Abstract
Exposure to environmental chemicals can have negative consequences for wildlife and even cause localized population extinctions. Resistance to chemical stress, however, can evolve and the mechanisms include desensitized target sites, reduced chemical uptake and increased metabolic detoxification and sequestration. Chemical resistance in wildlife populations can also arise independently of exposure and may be spread by gene flow between populations. Inbreeding—matings between closely related individuals—can have negative fitness consequences for natural populations, and there is evidence of inbreeding depression in many wildlife populations. In some cases, reduced fitness in inbred populations has been shown to be exacerbated under chemical stress. In chemical testing, both inbred and outbred laboratory animals are used and for human safety assessments, iso-genic strains (virtual clones) of mice and rats are often employed that reduce response variation, the number of animals used and associated costs. In contrast, for environmental risk assessment, strains of animals are often used that have been selectively bred to maintain heterozygosity, with the assumption that they are better able to predict adverse effects in wild, genetically variable, animals. This may not necessarily be the case however, as one outbred strain may not be representative of another or of a wild population. In this paper, we critically discuss relationships between genetic variation, inbreeding and chemical effects with the intention of seeking to support more effective chemical testing for the protection of wildlife.
Keywords: chemicals, inbreeding, resistance, populations, wildlife
1. Introduction
Consistent with the fundamental population genetics theory (Falconer 1989), an increasing number of studies find that wildlife populations with low genetic variation appear less able to adapt to changes in environmental conditions, such as physical climate change, biological threats, including disease outbreaks (O'Brien & Evermann 1988; Lande & Shannon 1996; Frankham 2003), transient or variable exposure involving combinations of different physico-chemical stressors (Reed et al. 2002) or novel chemical exposure (Bijlsma et al. 1999; Kovatch et al. 2000; Van Straalen & Timmermans 2002; DeSalle & Amato 2004; Kristensen et al. 2003). Furthermore, species threatened with extinction tend to have lower levels of genetic variation than related non-threatened species (Spielman et al. 2004). Environmental change is a source of strong selection on wildlife, and the exposure of organisms to novel, man-made chemicals is a relatively new pressure. Wildlife may be exposed to multiple chemicals with the potential to cause acute as well as long-term chronic toxicity via various physiological routes (reviewed in Escher & Hermens 2002), some of which may induce characteristic phenotypic (biomarker) responses. At the molecular level, each response involves the modulation of gene expression, and this includes interactions within and between gene loci and the environment. It has been shown that sustained chemical exposure can select on wildlife populations and promote the evolution of resistant genotypes. Studies of wildlife populations exposed to contaminants such as heavy metals (reviewed in Klerks & Weis 1987), persistent organic pollutants Meyer & Di Giulio (2002, 2003) or both (reviewed in Guttman 1994) provide evidence of this. More rapid adaptation has also been observed in wildlife with the evolution of pesticide and antibiotic resistance (reviewed in Roush & McKenzie 1987; Futuyma 1998; Palumbi 2001).
In considering how chemicals potentially affect evolutionary processes, some chemicals (genotoxins) can affect DNA integrity directly, leading to heritable changes. These genotoxic chemicals increase the frequency of DNA damage and thus increase the risk of replication and transcription of altered DNA sequences. The probability of DNA damage being converted into a permanent and/or heritable sequence alteration depends on the type of damage, the repair pathway recruited, the rate of repair and the fidelity and completeness of repair (Preston & Hoffman 2001). In some instances, mutagenic chemicals have been shown to increase genetic variation (albeit at neutral markers) (Chen et al. 2003; Berckmoes et al. 2005; Eeva et al. 2006), but in other cases, to reduce it. This reduction can have negative impacts on wildlife by increasing the frequency of deleterious mutations that are selected against, causing bottlenecks, impairing gene flow (reviewed in Van Straalen & Timmermans 2002) and, less dramatically, lead to cumulative erosion of reproductive fitness (reviewed in Bickham et al. 2000). Less obvious effects of chemicals on the genome are also possible but may be latent and may accumulate over generations. Some so-called endocrine-disrupting chemicals (EDCs), for example, can impose heritable reproductive effects via epigenetic processes affecting the germ line, without altering underlying gene sequences (Anway & Skinner 2006; Nilsson et al. 2008). Chemicals may also affect evolutionary processes indirectly by reducing effective (breeding) population size (Ne), with or without selection on adaptive traits and/or by reinforcing the reproductive isolation of exposed populations (Nacci & Hoffman 2008). Some EDCs have been shown to reduce effective breeding population size in wildlife populations (reviewed in Vos et al. 2000; Goodhead & Tyler 2008) and documented examples include population declines and local extinctions both in the dog whelk (Nucella lapillus) following tri-butyl tin exposure (Bryan et al. 1988) and in raptors owing to eggshell thinning resulting from the bioaccumulation of DDT and its metabolites (Ratcliffe 1967).
Populations are widely regarded as the minimum units for species conservation (UN Millennium Ecosystem Assessment and World Resources Institute 2005) and environmental protection (Barnthouse et al. 2008). It is generally accepted that a minimum Ne is required to maintain a genetically viable population (Frankham 1995a; Lynch & Lande 1998). Populations with low Ne contain low levels of genetic variation, lowering the chances for recombination and maintaining beneficial mutations (other than those under very strong selection) (Pasteur & Raymond 1996; Futuyma 1998). Other problems that can arise because of declining population size include increasing genetic drift where mildly deleterious alleles can reach appreciable frequencies (Lande 1994), and perhaps more significantly, the enhanced likelihood of inbreeding and inbreeding depression (Frankham 1995b; Amos & Balmford 2001; Brook et al. 2002). Ne is a key factor in considerations for conservation and environmental protection.
In this review, we evaluate the importance of genetic variation in wildlife populations for coping with environmental, including chemical, stressors and consider the implications for ecotoxicology. We focus on the importance of inbreeding as a factor affecting individual fitness and ultimately the viability of populations, and we assess potential interactions between inbreeding and chemical toxicity. Assessing the strength of these chemical–ecogenetic interactions is critical to the future refinement of the environmental risk assessment (ERA) process and for environmental realism in predicting the effects of chemicals in nature.
2. The genetic basis for chemical resistance
There are examples in the wild of selective effects of chemicals on populations. Numerous field studies have shown historical metal pollution has imposed selection on exposed wildlife populations, with directional evolution in neutral marker loci along spatial and temporal gradients of exposure (Klerks & Weis 1987; Groenendijk et al. 2002; Klerks 2002; Peles et al. 2003). Increasing levels of metal contamination are often associated with reduced genetic variation at the population level. At the same time, there is also evidence that individuals that are more genetically diverse (greater heterozygosity and lower ‘internal relatedness’) within populations derived from exposure areas are more able to tolerate higher levels of exposure than those from less contaminated areas (Bourret et al. 2008). Tolerance to mercury exposure in mosquitofish (Gambusia affinis) has been related specifically to genetic polymorphism at the glucose isomerase locus, and the distribution of genotypes in natural and artificial contaminated environments were shown to be consistent with this (Heagler et al. 1993; Tatara et al. 1999).
(a). Heritable variation and evolution of chemical resistance
Adaptation to organic pollutants also implies heritable variation in resistance, and this has been demonstrated in fish populations exposed long term to some organic pollutants (e.g. polyaromatic hydrocarbons (PAHs) and polychlorinated bi-phenyls (PCBs); Atlantic killifish, Fundulus heteroclitus; Meyer & Di Giulio 2002; Nacci et al. 1999). However, PAH resistance has been associated with reduced fitness when individuals were challenged with other environmental stressors (Meyer & Di Giulio 2003). Other studies of natural populations resistant to specific toxicants have also shown associations between resistance to one chemical and heightened sensitivities to other stressors, or sometimes a general reduction in performance in the absence of contaminants (Carrière et al. 1994; Shaw 1999; Nacci & Hoffman 2008). A further experimental study on the sheepshead minnow (Cyprinodon variegatus) found heritability of chemical resistance was reduced with exposure to increasing numbers of chemical pollutants, suggesting that resistance to any one chemical may develop more slowly during exposures to complex chemical mixtures, as experienced by most wildlife in the natural environment (Klerks & Moreau 2001).
Exposure to more contemporary and emerging contaminants, including pesticides and antibiotics, has also been shown to lead to the evolution of chemical resistance. The resistance mechanisms in these cases are often highly conserved across phyla (bacteria, fungi, higher plants, insects and vertebrates), despite the fact that these chemicals have been developed to selectively target specific groups of animals, and include target site insensitivity, reduced chemical uptake and increased metabolic detoxification and sequestration (Eckert et al. 1986; Feyereisen 1995). In the case of synthetic chemical pesticides, resistance at the molecular level is conferred by point mutations in the ion-channel component of a GABA receptor subunit (for cyclodiene insecticides) or within the sodium-channel gene (for DDT and pyrethroid insecticides), via mutations in the region coding for the active site of acetylcholinesterase (organophosphorus and carbamate insecticide resistance), by mutations leading to the upregulation of detoxification enzymes such as cytochrome P450 and glutathione-S-transferases (for many classes of insecticides), or via amplification of esterase genes (for organophosphorus and carbamate insecticides) (Feyereisen 1995; Pasteur & Raymond 1996) (table 1).
Table 1.
Physiological mechanisms and genetic basis of antibiotic and pesticide resistance.
| compound/chemical class | resistant species | resistance mechanism and genetic basis | reference |
|---|---|---|---|
| antibiotics | |||
| spectinomycin (antibiotic) | Escherichia coli | inhibition of peptidyl translocation via a base transition C/G to T/A at position 1192 of a 16S RNA gene | Sigmund et al. (1984) |
| streptogramin, lincosamide, macrolide (type B antibiotics) | E. coli | inhibition of peptidyl translocation via a base transversion A/T to T/A at position 2058 of a 23S RNA gene | Sigmund et al. (1984) |
| fungicides | |||
| triadimenol (DMI fungicide) | grape powdery mildew fungus (Uncinula necator) | cytochrome P450-mediated resistance to demethylation and sterol biosynthesis inhibition linked to a phenylamine to tyrosine substitution mutation at codon 136 | Delye et al. (1997) |
| fluconazole and itraconazole (azole fungicides) | Candida albicans | reduced azole affinity of cytochrome P450 14-α-demethylase ERG11 (CYP51) gene via mutations at codon Y132H and 266–287. | Marichal et al. (1999) |
| cyclodiene insecticides | |||
| dieldrin (cyclodiene) | fruitfly (D. melanogaster) | insensitivity of the GABAA receptor-chloride ion channel via an unknown mutation at the Rdl gene locus | ffrench-Constant et al. (1993) |
| dieldrin (cyclodiene) | house fly (Musca domestica) red flour beetle (Tribolium castaneum) American cockroach (Periplaneta americana) |
insensitivity of the GABAA receptor-chloride ion channel via an alanine to serine substitution mutation at the Rdl (resistant to dieldrin) gene locus. | Thompson et al. (1993) |
| pyrethroid insecticides | |||
| pyrethroid and DDT (organophosphate) | house fly (M. domestica) | nerve insensitivity via two point mutations in sodium-channel domain II: leucine to phenylamine at IIS6; methionine to threonine at IIS4-S5 | Williamson et al. (1996) |
| α-cypermethrin (pyrethroid) | olive fruitfly (Bactrocera oleae) | cytochrome P450 mono-oxygenase upregulation via an iAChE G4884 point mutation | Margaritopoulos et al. (2008) |
| organophosphate insecticides | |||
| diazinon (organophosphate) | house fly (M. domestica) | conversion of carboxyl esterase to OP hydrolase via a LcαE7 point mutation | Claudianos et al. (1999) |
| diazinon (organophosphate) | blow fly (Lucilia cuprina) | conversion of carboxyl esterase to OP hydrolase via MdαE7 point mutation | Claudianos et al. (1999) |
| insecticide mixtures (cross resistance) | |||
| organophosphates and carbamates (various) | lower insects e.g. Myzus sp., Anopheles sp., Aphis sp. | AChE receptor insensitivity in carbamates is greater than OPs owing to Pattern I resistance in AChE-1 gene—several possible mutations at two sites (119 and 331) | Russell et al. (2004) |
| organophosphates and carbamates (various) | higher diptera e.g. Musca sp., Bactrocera sp., Drosophila sp. | AChE receptor insensitivity in carbamates is equal to OPs owing to Pattern II resistance in AChE-2 gene—11 possible mutations at six sites | Russell et al. (2004) |
| permethrin (pyrethroid) carbaryl (carbamate) malathion (organophosphate) | Drosophila simulans | cross resistance owing to AChE receptor insensitivity. Resistant allele at the AChE locus, position 299 encoding a protein with at least one amino acid leucine is less than methionine substitution near the active site of AChE enzyme | Cochrane et al. (1998) |
Resistance to some chemicals evolves via mutations in a single gene (monogenic resistance). For example, cyclodiene resistance in insects is conferred by a single base pair substitution (ffrench-Constant et al. 2000). The finding that cyclodiene resistance in pest insects accounts for over 60 per cent of reported cases of insecticide resistance (Georghiou 1986) indicates just how important monogenic resistance can be. Single-point substitutions also confer some herbicide and fungicide resistance, and again the resistance-associated mutations are often highly conserved across species (Gressel 1986). Although the evolution of resistance to synthetic pesticides generally begins with single-point mutations, some of which may confer cross resistance to multiple classes of compounds (Cochrane et al. 1998), high-level resistance generally requires combinations of such mutations (Mutero et al. 1994). Indeed, pesticide resistance mechanisms conferred by elevated carboxylesterases (in aphids (Devonshire & Field 1991) and mosquitoes (Mouches et al. 1986)), and target site-mediated resistance, including insensitive acetylcholinesterases (Russell et al. 2004), are associated with a series of distinct mutations, each one increasing resistance. Resistance to fungicidal ergosterol biosysnthesis inhibitors and polyene antibiotics are also both polygenic (Eckert et al. 1986). In addition to physiological adaptation, altered morphology may also contribute to increased pesticide resistance. For example, DDT resistance in mosquitoes is facilitated by the evolution of a protective footpad cuticle, reducing DDT uptake (Guillaumot 2006). Recent work in evolutionary developmental biology suggests that adaptive mutations affecting morphology may occur in protein-coding regions as well as in cis-regulatory regions of genes (Hoekstra & Coyne 2007), which helps explain the apparent multi-trait DDT adaptation seen in mosquitoes. These examples highlight the importance of genetic variation at multiple loci in the evolution of chemical resistance.
(b). Adaptation to chemicals in wild populations
The work described above has been derived from laboratory or controlled field studies. Fewer investigations document direct evidence of adaptation to chemical pesticides in wild populations, and in those that do, phenotypic evolution is generally all that is assessed, with the specific genes and molecular mechanisms underlying these adaptive changes largely unknown (Merila & Crnokrak 2001). The evolution of resistance to Bacillus thuringiensis (Bt) toxins in insects is an exception to this. Bt toxins are used widely in insect pest management and kill insects by creating pores in mid-gut membranes. In the diamondback moth (Plutella xylostella), resistance to Bt toxins Cry1Aa, Cry1Ab, Cry1Ac and Cry1F has been shown to occur through a single autosomal recessive gene (Tabashnik et al. 1997).
It appears to be the case, however, that emergence of pesticide resistance from de novo mutations in susceptible, wild-type populations is rare (Pasteur & Raymond 1996; Tabashnik et al. 2003). Resistance is more likely to develop owing to emigration (gene flow) from pre-existing resistant populations (Pasteur & Raymond 1996; Futuyma 1998). Between-population gene flow and the resulting recombination, which increases genetic variation, are considered to be primary mechanisms for the development and spread of chemical resistance in wild populations (Leslie & Watt 1986). Recombination is also more frequent, thus, more likely, than de novo point mutation: King & Jukes (1969) estimated the substitution (mutation) rate at between 10−8 and 10−9 per codon per generation, whereas the crossing-over frequency (recombination) between two existing mutations in acetylcholine esterase has been shown to reach 10−5 (in Drosophila; Nagoshi & Gelbart 1987).
The evolution of pesticide resistance requires a number of conditions to be satisfied and depends on the initial frequencies of the resistance allele(s), the dominance of the allele(s), the extent and duration of chemical or toxin exposure and the intensity of the chemical effect (Nacci & Hoffman 2008). This is likely also to be the case for other chemical toxicants. Furthermore, not all traits associated with selected resistant phenotypes are beneficial, and adaptive sweeps can drag deleterious alleles linked to the locus under positive selection (hitch-hiking), resulting in fitness costs unassociated with resistance. Nevertheless, significant life-history costs associated with pesticide resistance (Carrière et al. 1994) and chemical resistance in general (Van Straalen & Timmermans 2002) appear to be relatively rare. When they do occur, they may be due to negative gene interaction (pleiotrophy and epistasis) and not simply the diversion of energy budgets, as is often suggested (Taylor & Feyereisen 1996).
In addition to genotypes that confer susceptibility or resistance to chemicals with specific modes of action, there are a number of cases where particular sets of genes produce phenotypes resistant to general toxicity (so-called genes of ‘major effect’), and these are also conserved across taxa (Hoffman & Parsons 2002). In many cases, genetic variation (polymorphism) at these loci confers significant ecological benefits in coping with a wide range of stressors and/or stress gradients. Initial chemical stress responses often involve metabolic regulation through various enzyme variants (allozymes) (Guttman 1994; Hoffman & Parsons 2002) and allozymes feature in a number of pathways initiated in general response to environmental stress. They include: (i) alcohol dehydrogenase (Adh) associated with the metabolism and detoxification of environmental ethanol during the growth and germination of plants under variable climatic conditions (Brown et al. 1976), and also with the synthesis of heat-shock proteins conferring high-temperature resistance in insects (Alahiotis 1982); (ii) heat-shock protein genes (e.g. hsp70), which are essential for cellular survival in prokaryotes and eukaryotes, performing a multitude of house-keeping functions via their ability to interact with a wide range of proteins and peptides, a property that is shared by major histocompatibility complex (MHC) molecules (Srivastava 2002); (iii) glutamate pyruvate transaminase (Gpt) and leucine amino-peptidase (Lap94s) involved in the regulation of hyper-osmotic stress in the estuarine copepod Tigriopus californicus (Burton & Feldman 1983) and the blue mussel Mytilus edulis (Beaumont et al. 1988); (iv) glucose isomerase (Gpi) associated with resistance in mosquitofish and fathead minnows (Pimephales promelas) to multiple environmental contaminants, including metals (Heagler et al. 1993; Schlueter et al. 1997) and fluoranthene (Schlueter et al. 2000); (v) cytochrome P450 genes, which encode a super-family of enzymes in prokaryotes and eukaryotes involved in the oxidative, peroxidative and reductive metabolism of numerous endogenous compounds and a wide range of environmental chemicals (Nebert & Nelson 2006). Accordingly, the physiological genotype or ‘physiotype’ of an organism will determine its ability to regulate environmental, including chemical, stress (Depledge 1990; Guttman 1994).
3. Inbreeding, and evidence of inbreeding depression in wildlife and its significance
Inbreeding can create problems for natural populations because it frequently leads to inbreeding depression. Moreover, traits closely related to fitness appear especially susceptible to inbreeding depression (e.g. Wright et al. 2008), and as a result, inbreeding can reduce effective population size, further increasing the likelihood of inbreeding and genetic drift (Lande 1976; Falconer 1989). Inbreeding depression occurs either because more homozygous individuals have increased expression of deleterious, recessive alleles (partial dominance hypothesis) and/or from the loss of heterozygote superiority (overdominance hypothesis) (Charlesworth & Charlesworth 1987). An inbreeding coefficient of F = 0.33 seems to represent a threshold marking the onset of significant inbreeding depression in populations in the laboratory (Frankham 1995b); however, inbreeding in the wild may progress more slowly, and hence selection may be able to purge the most deleterious mutations (Lande 1995; Brook et al. 2002; Keller & Waller 2002). In a changing environment, however, purging will be more limited (Bijlsma et al. 1999; Miller & Hedrick 2001) and in any event will not prevent erosion of heterozygosity. Furthermore, inbreeding depression may occur owing to large numbers of mildly deleterious alleles (Charlesworth & Charlesworth 1987), which may evade selection (Lande 1994; Lynch et al. 1995; see below).
(a). Inbreeding in wildlife populations
Most of our knowledge concerning inbreeding depression in animals has been derived from domestic populations and wildlife maintained in captivity. It has been argued that inbreeding depression is generally not a significant issue for wild-animal populations because inbreeding avoidance occurs via selective mate choice, multiple mating (Hosken & Blanckenhorn 1999), delayed maturation/reproductive suppression and dispersal (reviewed in Greenwood 1980; Pusey & Wolf 1996). However, there is a significant body of evidence to the contrary. Indeed, inbreeding can be pronounced in small wildlife populations that result following bottlenecks caused, for example, by a disease outbreak (O'Brien 1994) or pollution incident (Bickham et al. 2000). Inbreeding may also be promoted by specific behaviours, such as reproductive homing (philopatry), where animals, including some species of fish, amphibians and reptiles, return to their natal spawning grounds to breed (reviewed in Waldman & McKinnon 1993). In some small mammals (e.g. Sorex araneus, Peromyscus leucopus; Stockley et al. 1993), inbreeding occurs because individuals do not disperse. In fact, there is now extensive evidence of inbreeding and inbreeding depression in a wide range of wildlife populations (Wright 1984; Ralls et al. 1988; Frankham 1995a; Crnokrak & Roff 1999; Keller & Waller 2002; Frankham 2003). Crnokrak & Roff (1999), examining inbreeding depression in 137 traits in 35 wildlife species, found significant inbreeding depression, and in most cases, estimates were sufficiently high (F > 0.1–0.33) to be considered to have significant fitness implications (Frankham 1995c; Halverson et al. 2006).
Estimates for the minimum Ne for avoidance of the adverse consequences of genetic drift and inbreeding and for maintenance of a self sustaining, genetically viable population vary between 500 and 5000 (500–1000, Franklin & Frankham 1998; 1000–5000, Lande 1995; Lynch 1996; Lynch & Lande 1998) and depend on a range of demographic and environmental factors (Gilpin & Soulé 1986). Many wildlife populations do not meet this minimum Ne. As an example, many of the world's birds (ca. 1000 of the 9000+ species) have population sizes well below 1000 individuals. Two hundred bird species have census population sizes of less than 100 individuals (Green & Hirons 1991), and Ne may be considerably smaller. Some fish populations can also contain fewer than 200 individuals (e.g. sockeye salmon (Oncorhynchus nerka); Altukhov 1982), and reproductive homing adds further to the risk of inbreeding (FAO/UNEP 1981). It is recognized, however, that most genetic problems in endangered populations have accumulated over tens or hundreds of generations and a low Ne for several generations will not necessarily lead to irreversible genetic damage (Amos & Balmford 2001). Consequently World Conservation Union criteria (IUCN 2008) include demographic population structure, geographical distribution, temporal trends and generation time as well as breeding population numbers for defining endangered (Ne = 250–2500) and critically endangered species (Ne = 50–250).
(b). Reduced genetic variation and health impacts in wildlife populations
Reduced genetic variation that results from inbreeding and small population size has also been found to correlate with a range of defects, many of which are associated with reproductive traits. This has been demonstrated in African lion (Panthera leo krugeri) and Asiatic lion populations (Panthera leo persica), where males from small founder populations have reduced sperm count and motility, lower levels of male sex hormones and a greater proportion of abnormal sperm compared with larger populations having experienced less severe or no such bottlenecks (O'Brien 1994). In African cheetah (Acinonyx jubatus) populations too, low heterozygosity (at MHC loci and loci more widely) is associated with impaired immunocompetence and congenital defects including low sperm counts and viability in males (O'Brien 1994; Roldan & Gomendio 2009). A similar associated decline in genetic diversity and fitness has been shown in the Florida panther, a species on the brink of extinction with a remaining population of only 30 individuals (Roelke et al. 1993). In the case of the Florida panther, inbreeding may be responsible for several deleterious developmental and immunological impairments, including a high prevalence of cryptorchidism (undescended testes, now affecting over 90% of males) a significantly (fourfold) reduced sperm count, malformations in 90 per cent of the sperm and in many cases (20%) sterility, congenital cardiac defects and impaired immune system (Roelke et al. 1993). Similar findings have also been reported for rabbit populations (Gage et al. 2006). Examples of inbreeding depression in wild birds includes the song sparrow (Melospiza melodia) of Mandarte Island, British Columbia, and this has resulted in reduced life-time fecundity in females and reduced offspring survival (Keller 1998). Conversely, a male bias on reduced fitness, especially fertility, is displayed in the house mouse (Mus domesticus) (Meagher et al. 2000) and Drosphila melanogaster (Hughes 1995). Male fitness appears to be especially affected, possibly because male sexual fitness is under stronger selection than females' (there is greater variance in male reproductive success).
Most field or semi-field studies have focused on juvenile fitness and may have overlooked the effects of deleterious alleles expressed in later life (Meagher et al. 2000). It is likely that fitness differences will accumulate throughout an individual's life (Clutton-Brock 1988) and late-acting deleterious alleles are less likely to be removed by natural selection than early-acting mutations. This could lead to increased inbreeding depression in later life, and this is one explanation for why organisms senesce (Charlesworth & Hughes 1996). For example, in red deer, there is inbreeding depression in male breeding success (Slate et al. 2000), but none in neonatal survival or birth weight (Coulson et al. 1998), while in the house mouse there is significant inbreeding depression in adult male survivorship, paternity and male territoriality (Meagher et al. 2000).
Most studies that have shown inbreeding increases the risk of population extinction have been conducted on experimental populations (e.g. Wright et al. 2008), but there is also evidence that inbreeding can contribute significantly to population decline and extinction in the wild (Frankham 2003). This has been documented in mammals (e.g. Isle Royale wolf (Wayne et al. 1991); Florida panther (Roelke et al. 1993)), birds (e.g. heath hen; Simberloff 1988), reptiles (e.g. adder; Madsen et al. 1999), fish (e.g. topminnow; Vriejenhoek 1994) and invertebrates (e.g. colonial spiders; Riechert & Roeloffs 1993), although in each case genetic and non-genetic causes of extinction or local extirpation were not delineated. One of the most comprehensive examples of the impact of inbreeding and the importance of genetic diversity for population viability in the wild comes from studies on the Sonoran topminnow (Poeciliopsis monacha) (Vriejenhoek 1994). Here, a population decline coincided with reduced genetic diversity following a history of repeated bottlenecking and recolonization owing to alternating periods of drought and flooding. Following the translocation of individuals from a more genetically diverse upstream population, heterozygosity (allozyme heterozygosity at 4 loci (Idh-2, Ldh-1, Pgd, Ck-A) and fitness were reinstated and the population recovered. The initial fitness reduction was attributed to inbreeding depression in tolerance to physical extremes, in resistance to parasitism and in reproductive output (Vriejenhoek 1994). More recently, a single migrant wolf credited with introducing new allelic variation into an inbred population was found to coincide with a major population recovery (Vilà et al. 2003).
A major complication for studies of inbreeding in wild populations is that estimating inbreeding coefficients from heterozygosity estimates is problematic. This is demonstrated in a study in sheep of 138 microsatellite markers spread across all their 26 autosomes (Slate et al. 2004). Only a very weak association was found between heterozygosity and the inbreeding coefficient estimated from a known pedigree, and inbreeding depression was not predicted by multilocus heterozygosity (Slate et al. 2004). The low correlation between individual heterozygosity and inbreeding stems from the low variance of inbreeding in natural populations and the high stochasticity of individual genetic markers. Some gains could be achieved by estimating inbreeding coefficient rather than some of its proxies (see appendix A for the correct derivation).
4. Combined effects of inbreeding and chemical toxicity
Given the widespread occurrence of inbreeding, the resultant loss of heterozygosity and subsequent inbreeding depression, inbreeding can be an important determinant of the vulnerability of populations to environmental stressors (Armbruster & Reed 2005). Indeed, several controlled laboratory studies have demonstrated interactive effects of inbreeding and chemical exposure. Miller (1994) showed that fitness was reduced in a laboratory strain of D. melanogaster when exposed to lead contamination after periods of inbreeding. In another study on D. melanogaster, where isolines were exposed to chemicals during eight consecutive generations of full-sib matings (generating a theoretical inbreeding coefficient of F = 0.83) more inbred lines failed (became extinct) under chemical exposure (copper sulphate, 76% failure; methanol, 83% failure) than inbred controls (63%), while extinction rates for outbred controls with and without exposure ranged between only 3.1 and 4.4 per cent (Reed et al. 2002). Similarly, Nowak et al. (2007a) found amplified inbreeding depression in midges (Chironomus riparius) exposed to cadmium. Here there was a highly significant interaction between exposure concentration (0–0.72 mg kg−1 in sediment) and level of inbreeding (F = 0, 0.125 and 0.375) influencing development time and survival of homozygotes. The above laboratory studies generally indicate that environmentally realistic levels of inbreeding (F = 0.1–0.33) and chemical exposure can combine to severely reduce fitness.
Similar interactions between inbreeding and chemical stress are likely to occur in the wild but evidence for this is extremely limited. Correlative evidence for an interaction between inbreeding and chemical exposure in the wild comes from studies on the Florida panther. This species is now significantly inbred in Florida and many reproductive traits are impaired (described above). These animals also contain high levels of a range of EDCs, including mercury and PCBs, known to be able to cause reproductive impairment at the detected levels (Roelke et al. 1992; Facemire et al. 1995).
It is possible that combined effects of inbreeding and environmental chemical contamination could cause declines in some wildlife populations, and indeed may have already done so. However, until recently, this possibility has largely been ignored (Liao & Reed 2009). Interactive effects are arguably most likely for populations exposed to chemicals that cause reproductive impairment, as life-history characters related to reproductive output show most inbreeding depression. One example where this may have occurred is in North Sea seal populations. Here there is compelling evidence that exposure to a range of environmental chemicals including PCBs, DDT, polychlorinated dibenzofurans and polychlorinated dibenzo-para-dioxins led to reproductive failure and immunosuppression (Van Loveren et al. 2000; Vos et al. 2000), and ultimately contributed to mass mortality during a morbillivirus outbreak in the 1980s. The chemical effects were verified in a controlled field experiment in which common (harbour) seals (Phoca vitulina) fed with PCB-contaminated fish showed lower reproductive success than those fed with fish from a less polluted area of the Wadden Sea (Reijnders 1986). Inbreeding levels of harbour seal parents were not reported in that study; however, European populations of this species are apparently somewhat inbred, and inbreeding depression in pup survivorship is well documented (Coltman et al. 1998).
5. Genetic variability, inbreeding and chemical testing
Chemical Safety Assessment and ERA, set to protect humans and wildlife, respectively, require testing with laboratory animals. Testing guidelines have for several decades employed both inbred and outbred laboratory strains (here we define a strain as a stock or line derived from a closed population with distinct genetic characteristics that distinguish them from other groups within a species). In toxicology studies employed in human safety assessment, iso-genic strains of mice and rats—animals derived from at least 20 consecutive generations of full-sib matings—are often used (they are virtual clones) and are effectively purged of deleterious recessive alleles (Kacew 2001). The principle of using domesticated laboratory strains of animals in ecotoxicology studies supporting ERA, however, and their ability to predict and prevent adverse effects in wild animals has been questioned (Schaeffer & Beasley 1989; reviewed by Hill 1994). Thus, for ERA, strains of animals are often used that have been selectively bred to maintain heterozygosity with the intention that they are representative of wild populations. In the final section of this paper, we critically analyse the use of inbred versus outbred animals in ecotoxicology and for ERA.
(a). Use of inbred animals for human safety assessment
Two US Government agency white papers on toxicity testing (US EPA 2003; US FDA 2004) have recognized that the field of toxicology has been hampered by the wide variation in strains of animals used across different laboratories because strains can vary in their sensitivity to chemical effects. As an example of this, the sensitivity of outbred stains of rat to the model carcinogen TCDD has been shown to vary by almost 1000-fold (Kacew & Festing 1996). For human chemical safety assessments, the US National Academy of Science (NAS 2007) has now advocated the use of inbred isogenic strains in preference to outbred strains. This approach is also advocated in animal-based research in the UK (Festing et al. 2002). The compelling case presented is that side effects, such as carcinogenicity of a chemical, can be better evaluated by choosing a sensitive strain. Strains can also be genotyped accurately to both help inform on the intended mode of action of the chemical and authenticate the strains themselves. Phenotypic variation is also minimized, enabling increased statistical power based on fewer test organisms, with obvious financial and ethical benefits.
(b). Inbred or outbred animals for use in environmental risk assessment?
Ecotoxicology is a rather different process for the majority of chemicals under consideration. Here the process is a ‘risk assessment’, a balanced consideration of potential risk, as opposed to a ‘safety assessment’ that implies a higher level of knowledge. For the majority of chemicals entering the environment, little is known about the toxicology or potential adverse effects they may have at the concentrations that are present or predicted. In ecotoxicology, however, inbred strains may also have some merit, as they can have increased susceptibility to chemical stressors. This has been shown in the fruitfly Drosophila buzzatti for exposures to the organophosphorus insecticide dimethoate (Kristensen et al. 2003), in the midge (C. riparius) for cadmium exposure (Nowak et al. 2007a) and in inbred strains of mice for sensitivity to oestrogen (Spearow et al. 1999; Spearow 2004). Conversely, there is the possibility of pre-existing, heritable resistance to certain chemicals in wild populations (Pasteur & Raymond 1996; Futuyma 1998) compared with inbred laboratory strains, as was shown for DDT resistance in a wild-type strain of D. melanogaster (Bijlsma et al. 1999).
In addition to offering increased biological sensitivity, the use of inbred, isogenic strains increases statistical power since variability in phenotypic response to chemical exposure is minimized. However, by the same token, they may fail to take into account the full spectrum of genetic variation and responses in natural populations. Therefore, although inbred strains may well be protective in many cases, they should not be considered wholly representative of wild populations. By definition, outbred strains should be more representative of wild populations. However, details of the origin and breeding history of strains are rarely reported for animals used in ecotoxicology and when they are, they are typically described as ‘outbred’ or ‘genetically diverse’. Furthermore, this assumption is often untested and in cases where it has been, genetic variation has been shown to differ widely between strains and between populations derived from the same strain, as shown in laboratory rats (Kacew 2001) and zebrafish (Guryev et al. 2006; Coe et al. 2009). Genomic variation in zebrafish may be particularly high: there are an estimated 425 000 coding single-nucleotide polymorphisms, four times greater than in outbred strains of rat and higher than in any vertebrate genome sequenced to date (Guryev et al. 2006). Nevertheless, even the most outbred laboratory strains are likely to be significantly less genetically diverse than wild populations, as has been indicated by neutral markers of heterozygosity in trematode worms (Stohler et al. 2004), dipteran insects (Nowak et al. 2007b), amphipod crustaceans (Duan et al. 1997), rodents (Razzoli et al. 2003) and fish (Coe et al. 2009). It is likely that the comparatively lower genetic diversity in these outbred laboratory strains is because their outbreeding is normally limited to introgression with other laboratory strains or substrains and they lack the gene flow required to counterbalance genetic drift, resulting in the loss of alleles over time. Even the generation of new wild-type laboratory strains may be hampered at the outset by founder events or genetic bottlenecks in which the founders carry only a limited proportion of the initial allelic variation in the source populations (Nowak et al. 2007b).
There are also difficulties associated with characterizing outbred strains as highlighted by the US FDA (2004). Genome-wide characterization is not practical or useful because every individual will be different. Instead, a surrogate measure of genetic diversity, such as heterozygosity or allelic richness, for example, at highly variable microsatellite loci can indicate how representative a strain is of wild populations (Nowak et al. 2007b; Coe et al. 2009). Setting acceptance criteria for such measures, based on neutral markers of genetic diversity, however, needs careful consideration, since in some cases their correlation with fitness traits has been shown to be limited (Lynch 1996; Reed & Frankham 2003; Kohn et al. 2006). The targeted assessment of heterozygosity at quantitative trait loci (Falconer et al. 1996) may provide a better, practical solution. However, in the future, genomic measures of genetic diversity being advocated for conservation (Kohn et al. 2006), evolutionary biology (McKay & Stinchcombe 2008), pharmacology (Guryev et al. 2006) and ecotoxicology (Ankley et al. 2006) may also prove to be useful. Further evidence to support the contention that outbred strains are more representative of wild populations could be gained by comparing their respective levels of inbreeding, based on markers of heterozygosity (Goodnight & Queller 1999) or, better still, this in combination with pedigree information (Pemberton 2004). Alternatively, see appendix A.
(c). Critical consideration on genetic variation and standard test guidelines in ecotoxicology
Standard test guidelines for ecotoxicology studies including maximum tolerable limits for mortality in controls are probably not sufficient to prevent potential bias owing to genetic drift, mutational accumulation and inbreeding depression, which may affect other, perhaps more sensitive fitness components. These potential problems stem from limited genetic diversity and population numbers constituting laboratory strains. These problems are not easily overcome and, in practice, the use of robust outbred laboratory strains in ecotoxicology, which are closely representative of wild populations, is an ideal that is hard to achieve for the following reasons:
Some control of genetic variability is required to maintain consistency within strains and therefore between tests and laboratories. Some degree of restricted breeding will be required to achieve this, and this may be justifiable since it occurs in a wide range of species in the wild. However, it is important to avoid repeated inbreeding as this could lead to inbreeding depression, the over-sensitization of laboratory strains to chemical exposure and, ultimately, potentially over conservatism in ERA. Conversely, introgressions involving other outbred strains and wild-types will certainly lead to wider genotypic variation, potentially affecting phenotypic responses within and between treatments and studies. However, basic data quantifying phenotypic variation in apical endpoints concerning growth, development and reproduction in relation to genotypic variation in laboratory strains under control conditions is lacking.
In order to maintain genetic diversity in laboratory strains, sizeable breeding populations in excess of 1000 animals would ideally be required in a self-sustaining group-breeding situation. This may be impractical for some species; however, a more structured breeding programme (e.g. pair breeding) based on smaller numbers of individuals could possibly achieve similar results. Ethical and practical considerations will dictate the final solution, but the alternative solutions for maintaining genetic diversity in strains (suggested above) need to be tested first.
Before attempting to devise quality control guidelines with respect to genetic variation and inbreeding in laboratory strains, two key questions need to be addressed: (i) What is an acceptable level of genetic variation within a test organism/ batch of test organisms? (ii) What level of inbreeding significantly reduces fitness? For outbred strains traditionally used in ecotoxicology, there is no immediate answer to the first question, since fundamentally a universal, quantitative relationship between the erosion of genetic diversity, reduced adaptation and fitness is lacking (Nacci & Hoffman 2008) and establishing this relationship may be particularly challenging where novel chemical stressors with multiple modes of action are concerned. However, in answer to question (ii), the threshold inbreeding coefficient of F = 0.33 proposed by Frankham (1995b) provides a guideline maximum tolerable limit for inbreeding under laboratory conditions.
In this final section, we have raised the question of how useful or representative inbred laboratory strains are in ecotoxicology for the protection of wildlife. Although we present evidence that laboratory strains of animals can be conservative predictors of adverse effects, there may be instances in which they fail to take into account the full spectrum of genetic variation and responses in wildlife populations. Therefore, there is a possibility that they may sometimes be over-protective and at other times under-protective. As we move forward in ERA in our attempts to develop more realistic and intelligent testing strategies, we need to consider the fundamental assumption that our test subjects are representative of the wild populations that we aim to protect. It therefore would seem logical that effort is put into assessing the diversity of our laboratory animals and the populations that they are meant to represent.
Acknowledgements
Co-authors on this paper were supported by AstraZeneca's SHE Research Programme, the UK Natural Environmental Research Council (NE/F008 104/1, NE/F007 787/1, NE/G011 133/1and NE/D002 818/1, NER/T/S/2002/00 182) and The Department for Environment, Food and Rural Affairs (Defra) as part of the UK-Japan Partnership for Research into Endocrine Disruption to CRT.
Appendix A
An individual can be homozygote at any given allele pi, through identity by descent with probability F(pi), and through identity by state (IBS) with probability (1 − F)( ), F being the inbreeding coefficient of an individual.
), F being the inbreeding coefficient of an individual.
Summing over all alleles at a locus, the probability of being homozygote at any allele yields
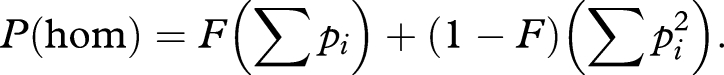 |
As ∑ pi = 1, this simplifies to
 |
This can be rearranged as
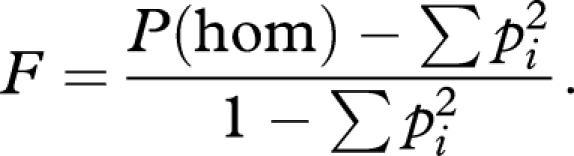 |
When considering a single individual, p(hom) can take only two values 0 and 1, so that
 |
For a multi-locus estimator, each locus must be weighted by the term  , as this represents the amount of information for each locus. Note that
, as this represents the amount of information for each locus. Note that  is not an unbiased estimator of
is not an unbiased estimator of  , but the difference is trivial. One could also consider a weighting for the number of alleles rather than their frequencies.
, but the difference is trivial. One could also consider a weighting for the number of alleles rather than their frequencies.
A multi-locus inbreeding estimator at l loci should thus read
 |
Footnotes
One contribution of 11 to a Theme Issue ‘Impacts of environmental change on reproduction and development in wildlife’.
References
- Alahiotis S. N.1982Adaptation of Drosophila enzymes to temperature IV. Natural selection at the alcohol-dehydrogenase locus. Genetica 59, 81–87 (doi:10.1007/BF00133290) [Google Scholar]
- Altukhov Y. P.1982Biochemical population genetics and speciation. Evolution 36, 1168–1181 (doi:10.2307/2408151) [DOI] [PubMed] [Google Scholar]
- Amos W., Balmford A.2001When does conservation genetics matter? Heredity 87, 257–265 (doi:10.1111/j.1365-2540.2001.00940.pp.x) [DOI] [PubMed] [Google Scholar]
- Ankley G. T., et al. 2006Toxicogenomics in regulatory ecotoxicology. Environ. Sci. Technol. 40, 4055–4065 (doi:10.1021/es0630184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway M. D., Skinner M. K.2006Epigenetic transgenerational actions of endocrine disruptors. Endocrinology 147, 1466–1469 (doi:10.1210/en.2005-1058) [DOI] [PubMed] [Google Scholar]
- Armbruster P., Reed D. H.2005Inbreeding depression in benign and stressful environments. Heredity 95, 235–242 (doi:10.1038/sj.hdy.6800721) [DOI] [PubMed] [Google Scholar]
- Barnthouse L. W., Munns W. R., Jr, Sorenson M. T. Population-level ecological risk assessment. New York: CRC Press; 2008. [Google Scholar]
- Beaumont A. R., Beveridge C. M., Baret E. A., Budd M. D., Smyth-Chamosa M.1988Genetic studies of laboratory reared Mytilus edulis. I. Genotype specific selection in relation to salinity. Heredity 61, 389–400 (doi:10.1038/hdy.1988.129) [Google Scholar]
- Berckmoes V., Scheirs J., Jordaens K., Blust R., Backeljau T., Verhagen R.2005Effects of environmental pollution on microsatellite DNA diversity in wood mouse (Apodemus sylvaticus) populations. Environ. Toxicol. Chem. 24, 2898–2907 (doi:10.1897/04-483R.1) [DOI] [PubMed] [Google Scholar]
- Bickham J. W., Sandhu S., Hebert P. D. N., Chikhi L., Athwal R.2000Effects of chemical contaminants on genetic diversity in natural populations: implications for biomonitoring and ecotoxicology. Mutat. Res. Rev. Mutat. Res. 463, 33–51 (doi:10.1016/S1383-5742(00)00004-1) [DOI] [PubMed] [Google Scholar]
- Bijlsma R., Bundgaard J., Van Putten W. F.1999Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. J. Evol. Biol. 12, 1125–1137 (doi:10.1046/j.1420-9101.1999.00113.x) [Google Scholar]
- Bourret V., Couture P., Campbell P. G. C., Bernatchez L.2008Evolutionary ecotoxicology of wild yellow perch (Perca flavescens) populations chronically exposed to a polymetallic gradient. Aquat. Toxicol. 86, 76–90 (doi:10.1016/j.aquatox.2007.(doi:10.003) [DOI] [PubMed] [Google Scholar]
- Brook B. W., Tonkyn D. W., O'Grady J. J., Frankham R.2002Contribution of inbreeding to extinction risk in threatened species. Conserv. Ecol. 6(1) 16. See http://www.consecol.org/vol6/iss1/art16/ [Google Scholar]
- Brown A. D. H., Marshall D. R., Munday J.1976Adaptedness of variants at an alcohol dehydrogenase locus in Bromus mollis L. (Soft Bromegrass). Aust. J. Biol. Sci. 29, 389–396 [Google Scholar]
- Bryan G. W., Gibbs P. E., Burt G. R.1988Comparison of the effectiveness of tri-N-butyltin chloride and five other organotin compounds in promoting the development of imposex in the dog-whelk, Nucella lapillus. J. Mar. Biol. Assoc. UK 68, 733–744 (doi:10.1017/S0025315400028836) [Google Scholar]
- Burton R. S., Feldman M. W.1983Physiological effects of an allozyme polymorphism: glutamate-pyruvate transaminase and response to hyperosmotic stress in the copepod Tigriopus californicus. Biochem. Genet. 21, 239–251 (doi:10.1007/BF00499136) [DOI] [PubMed] [Google Scholar]
- Carrière Y., Roff D. A., Vincent C.1994Life-history costs associated with the evolution of insecticide resistance. Proc. R. Soc. Lond. B 258, 35–40 (doi:10.2307/49970) [Google Scholar]
- Charlesworth D., Charlesworth B.1987Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18, 237–268 (doi:10.1146/annurev.es.18.110187.001321) [Google Scholar]
- Charlesworth B., Hughes K. A.1996Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl Acad. Sci. USA 93, 6140–6145 See http://www.pnas.org/content/93/12/6140.abstract (doi:10.1073/pnas.93.12.6140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li N., Shen L., Li Y.2003Genetic structure along a gaseous organic pollution gradient: a case study with Poa annua L. Environ. Pollut. 124, 449–455 (doi:10.1016/S0269-7491(03)00042-3) [DOI] [PubMed] [Google Scholar]
- Claudianos C., Russell R. J., Oakeshott J. G.1999The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochem. Mol. Biol. 29, 675–686 (doi:10.1016/S0965-1748(99)00035-1) [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T. H. Reproductive success: studies of individual variation in contrasting breeding systems. Chicago, IL: University of Chicago Press; 1988. [Google Scholar]
- Cochrane B. J., Windelspecht M., Brandon S., Morrow M., Dryden L.1998Use of recombinant inbred lines for the investigation of insecticide resistance and cross resistance in Drosophila simulans. Pest. Biochem. Physiol. 61, 95–114 (doi:10.1006/pest.1998.2355) [Google Scholar]
- Coe T. S., Hamilton P. B., Griffiths A. M., Hodgson D. J., Wahab M. A., Tyler C. R.2009Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology 18, 144–150 (doi:10.1007/s10646-008-0267-0) [DOI] [PubMed] [Google Scholar]
- Coltman D. W., Bowen W. D., Wright J. M.1998Birth weight and neonatal survival of harbour seal pups are positively correlated with genetic variation measured by microsatellites. Proc. R. Soc. Lond. B 265, 803–809 (doi:10.1098/rspb.1998.0363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson T. N., Pemberton J. M., Albon S. D., Beaumont M., Marshall T. C., Slate J., Guiness F. E., Clutton-Brock T. H.1998Microsatellites measure inbreeding depression and heterosis in red deer. Proc. R. Soc. Lond. B 265, 489–495 (doi:10.1098/rspb.1998.0321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnokrak P., Roff D. A.1999Inbreeding depression in the wild. Heredity 83, 260–270 (doi:10.1038/sj.hdy.6885530) [DOI] [PubMed] [Google Scholar]
- Delye C., Laigret F., Corio-Costet M.1997A mutation in the 14 alpha-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl. Environ. Microbiol. 63, 2966–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge M. H.1990New approaches in ecotoxicology: can interindividual physiological variability be used as a tool to investigate pollution effects? Ambio 19, 251–252 [Google Scholar]
- DeSalle R., Amato G.2004The expansion of conservation genetics. Nat. Rev. Genet. 5, 702–712 (doi:10.1038/nrg1425) [DOI] [PubMed] [Google Scholar]
- Devonshire A. L., Field L. M.1991Gene amplification and insecticide resistance. Annu. Rev. Entomol. 36, 1–21 (doi:10.1146/annurev.en.36.010191.000245) [DOI] [PubMed] [Google Scholar]
- Duan Y., Guttman S. I., Oris J. T.1997Genetic differentiation among laboratory populations of Hyalella azteca: implications for toxicology. Environ. Toxicol. Chem. 16, 691–695 (doi:10.1897/1551-5028(1997)016) [Google Scholar]
- Eckert J. W., et al. 1986Pesticide resistance: strategies and tactics for management Washington, DC: National Academy Press; See http://www.nap.edu/openbook.php?isbn=0309036275 [Google Scholar]
- Eeva T., Belskii E., Kuranov B.2006Environmental pollution affects genetic diversity in wild bird populations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 608, 8–15 (doi:10.1016/j.mrgentox.2006.04.021) [DOI] [PubMed] [Google Scholar]
- Escher B. I., Hermens J. L. M.2002Modes of action in ecotoxicology: their role in body burdens, species sensitivity, QSARs, and mixture effects. Environ. Sci. Technol. 36, 4201–4217 (doi:10.1021/es015848h) [DOI] [PubMed] [Google Scholar]
- Facemire C. F., Gross T. S., Guillette L. J., Jr1995Reproductive impairment in the Florida panther: nature or nurture? Environ. Health Perspect. 103Suppl. 4, 79–86 See http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1519283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S.1989Introduction to quantitative genetics, 3rd edn Harlow, UK: Longman [Google Scholar]
- Falconer D. S., Mackay T. F. C., Bulmer M.1996Introduction to quantitative genetics New York, NY: Longman [Google Scholar]
- FAO/UNEP. Conservation of the genetic resources of fish: problems and recommendations. Rome: FAO/UNEP; 1981. See http://www.fao.org/docrep/005/AD013E/AD013E00.htm . [Google Scholar]
- Festing M. F. W., Overend P., Gaines Das R., Cortina Borja M., Berdoy M.2002The design of animal experiments: reducing the number of animals in research through better experimental design. Laboratory Animal Handbooks Number 14. London, UK: The Royal Society of Medicine Press [Google Scholar]
- Feyereisen R.1995Molecular biology of insecticide resistance. Toxicol. Lett. 82, 83–90 (doi:10.1016/0378-4274(95)03470-6) [DOI] [PubMed] [Google Scholar]
- ffrench-Constant R. H., Rocheleau T. A., Steichen J. C., Chalmers A. E.1993A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature 363, 449–451 (doi:10.1038/363449a0) [DOI] [PubMed] [Google Scholar]
- ffrench-Constant R. H., Anthony N., Aronstein K., Rocheleau T., Stilwell G.2000Cyclodiene insecticide resistance: from molecular to population genetics. Annu. Rev. Entomol. 45, 449–466 (doi:10.1146/annurev.ento.45.1.449) [DOI] [PubMed] [Google Scholar]
- Frankham R.1995aEffective population size/adult population size ratios in wildlife: a review. Genet. Res. Camb. 66, 95–107 [DOI] [PubMed] [Google Scholar]
- Frankham R.1995bConservation genetics. Annu. Rev. Genet. 29, 305–327 (doi:10.1146/annurev.ge.29.120195.001513) [DOI] [PubMed] [Google Scholar]
- Frankham R.1995cInbreeding and extinction: a threshold effect. Conserv. Biol. 9, 792–799 (doi:10.2307/2386988) [Google Scholar]
- Frankham R.2003Genetics and conservation biology. C. R. Biol. 326Suppl. 1, 22–29 (doi:10.1016/S1631-0691(03)00023-4) [DOI] [PubMed] [Google Scholar]
- Franklin I. R., Frankham R.1998How large must populations be to retain evolutionary potential? Anim. Conserv. 1, 69–70 (doi:10.1017/S1367943098211103) [Google Scholar]
- Futuyma D. J.1998Evolutionary biology, 3rd edn Sunderland, MA: Sinauer Associates [Google Scholar]
- Gage M. J. G., Surridge A. K., Tomkins J. L., Green E., Wiskin L., Bell D. J., Hewitt G. M.2006Reduced heterozygosity depresses sperm quality in wild rabbits Oryctolagus cuniculus. Curr. Biol. 16, 612–617 (doi:10.1016/j.cub.2006.02.059) [DOI] [PubMed] [Google Scholar]
- Georghiou G. P.1986The magnitude of the resistance problem in. In Pesticide resistance: strategies and tactics for management (eds Georghiou G. P., Taylor C. E.), pp. 14–43 Washington, DC: National Academy Press [Google Scholar]
- Gilpin M. E., Soulé M. E.1986Minimum viable populations: processes of species extinction. In Conservation biology: the science of scarcity and diversity (ed. Soulé M. E.), pp. 19–34 Sunderland, MA: Sinauer Associates [Google Scholar]
- Goodhead R. M., Tyler C. R.2008Endocrine disrupting chemicals and their environmental impacts. In Organic pollutants (ed. Walker C.), pp. 265–292 Boca Raton, FL: CRC Press, Taylor and Francis [Google Scholar]
- Goodnight K. F., Queller D. C.1999Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol. Ecol. 8, 1231–1234 (doi:10.1046/j.1365-294x.1999.00664.x) [DOI] [PubMed] [Google Scholar]
- Green R. E., Hirons G. J. M.1991The relevance of population studies to the conservation of threatened birds. In Bird population studies: relevance to conservation and management (eds Perrins J., Lebreton D., Hirons G. J. M.), pp. 594–633 Oxford, UK: Oxford University Press [Google Scholar]
- Greenwood P. J.1980Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 28, 1140–1162 (doi:10.1016/S0003-3472(80)80103-5) [Google Scholar]
- Gressel J.1986Modes and genetics of herbicide resistance in plants. In Pesticide resistance: strategies and tactics for management (eds Georghiou G. P., Taylor C. E.), pp. 54–73 Washington, DC: National Academy Press [Google Scholar]
- Groenendijk D., Lücker S. M. G., Plans M., Kraak M. H. S., Admiraal W.2002Dynamics of metal adaptation in riverine chironomids. Environ. Pollut. 117, 101–109 (doi:10.1016/S0269-7491(01)00154-3) [DOI] [PubMed] [Google Scholar]
- Guillaumot L. Mosquitoes and disease transmission. 2006 (Homepage of Institut Pasteur de Nouvelle-Calédonie). See http://www.institutpasteur.nc/article.php3?id_article=80. (13 March 2009). 5 September 2006, last update (Online) [Google Scholar]
- Guryev V., Koudijs M. J., Berezikov E., Johnson S. L., Plasterk R. H. A., van Eeden F. J. M., Cuppen E.2006Genetic variation in the zebrafish. Genome Res. 16, 491–497 (doi:10.1101/gr.4791006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman S. I.1994Population genetic structure and ecotoxicology. Environ. Health Perspect. 102Suppl. 12, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson M. A., Skelly D. K., Caccone A.2006Inbreeding linked to amphibian survival in the wild but not in the laboratory. J. Hered. 97, 499–507 (doi:10.1093/jhered/esl019) [DOI] [PubMed] [Google Scholar]
- Heagler M. G., Newman M. C., Mulveym M., Dixon P. M.1993Allozyme genotype in mosquitofish, Gambusia holbrooki, during mercury exposure: temporal stability, concentration effects and field verification. Environ. Toxicol. Chem. 12, 385–395 (doi:10.1897/1552-8618(1993)12[385:AGIMGH]2.0.CO) [Google Scholar]
- Hill E. F.1994Acute and subacute toxicology in evaluation of pesticide hazard to avian wildlife. In Wildlife toxicology and population modeling: integrated studies of agroecosystems (eds Kendall R. J., Lacher T. E. J.), pp. 219–238 Chelsea, MI: Lewis Publishing [Google Scholar]
- Hoekstra H. E., Coyne J. A.2007The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (doi:10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Parsons P. A. Evolutionary genetics and environmental stress. New York: Oxford University Press; 2002. [Google Scholar]
- Hosken D. J., Blanckenhorn W. U.1999Female multiple mating, inbreeding avoidance and fitness: it's not only the magnitude of costs and benefits that count. Behav. Ecol. 10, 462–464 (doi:10.1093/beheco/10.4.462) [Google Scholar]
- Hughes K. A.1995The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution 49, 521–537 (doi:10.2307/2410276) [DOI] [PubMed] [Google Scholar]
- IUCN. IUCN list of threatened species 2008: 2001 categories and criteria (version 3.1) 2008 See http://www.iucnredlist.org/static/categories_criteria_3_1. (13 March 2009) 1 January 2008, last update. [Google Scholar]
- Kacew S.2001Confounding factors in toxicity testing. Toxicology 160, 87–96 (doi:10.1016/S0300-483X(00)00440-6) [DOI] [PubMed] [Google Scholar]
- Kacew S., Festing M. F. W.1996Role of rat strain in the differential sensitivity of pharmaceutical agents and naturally occurring substances. J. Toxicol. Environ. Health 47, 1–30 See http://www.ingentaconnect.com/content/tandf/utehold/1996/00000047/00000001/art00001 [DOI] [PubMed] [Google Scholar]
- Keller L. F.1998Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia). Evolution 52, 240–250 (doi:10.2307/2410939) [DOI] [PubMed] [Google Scholar]
- Keller L. F., Waller D. M.2002Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (doi:10.1016/S0169-5347(02)02489-8) [Google Scholar]
- King J. L., Jukes T. H.1969Non-Darwinian evolution. Science 164, 788–798 (doi:10.1126/science.164.3881.788) [DOI] [PubMed] [Google Scholar]
- Klerks P.2002Adaptation, ecological impacts, and risk assessment: insights from research at Foundry Cove, Bayou Trepagnier, and Pass Fourchon. Hum. Ecol. Risk Assess. 8, 971–982 (doi:10.1080/1080-700291905774) [Google Scholar]
- Klerks P. L., Moreau C. J.2001Heritability of resistance to individual contaminants and contaminant mixtures in the sheepshead minnow (Cyprinodon variegatus). Environ. Toxicol. Chem. 20, 1746–1751 (doi:10.1897/1551-5028(2001)020<1746:HORTIC>2.0.CO;2) [PubMed] [Google Scholar]
- Klerks P. L., Weis J. S.1987Genetic adaptation to heavy metals in aquatic organisms: a review. Environ. Pollut. 45, 173–205 (doi:10.1016/0269-7491(87)90057-1) [DOI] [PubMed] [Google Scholar]
- Kohn M. H., Murphy W. J., Ostrander E. A., Wayne R. K.2006Genomics and conservation genetics. Trends Ecol. Evol. 21, 629–637 (doi:10.1016/j.tree.2006.08.001) [DOI] [PubMed] [Google Scholar]
- Kovatch C. E., Schizas N. V., Thomas Chandler G., Coull B. C., Quattro J. M.2000Tolerance and genetic relatedness of three meiobenthic copepod populations exposed to sediment-associated contaminant mixtures: role of environmental history. Environ. Toxicol. Chem. 19, 912–919 (doi:10.1897/1551-5028(2000)019<0912:TAGROT>2.3.CO;2) [Google Scholar]
- Kristensen T. N., Dahlgaard J., Loeschcke V.2003Effects of inbreeding and environmental stress on fitness—using Drosophila buzzatii as a model organism. Conserv. Genet. 4, 453–465 (doi:10.1023/A:1024763013798) [Google Scholar]
- Lande R.1976Natural selection and random genetic drift in phenotypic evolution. Evolution 30, 314–334 (doi:10.2307/2407703) [DOI] [PubMed] [Google Scholar]
- Lande R.1994Risk of population extinction from fixation of new deleterious mutations. Evolution 48, 1460–1469 (doi:10.2307/2410240) [DOI] [PubMed] [Google Scholar]
- Lande R.1995Mutation and conservation. Conserv. Biol. 9, 782–791 (doi:10.2307/2386987) [Google Scholar]
- Lande R., Shannon S.1996The role of genetic variation in adaptation and population persistence in a changing environment. Evolution 50, 434–437 (doi:10.2307/2410812) [DOI] [PubMed] [Google Scholar]
- Leslie J. F., Watt W. B.1986Some evolutionary consequences of the molecular recombination process. Trends Genet. 2, 288–291 (doi:10.1016/0168-9525(86)90271-4) [Google Scholar]
- Liao W., Reed D. H.2009Inbreeding–environment interactions increase extinction risk. Anim. Conserv. 12, 54–61 [Google Scholar]
- Lynch M.1996A quantitative-genetic perspective on conservation issues. In Conservation genetics: case histories from nature (eds Avise J., Hamrick J.), pp. 471–501 London, UK: Chapman & Hall [Google Scholar]
- Lynch M., Lande R.1998The critical effective size for a genetically secure population. Anim. Conserv. 1, 70–72 (doi:10.1017/S136794309822110X) [Google Scholar]
- Lynch M., Conery J., Burger R.1995Mutation accumulation and the extinction of small populations. Am. Nat. 146, 489–518 (doi:10.1086/285812) [Google Scholar]
- Madsen T., Shine R., Olsson M., Wittzell H.1999Conservation biology: restoration of an inbred adder population. Nature 402, 34–35 (doi:10.1038/46941) [Google Scholar]
- Margaritopoulos J. T., Skavdis G., Kalogiannis N., Nikou D., Morou E., Skouras P. J., Tsitsipis J. A., Vontas J.2008Efficacy of the pyrethroid alpha-cypermethrin against Bactrocera oleae populations from Greece, and improved diagnostic for an iAChE mutation. Pest. Manag. Sci. 64, 900–908 (doi:10.1002/ps.1580) [DOI] [PubMed] [Google Scholar]
- Marichal P., et al. 1999Contribution of mutations in the cytochrome P450 14{alpha}-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145, 2701–2713 [DOI] [PubMed] [Google Scholar]
- McKay J. K., Stinchcombe J. R.2008Ecological genomics of model eukaryotes 1. Evolution 62, 2953–2957 (doi:10.1111/j.1558-5646.2008.00536.x) [DOI] [PubMed] [Google Scholar]
- Meagher S., Penn D. J., Potts W. K.2000Male–male competition magnifies inbreeding depression in wild house mice. Proc. Natl Acad. Sci. 97, 3324–3329 (doi:10.1073/pnas.060284797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merila J., Crnokrak P.2001Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 14, 892–903 (doi:10.1046/j.1420-9101.2001.00348.x) [Google Scholar]
- Meyer J., Di Giulio R.2002Patterns of heritability of decreased EROD activity and resistance to PCB 126-induced teratogenesis in laboratory-reared offspring of killifish (Fundulus heteroclitus) from a creosote-contaminated site in the Elizabeth River, VA, USA. Mar. Environ. Res. 54, 621–626 (doi:10.1016/S0141-1136(02)00170-8) [DOI] [PubMed] [Google Scholar]
- Meyer J. N., Di Giulio R. T.2003Heritable adaptation and fitness costs in killifish (Fundulus heteroclitus) inhabiting a polluted estuary. Ecol. Appl. 13, 490–503 (doi:10.1890/1051-0761(2003)013[0490:HAAFCI]2.0.CO;2) [Google Scholar]
- Miller P. S.1994Is inbreeding depression more severe in a stressful environment? Zoo Biol. 13, 195–208 (doi:10.1002/zoo.1430130302) [Google Scholar]
- Miller P. S., Hedrick P. W.2001Purging of inbreeding depression and fitness decline in bottlenecked populations of Drosophila melanogaster. J. Evol. Biol. 14, 595–601 (doi:10.1046/j.1420-9101.2001.00303.x) [Google Scholar]
- Mouches C., Pasteur N., Berge J. B., Hyrien O., Raymond M., de Saint Vincent B. R., De Silvestri M., Georghiou G. P.1986Amplification of an esterase gene is responsible for insecticide resistance in a California Culex mosquito. Science 233, 778–780 (doi:10.1126/science.3755546) [DOI] [PubMed] [Google Scholar]
- Mutero A., Pralavorio M., Bride J., Fournier D.1994Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl Acad. Sci. USA 91, 5922–5926 See http://www.pnas.org/content/91/13/5922.abstract (doi:10.1073/pnas.91.13.5922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacci D., Hoffman G. R.2008Genetic variation in population-level ecological risk assessment. In Population-level ecological risk assessment (eds Barnthouse L. W., Munns W. R., Jr, Sorensen M. T.), pp. 93–112 New York, NY: Taylor & Francis [Google Scholar]
- Nacci D., Coiro L., Champlin D., Jayaraman S., McKinney R., Gleason T. R., Munns W. R., Jr, Specker J. L., Cooper K. R.1999Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar. Biol. 134, 9–17 (doi:10.1007/s002270050520) [Google Scholar]
- Nagoshi R. N., Gelbart W. M.1987Molecular and recombinational mapping of mutations in the Ace locus of Drosophila melanogaster. Genetics 117, 487–502 See http://www.genetics.org/cgi/content/abstract/117/3/487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academy of Sciences (NAS) 2007 Toxicity testing in the 21st century—a vision and a strategy Washington, DC: National Academies Press; See http://www.nap.edu/catalog.phprecord_id=11970 [Google Scholar]
- Nebert D. W., Nelson D. R.2006Cytochrome P450 (CYP) gene superfamily. Encyclopedia of life sciences, pp. 1–9 Hoboken, NJ: John Wiley & Sons: See http://mrw.interscience.wiley.com/emrw/9780470015902/els/article/a0005667/current/pdf (31 March 2009) (Online) [Google Scholar]
- Nilsson E., Anway M., Stanfield J., Skinner M.2008Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction 135, 713–721 (doi:10.1210/en.2005-1058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak C., Jost D., Vogt C., Oetken M., Schwenk K., Oehlmann J.2007aConsequences of inbreeding and reduced genetic variation on tolerance to cadmium stress in the midge Chironomus riparius. Aquat. Toxicol. 85, 278–284 (doi:10.1016/j.aquatox.2007.04.015) [DOI] [PubMed] [Google Scholar]
- Nowak C., Vogt C., Barateiro Diogo J., Schwenk K.2007bGenetic impoverishment in laboratory cultures of the test species Chironomus riparius. Environ. Toxicol. Chem. 26, 118–122 (doi:10.1897/06-349R.1) [DOI] [PubMed] [Google Scholar]
- O'Brien S. J.1994A role for molecular genetics in biological conservation. Proc. Natl Acad. Sci. 91, 5748–5755 See http://www.pnas.org/content/91/13/5748.abstract (doi:10.1073/pnas.91.13.5748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien S. J., Evermann J. F.1988Interactive influence of infectious disease and genetic diversity in natural populations. Trends Ecol. Evol. 3, 254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi S. R.2001Humans as the world's greatest evolutionary force. Science 293, 1786–1790 (doi:10.1126/science.293.5536.1786) [DOI] [PubMed] [Google Scholar]
- Pasteur N., Raymond M.1996Insecticide resistance genes in mosquitoes: their mutations, migration, and selection in field populations. J. Hered. 87, 444–449 See http://jhered.oxfordjournals.org/cgi/content/abstract/87/6/444 [DOI] [PubMed] [Google Scholar]
- Peles J. D., Towler W. I., Guttman S. I.2003Population genetic structure of earthworms (Lumbricus rubellus) in soils contaminated by heavy metals. Ecotoxicology 12, 379–386 (doi:10.1023/A:1026269804938) [DOI] [PubMed] [Google Scholar]
- Pemberton J.2004Measuring inbreeding depression in the wild: the old ways are the best. Trends Ecol. Evol. 19, 613–615 (doi:(doi:10.1016/j.tree.2004.09.010) [DOI] [PubMed] [Google Scholar]
- Preston R. J., Hoffman G. R.2001Genetic toxicology. In Casarett and Doull's toxicology: the basic science of poisons (ed. Klaassen C. D.), pp. 321–350, 6th edn New York, NY: McGraw-Hill [Google Scholar]
- Pusey A., Wolf M.1996Inbreeding avoidance in animals. Trends Ecol. Evol. 11, 201–206 (doi:10.1016/0169-5347(96)10028-8) [DOI] [PubMed] [Google Scholar]
- Ralls K., Ballou J. D., Templeton A.1988Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv. Biol. 2, 185–193 (doi:10.2307/2386104) [Google Scholar]
- Ratcliffe D. A.1967Decrease in eggshell weight in certain birds of prey. Nature 215, 208–210 (doi:10.1038/215208a0) [DOI] [PubMed] [Google Scholar]
- Razzoli M., Papa R., Valsecchi P., Nonnis Marzano F.2003AFLP to assess genetic variation in laboratory gerbils (Meriones unguiculatus). J. Hered. 94, 507–511 (doi:10.1093/jhered/esg097) [DOI] [PubMed] [Google Scholar]
- Reed D. H., Frankham R.2003Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237 (doi:10.1046/j.1523-1739.2003.01236.x) [Google Scholar]
- Reed D. H., Briscoe D. A., Frankham R.2002Inbreeding and extinction: the effect of environmental stress and lineage. Conserv. Genet. 3, 301–307 (doi:10.1023/A:1019948130263) [Google Scholar]
- Reijnders P. J. H.1986Reproductive failure in common seals feeding on fish from polluted coastal waters. Nature 324, 456–457 (doi:10.1038/324456a0) [DOI] [PubMed] [Google Scholar]
- Riechert S. E., Roeloffs R. M.1993Evidence for and consequences of inbreeding in the cooperative spiders. In The natural history of inbreeding and outbreeding: theoretical and empirical perspectives (ed. Thornhill N. W.), pp. 283–303 Chicago, IL: University of Chicago Press [Google Scholar]
- Roelke M. E., Schultz D. P., Facemire C. F., Sundolf S. F., Royals H. E.1992Mercury contamination in the Florida panthers. A report of the Florida panther technical sub-committee to the Florida panther interagency committee Gainesville, FL: Gainesville Florida Game and Freshwater Fish Commission [Google Scholar]
- Roelke M. E., Martenson J. S., O'Brien S. J.1993The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Curr. Biol. 3, 340–350 (doi:10.1016/0960-9822(93)90197-V) [DOI] [PubMed] [Google Scholar]
- Roldan E. R. S., Gomendio M.2009Sperm and conservation. In Biology of sperm: an evolutionary perspective (eds Birkhead T. R., Hosken D. J., Pitnick S.), pp. 539–564 London, UK: Academic Press [Google Scholar]
- Roush R. T., McKenzie J. A.1987Ecological genetics of insecticide and acaricide resistance. Annu. Rev. Entomol. 32, 361–380 (doi:10.1146/annurev.en.32.010187.002045) [DOI] [PubMed] [Google Scholar]
- Russell R. J., Claudianos C., Campbell P. M., Horne I., Sutherland T. D., Oakeshott J. G.2004Two major classes of target site insensitivity mutations confer resistance to organophosphate and carbamate insecticides. Pest. Biochem. Physiol. 79, 84–93 (doi:10.1016/j.pestbp.2004.03.002) [Google Scholar]
- Schaeffer D. J., Beasley V. R.1989Ecosystem health. II. Quantifying and predicting ecosystem effects of toxic chemicals: can mammalian testing be used for lab-to-field and field-to-lab extrapolations? Regul. Toxicol. Pharmacol. 9, 296–311 (doi:10.1016/0273-2300(89)90068-8) [DOI] [PubMed] [Google Scholar]
- Schlueter M. A., Guttman S. I., Oris J. T., Bailer A. J.1997Differential survival of fathead minnows, Pimephales promelas, as affected by copper exposure, prior population stress, and allozyme genotypes. Environ. Toxicol. Chem. 16, 939–947 (doi:10.1897/1551-5028(1997)016<0939:DSOFMP>2.3.CO;2) [Google Scholar]
- Schlueter M. A., Guttman S. I., Duan Y., Oris J. T., Huang X., Burton G. A.2000Effects of acute exposure to fluoranthene-contaminated sediment on the survival and genetic variability of fathead minnows (Pimephales promelas). Environ. Toxicol. Chem. 19, 1011–1018 (doi:10.1897/1551-5028(2000)019<1011:EOAETF>2.3.CO;2) [Google Scholar]
- Shaw A. J.1999The evolution of heavy metal tolerance in plants: adaptations, limits, and costs. In Genetics and ecotoxicology (ed. Forbes V. E.), pp. 9–30 New York: Taylor & Francis [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A.1984Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12, 4653–4664 (doi:10.1093/nar/12.11.4653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simberloff D.1988The contribution of population and community biology to conservation science. Annu. Rev. Ecol. Syst. 19, 473–511 (doi:10.1146/annurev.es.19.110188.002353) [Google Scholar]
- Slate J., Kruuk L. E. B., Marshall T. C., Pemberton J. M., Clutton-Brock T. H.2000Inbreeding depression influences lifetime breeding success in a wild population of red deer (Cervus elaphus). Proc. R. Soc. Lond. B 267, 1657–1662 (doi:10.1098/rspb.2000.1192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J., David P., Dodds K. G., Veenvliet B. A., Glass B. C., Broad T. E., McEwan J. C.2004Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity 93, 255–265 (doi:10.1038/sj.hdy.6800485) [DOI] [PubMed] [Google Scholar]
- Spearow J. L.2004Reviewer's appendix to the white paper on species/strain/stock/ in endocrine disruptor assays. See http://www.epa.gov/sciploy/oscpendo/pubs/edt02wdisc.pdf (6 March 2009) [Google Scholar]
- Spearow J. L., Doemeny P., Sera R., Leffler R., Barkley M.1999Genetic variation in susceptibility to endocrine disruption by estrogen in mice. Science 285, 1259–1261 (doi:10.1126/science.285.5431.1259) [DOI] [PubMed] [Google Scholar]
- Spielman D., Brook B. W., Frankham R.2004Most species are not driven to extinction before genetic factors impact them. Proc. Natl Acad. Sci. USA 101, 15 261–15 264 (doi:10.1073/pnas.0403809101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P.2002Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2, 185–194 (doi:10.1038/nri749) [DOI] [PubMed] [Google Scholar]
- Stockley P., Searle J. B., Macdonald D. W., Jones C. S.1993Female multiple mating behaviour in the common shrew as a strategy to reduce inbreeding. Proc. R. Soc. Lond. B 254, 173–179 (doi:10.1098/rspb.1993.0143) [DOI] [PubMed] [Google Scholar]
- Stohler R. A., Curtis J., Minchella D. J.2004A comparison of microsatellite polymorphism and heterozygosity among field and laboratory populations of Schistosoma mansoni. Int. J. Parasitol. 34, 595–601 (doi:10.1016/j.ijpara.2003.11.026) [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E., Liu Y. B., Finson N., Masson L., Heckel D. G.1997One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc. Natl Acad. Sci. USA 94, 1640–1644 See http://www.pnas.org/content/94/5/1640.abstract (doi:10.1073/pnas.94.5.1640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabashnik B. E., Carrière Y., Dennehy T. J., Morin S., Sisterson M. S., Roush R. T., Shelton A. M., Zhao J. Z.2003Insect resistance to transgenic Bt crops: lessons from the laboratory and field. J. Econ. Entomol. 96, 1031–1038 (doi:10.1603/0022-0493(2003)096[1031:IRTTBC]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Tatara C. P., Mulvey M., Newman M. C.1999Genetic and demographic responses of mosquitofish (Gambusia holbrooki) populations exposed to mercury for multiple generations. Environ. Toxicol. Chem. 18, 2840–2845 (doi:10.1897/1551-5028(1999)018<2840:GADROM>2.3.CO;2) [PubMed] [Google Scholar]
- Taylor M., Feyereisen R.1996Molecular biology and evolution of resistance to toxicants. Mol. Biol. Evol. 13, 719–734 [DOI] [PubMed] [Google Scholar]
- Thompson M., Steichen J. C., ffrench-Constant R. H.1993Conservation of cyclodiene insecticide resistance-associated mutations in insects. Insect Mol. Biol. 2, 149–154 (doi:10.1111/j.1365-2583.1993.tb00134.x) [DOI] [PubMed] [Google Scholar]
- UN Millennium Ecosystem Assessment and World Resources Institute. Ecosystems and human well-being. Washington, DC: Island Press; 2005. [Google Scholar]
- US EPA. White paper on species/strain/stock in endocrine disrupter assays. 2003 (Homepage of US EPA) (Online). See http://www.epa.gov/endo/pubs/strainswhitepaper072503.pdf. (17 March 2009) [Google Scholar]
- US FDA. Innovation or stagnation: challenge and opportunity on the critical path to new medicinal products. 2004 (Homepage of US Department of Health and Human Services, Food and Drug Administration) (Online). See http://www.fda.gov/oc/initiative/criticalpath/whitepaper.html. (17 March 2009) [Google Scholar]
- Van Loveren H., Ross P. S., Osterhaus A. D. M. E., Vos J. G.2000Contaminant-induced immunosuppression and mass mortalities among harbor seals. Toxicol. Lett. 112, 319–324 (doi:10.1016/S0378-4274(99)00198-8) [DOI] [PubMed] [Google Scholar]
- Van Straalen N., Timmermans M.2002Genetic variation in toxicant-stressed populations: an evaluation of the genetic erosion hypothesis. Hum. Ecol. Risk Assess. 8, 983–1002 (doi:10.1080/1080_700291905783) [Google Scholar]
- Vilà C., Sundqvist A.-K., Flagstad Ø., Seddon J., Björnerfeldt S., Kojola I., Casulli A., Sand H., Wabakken P., Ellegren H.2003Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc. R. Soc. Lond. B 270, 91–97 (doi:10.1098/rspb.2002.2184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. G., Dybing E., Greim H. A., Ladefoged O., Lambre C., Tarazona J. V., Brandt I., Vethaak A. D.2000Health effects of endocrine-disrupting chemicals on wildlife, with special reference to the European situation. Crit. Rev. Toxicol. 30, 71–133 (doi:10.1080/10408440091159176) [DOI] [PubMed] [Google Scholar]
- Vriejenhoek R. C.1994Genetic diversity and fitness in small populations. In Conservation genetics (eds Loeschcke V., Tomiuk J., Jain S. K.), pp. 37–53 Berlin, Germany: Birkhauser [Google Scholar]
- Waldman B., McKinnon J. S.1993Inbreeding and outbreeding in fishes, amphibians, and reptiles. In The natural history of inbreeding and outbreeding: theoretical and empirical perspectives (ed. Thornhill N. W.), pp. 250–282 Chicago, IL: University of Chicago Press [Google Scholar]
- Wayne R. K., et al. 1991Conservation genetics of the endangered Isle Royale gray wolf. Conserv. Biol. 5, 41–51 (doi:10.2307/2386337) [Google Scholar]
- Williamson M. S., Martinez-Torres D., Hick C. A., Devonshire A. L.1996Identification of mutations in the housefly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol. Gen. Genet. 252, 51–60 [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations, volume 3: experimental results and evolutionary deductions. Chicago, IL: University of Chicago Press; 1984. [Google Scholar]
- Wright L. I., Tregenza T., Hosken D. J.2008Inbreeding, inbreeding depression and extinction. Conserv. Genet. 9, 833–843 (doi:10.1007/s10592-007-9405-0) [Google Scholar]


