42. doi: 10.1200/JCO.2005.08.409. [DOI] [PubMed] [Google Scholar]
- Giaccone G. HER1/EGFR-targeted agents: predicting the future for patients with unpredictable outcomes to therapy. Ann Oncol. 2005;16:538–48. doi: 10.1093/annonc/mdi129. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–68. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–41. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Bell DW, Lynch TJ, Haserlat SM, Harris PL, Okimoto RA, Brannigan BW, Sgroi DC, Muir B, Riemenschneider MJ, Iacona RB, Krebs AD, Johnson DH, Giaccone G, Herbst RS, Manegold C, Fukuoka M, Kris MG, Baselga J, Ochs JS, Haber DA. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–92. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn PA Jr, Varella-Garcia M. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, Bunn PA Jr, Franklin WA, Crowley J, Gandara DR. Southwest Oncology Group. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- Bell DW, Lynch TJ, Haserlat SM, Harris PL, Okimoto RA, Brannigan BW, Sgroi DC, Muir B, Riemenschneider MJ, Iacona RB, Krebs AD, Johnson DH, Giaccone G, Herbst RS, Manegold C, Fukuoka M, Kris MG, Baselga J, Ochs JS, Haber DA. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–92. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Ligorio C, Jänne PA, Toschi L, Rossi E, Trisolini R, Paioli D, Holmes AJ, Magrini E, Finocchiaro G, Bartolini S, Cancellieri A, Ciardiello F, Patelli M, Crino L, Varella-Garcia M. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-Akt-positive or never smoker patients with advanced non-small-cell lung cancer: the ONCOBELL trial. J Clin Oncol. 2007;25:2248–55. doi: 10.1200/JCO.2006.09.4300. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Ligorio C, Toschi L, Rossi E, Trisolini R, Paioli D, Magrini E, Finocchiaro G, Bartolini S, Cancellieri A, Hirsch FR, Crino L, Varella-Garcia M. EGFR and HER2 gene copy number and response to first-line chemotherapy in patients with advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2007;2:423–9. doi: 10.1097/01.JTO.0000268676.79872.9b. [DOI] [PubMed] [Google Scholar]
- Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire JA, Kamel-Reid S, Seymour L, Shepherd FA, Tsao MS. National Cancer Institute of Canada Clinical Trials Group Study BR.21. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Shibata T, Sakiyama T, Yoshida T, Tamura T. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–37. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, Cappuzzo F, McCoy J, Bemis L, Xavier AC, Dziadziuszko R, Gumerlock P, Chansky K, West H, Gazdar AF, Crino L, Gandara DR, Franklin WA, Bunn PA Jr. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–60. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–94. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridisation assay. Diagn Pathol. 2006;1:19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch FR, Witta S. Biomarkers for prediction of sensitivity to EGFR inhibitors in non-small cell lung cancer. Curr Opin Oncol. 2005;17:118–22. doi: 10.1097/01.cco.0000155059.39733.9d. [DOI] [PubMed] [Google Scholar]
- Metro G, Finocchiaro G, Cappuzzo F. Anti-cancer therapy with EGFR inhibitors: factors of prognostic and predictive significance. Ann Oncol. 2006;17(Suppl 2):ii42–45. doi: 10.1093/annonc/mdj920. [DOI] [PubMed] [Google Scholar]
- Toschi L, Cappuzzo F. Understanding the new genetics of responsiveness to epidermal growth factor receptor tyrosine kinase inhibitors. Oncologist. 2007;12:211–20. doi: 10.1634/theoncologist.12-2-211. [DOI] [PubMed] [Google Scholar]
- Hann CL, Brahmer JR. "Who should receive epidermal growth factor receptor inhibitors for non-small cell lung cancer and when?". Curr Treat Options Oncol. 2007;8:28–37. doi: 10.1007/s11864-007-0024-2. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21(Suppl 2):S16–22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- Dacic S. EGFR assays in lung cancer. Adv Anat Pathol. 2008;15:241–7. doi: 10.1097/PAP.0b013e31817bf5a9. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Jänne PA. Selecting patients for epidermal growth factor receptor inhibitor treatment: A FISH story or a tale of mutations? Clin Oncol. 2005;23:6813–6. doi: 10.1200/JCO.2005.97.008. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Endo K, Okuda K, Kawano O, Kitahara N, Tanaka H, Matsumura A, Iuchi K, Takada M, Kawahara M, Kawaguchi T, Yukiue H, Yokoyama T, Yano M, Fujii Y. Epidermal growth factor receptor gene amplification and gefitinib sensitivity in patients with recurrent lung cancer. J Cancer Res Clin Oncol. 2008;134:569–77. doi: 10.1007/s00432-007-0320-z. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Bebb G, Barclay L, Sutcliffe M, Fee J, Salski C, O'Connor R, Ho C, Murray N, Melosky B, English J, Vielkind J, Horsman D, Laskin JJ, Marra MA. Correlations of EGFR mutations and increases in EGFR and HER2 copy number to gefitinib response in a retrospective analysis of lung cancer patients. BMC Cancer. 2007;7:128. doi: 10.1186/1471-2407-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JR, Siegfried JM. Chromosome abnormalities in human non-small cell lung cancer. Cancer Res. 1992;52(Suppl 9):2702s–2706s. [PubMed] [Google Scholar]
- Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877–83. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- Feder M, Siegfried JM, Balshem A, Litwin S, Keller SM, Liu Z, Testa JR. Clinical relevance of chromosome abnormalities in non-small cell lung cancer. Cancer Genet Cytogenet. 1998;102:25–31. doi: 10.1016/S0165-4608(97)00274-4. [DOI] [PubMed] [Google Scholar]
- Bempt I Vanden, Vanhentenrijk V, Drijkoningen M, Wlodarska I, Vandenberghe P, De Wolf-Peeters C. Real-time reverse transcription-PCR and fluorescence in-situ hybridization are complementary to understand the mechanisms involved in HER-2/neu overexpression in human breast carcinomas. Histopathology. 2005;46:431–41. doi: 10.1111/j.1365-2559.2005.02112.x. [DOI] [PubMed] [Google Scholar]
- Bempt I Vanden, Van Loo P, Drijkoningen M, Neven P, Smeets A, Christiaens MR, Paridaens R, De Wolf-Peeters C. Polysomy 17 in Breast Cancer: Clinicopathologic Significance and Impact on HER-2 Testing. J Clin Oncol. 2008;26:4869–74. doi: 10.1200/JCO.2007.13.4296. [DOI] [PubMed] [Google Scholar]
- Okabe T, Okamoto I, Tamura K, Terashima M, Yoshida T, Satoh T, Takada M, Fukuoka M, Nakagawa K. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 2007;67:2046–53. doi: 10.1158/0008-5472.CAN-06-3339. [DOI] [PubMed] [Google Scholar]
- Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, Siegelman M, Feng Z, Kato H, Marchetti A, Shay JW, Spitz MR, Wistuba II, Minna JD, Gazdar AF. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–7. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Takahashi T, Mitsudomi T. Epidermal growth factor receptor gene amplification is acquired in association with tumor progression of EGFR-mutated lung cancer. Cancer Res. 2008;68:2106–11. doi: 10.1158/0008-5472.CAN-07-5211. [DOI] [PubMed] [Google Scholar]
- Soh J, Toyooka S, Ichihara S, Asano H, Kobayashi N, Suehisa H, Otani H, Yamamoto H, Ichimura K, Kiura K, Gazdar AF, Date H. Sequential molecular changes during multistage pathogenesis of small peripheral adenocarcinomas of the lung. J Thorac Oncol. 2008;3:340–7. doi: 10.1097/JTO.0b013e318168d20a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga R, Okamoto I, Fujita Y, Arao T, Sekijima M, Nishio K, Ito H, Fukuoka M, Kadota J, Nakagawa K. Association of epidermal growth factor receptor (EGFR) gene mutations with EGFR amplification in advanced non-small cell lung cancer. Cancer Sci. 2008;99:2455–60. doi: 10.1111/j.1349-7006.2008.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]