Abstract
Vertebrate cones and rods in several cases use separate but related components for their signal transduction (opsins, G-proteins, ion channels, etc.). Some of these proteins are also used differentially in other cell types in the retina. Because cones, rods and other retinal cell types originated in early vertebrate evolution, it is of interest to see if their specific genes arose in the extensive gene duplications that took place in the ancestor of the jawed vertebrates (gnathostomes) by two tetraploidizations (genome doublings). The ancestor of teleost fishes subsequently underwent a third tetraploidization. Our previously reported analyses showed that several gene families in the vertebrate visual phototransduction cascade received new members in the basal tetraploidizations. We here expand these data with studies of additional gene families and vertebrate species. We conclude that no less than 10 of the 13 studied phototransduction gene families received additional members in the two basal vertebrate tetraploidizations. Also the remaining three families seem to have undergone duplications during the same time period but it is unclear if this happened as a result of the tetraploidizations. The implications of the many early vertebrate gene duplications for functional specialization of specific retinal cell types, particularly cones and rods, are discussed.
Keywords: gene duplication, tetraploidization, eye, retina, phototransduction, opsin
1. Introduction
(a). Vertebrate phototransduction
Phototransduction is the process whereby light is converted to electrical signals in the eye. The signal transduction cascade of vertebrate rods and cones has been worked out in great detail and has been described in several excellent reviews (Fu & Yau 2007; Wensel 2008). The principal components of a vertebrate cone phototransduction machinery are shown schematically in figure 1. The cascade starts with the absorption of photons by the photoreceptive pigments, the opsins, which are G-protein-coupled receptors. The photon isomerizes the receptor's covalently bound 11-cis-retinal to all-trans-retinal which induces a structural change that activates the opsin. The signal is relayed by activation of transducin (a trimeric G protein) that stimulates the activity of phosphodiesterase (PDE), which reduces the level of the second messenger cGMP. This leads to closure of a cyclic nucleotide-gated (CNG) ion channel leading to hyperpolarization of the cell and reduced synaptic release of the neurotransmitter glutamate onto the bipolar and horizontal cells.
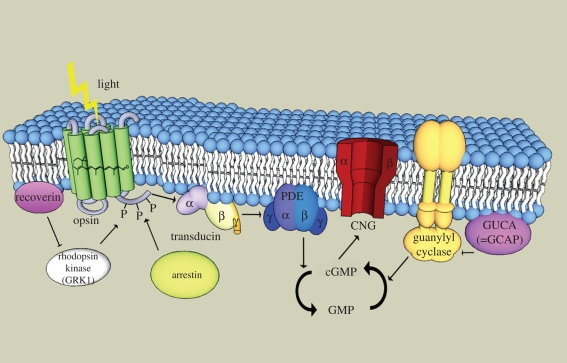
Figure 1.
Schematic outline of the phototransduction cascade in a vertebrate cone. All of the components are shown within or near the same membrane, the cell-surface membrane, as is assumed to have been the case in the ancestor of cones and rods (rods subsequently evolved an internalization mechanism for parts of the membrane to form intracellular discs whose membranes harbour some of the phototransduction components including rhodopsin). Upon absorption by a photon, 11-cis-retinal within the opsin is transformed to all-trans-retinal. This activates the opsin, which in turn activates the transducin (a G protein) consisting of three subunits (α, β and γ). Transducin in turn activates phophodiesterase 6 (PDE), which forms a dimer with α and β (or two α subunits), inhibited by two γ subunits. Active PDE hydrolyzes cGMP to GMP. cGMP keeps the cyclic nucleotide ion channel (CNG, a tetramer with α and β subunits) open. New cGMP is generated by guanylyl cyclase (GC), which is regulated by a GC-activating protein (GUCA or GCAP). Rhodopsin kinase (GRK) deactivates the opsin by phosphorylation, and is itself regulated by recoverin. Arrestin binds to the phosphorylated opsin to reduce its signalling to the G protein. GRK is anchored to the membrane and arrestin undergoes translocation to the membrane on activation (not shown in the figure).
Signalling is reduced and terminated by phosphorylation of the opsin by the G-protein-coupled receptor kinase (GRK) followed by binding of arrestin. The system is restored by synthesis of cGMP by guanylyl cyclase (GC) stimulated by GC-activating protein (abbreviated GUCA or GCAP). Recoverin reduces the activity of GRK. A separate complicated machinery involving non-photoreceptor cell types returns the all-trans-retinal to 11-cis-retinal for re-insertion into opsin.
To some extent the proteins are the same in rods and cones, but there are also several cases where cones use one variant of a protein and rods use a different but closely related member of the same protein family expressed from a separate gene. For instance, the transducins, like all other trimeric G proteins, consist of α, β and γ subunits, and rods and cones have been found to express distinct variants of all three subunits: rods express the α subunit gene GNAT1, whereas cones use GNAT2, rods express the β subunit gene GNB1, while cones use GNB3, and rods express the γ subunit gene GNGT1, whereas cones use GNGT2 (see Nordström et al. 2004 for references).
Some of the phototransduction genes are also expressed differentially in other retinal cell types. One example is the transducin β subunit just mentioned, where the GNB1 gene has been found to be expressed in amacrine cells, whereas the GNB3 gene product was detected in some bipolar cells in crab-eating macaque (cynomolgus monkey, Macaca fascicularis) as well as in rat, rabbit and cattle (Peng et al. 1992). The GNB3 gene is expressed in ON-bipolar cells in mouse (Huang et al. 2003). This shared use of specific gene family members by distinct retinal cell types may help shed light on the evolutionary and developmental relationships of the various cell types of the retina. Although several aspects of retinal development have been described (Dyer & Cepko 2001; Marquardt & Gruss 2002), many details remain to be resolved regarding the roles and the ontogeny of the many subtypes of bipolar, amacrine and ganglion cells.
The major cell types of the vertebrate retina, i.e. rods, cones, horizontal cells, bipolar cells, amacrine cells and ganglion cells, are thought to have originated in early vertebrate evolution (Lamb et al. 2007). Even subtypes of some of these cells arose in early vertebrates. Not only rods and cones are known to be present in lampreys, but also subtypes of bipolar cells and ganglion cells. Indeed, different categories of ganglion cells were first identified in a frog (Hartline 1938) and subtypes of bipolar cells were originally observed in a salamander (Dowling & Werblin 1969). Subsequently, such subtypes of cells have been found to exist in mammals, birds and teleost fishes. Therefore, it is of great interest to see if the genes that are differentially expressed in these cell types arose in the massive gene duplications that took place before the radiation of the jawed vertebrates, the gnathostomes (Miyata & Suga 2001), resulting from the recently confirmed tetraploidizations (Putnam et al. 2008).
We previously reported that many human phototransduction gene families are located in chromosomal regions that seem to have been duplicated in early vertebrate evolution, as based on sequence-based phylogenetic trees, supporting origin in the tetraploidizations (Nordström et al. 2004). We investigated nine gene families (opsins, transducins α, β and γ, PDE, CNG, GRK, arrestin and recoverin) and concluded that all nine of these received additional family members before the divergence of tetrapods and ray-finned fishes (albeit the recoverin family duplications did not at that time seem to have generated duplicates expressed specifically in rods and cones). In the light of our findings, concomitant duplication of genes for the entire phototransduction pathway (co-duplication) versus incorporation of individual components by co-option has been discussed (Plachetzki & Oakley 2007).
Now that the double tetraploidization in the gnathostome ancestor has been confirmed by whole-genome studies (Putnam et al. 2008), we have expanded our phototransduction data by studies of additional gene families and vertebrate species. We have investigated in total 13 phototransduction gene families and conclude from a combination of sequence-based phylogeny and comparative chromosome analyses that all 13 expanded in the time period of the two basal vertebrate tetraploidizations and that no less than 10 of them received additional members as a direct result of the tetraploidizations. We discuss how the new gene copies may have contributed to the functional specialization of specific retinal cell types, particularly cones and rods.
(b). Tetraploidizations in early vertebrate evolution (2R and 3R)
For several years the notion of tetraploidizations at the dawn of vertebrate evolution was highly controversial and widely discussed in the literature (Panopoulou & Poustka 2005). The concept of tetraploidizations was proposed already in 1970 by Susumu Ohno (Ohno 1970), but the time points for the events were rather vaguely discussed. The first hard evidence for extensive chromosome duplications was reported by Lars G. Lundin in 1993 who described four sets of chromosomes in the human and mouse genomes with similarities among the members of each set (Lundin 1993). Two of the sets consisted of four chromosomes indicating that the genome had been quadrupled through two tetraploidizations. The double tetraploidization scenario was later named 2R for two rounds of genome doubling and this abbreviation will be used in this article.
Detailed molecular genetic evidence for one quadrupled chromosome region was presented when the lancelet Branchiostoma floridae, also called amphioxus, a cephalochordate, was found to have a single gene cluster with Hox-type homeobox genes (Garcia-Fernàndez & Holland 1994), whereas tetrapods have four Hox clusters. But it was not until the long-awaited genome assembly of amphioxus was published in June 2008 (Putnam et al. 2008) that it was confirmed that many parts of its genome correspond to four paralogous regions in the genomes of Homo sapiens and other tetrapods. Also several previous studies had accumulated strong evidence for quadruplication of many large chromosome regions (Abi-Rached et al. 2002; Larhammar et al. 2002; Lundin et al. 2003; Vienne et al. 2003; Dehal & Boore 2005; Nakatani et al. 2007) in line with the double tetraploidization hypothesis. Related chromosome regions within a genome are said to be paralogous, and together they form a paralogon (Coulier et al. 2000), i.e. a set of related regions, usually a quartet.
Both of the basal vertebrate tetraploidizations have been confirmed to have taken place before the radiation of the gnathostomes (Vandepoele et al. 2004; figure 2). The first divergence among these led to Chondrichthyes (cartilaginous fishes) and Osteichthyes (ray-finned and lobe-finned fishes), and both of these lineages have at least four Hox clusters as concluded from the genome sequence of the elephant shark, Callorhinchus milii (Venkatesh et al. 2007) and all studied representatives for Osteichthyes (Hoegg & Meyer 2005). Recently, we reported that the elephant shark also possesses members in a gene family located on all four chromosomes of a different quartet in tetrapods, namely the neuropeptide Y (NPY) receptor family (Larsson et al. 2008), thereby supporting its shared genome quadruplication with Osteichthyes (Venkatesh et al. 2007). Studies in ray-finned fishes (Actinopterygii) have shown that euteleost fishes (Euteleostei) have undergone a third tetraploidization named 3R. This was suggested already by the zebrafish genome (Postlethwait et al. 2000) and persuasively concluded from the genomes of the two pufferfishes Takifugu rubripes (Christoffels et al. 2004) and Tetraodon nigroviridis (Jaillon et al. 2004).
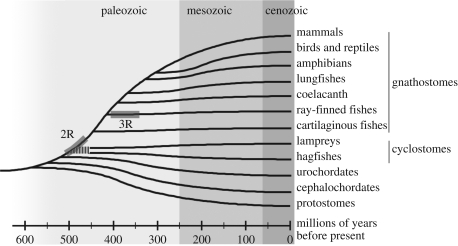
Figure 2.
Evolutionary tree for the different classes of gnathostomes (jawed vertebrates) in relation to cyclostomes (lampreys and hagfishes), urochordates (tunicates) and cephalochordates (lancelets). It is still unclear when the two genome-wide duplications (2R) took place in relation to the split of the cyclostomes from the gnathostomes. It is also still unclear whether lampreys and hagfishes are monophyletic or paraphyletic, which is why their deep branch is dashed in the figure. A third independent genome duplication (3R) took place in the teleost lineage.
It is still unclear when the two basal tetraploidizations occurred relative to the divergencies of the two cyclostome groups, the lampreys and the hagfishes (figure 2). It has been proposed that one tetraploidization took place before the cyclostomes branched off, whereupon the second tetraploidization took place in the lineage leading to the gnathostomes (Holland et al. 1994; Furlong & Holland 2002). Recently, it was suggested that both of the tetraploidizations happened before the divergence of cyclostomes (Kuraku et al. 2009), implying perhaps that more gene losses may have taken place in the cyclostome lineages. Another issue pertinent to this discussion is whether the cyclostome lineages are monophyletic or paraphyletic (Stadler et al. 2004; Lamb et al. 2007). Traditionally, the hagfishes have been considered to be the sister group of the vertebrates, and together with these comprise the craniates (animals with a skull). However, several studies suggest that the hagfishes and lampreys are monophyletic, together forming the sister group of the gnathostomes (Takezaki et al. 2003; Kuraku et al. 2009).
Nevertheless, the confirmation of the 2R scenario means that a huge number of gene duplicates was generated early in vertebrate evolution. A large proportion of the duplicates was presumably lost at an early stage after the tetraploidizations, and duplicates have continued to disappear in various lineages. However, it is likely that the extensive duplications resulting from the two more or less complete genome doublings gave rise to many new genes that facilitated the appearance of new functions (neo-functionalization) or more specific functions such as restricted gene expression (sub-functionalization), thereby providing raw material for emergence of new cell types with specialized functions (Arendt 2008).
Important for resolving the evolution of vertebrate genes is to have appropriate outgroups for comparison. For a long time the cephalochordates (lancelets, amphioxus) have been considered to be the sister group of craniates or vertebrates. The cephalochordates are comprised of more than 30 species of which the majority belongs to the genus Branchiostoma. Extensive sequence analyses have suggested that the cephalochordates are actually slightly more distantly related to the vertebrates than the urochordates (Delsuc et al. 2006). This was recently confirmed by the genome sequence of the Florida lancelet Branchiostoma floridae (Putnam et al. 2008).
2. Phototransduction Gene Families
Below we describe in detail each of the gene families involved in the phototransduction cascade. We primarily describe gene locations in the human genome. Chromosomes are abbreviated as Hsa for Homo sapiens and Gga for Gallus gallus, i.e. chicken. For phylogenetic trees we usually refer to our previous study (Nordström et al. 2004) or to trees available in the TreeFam database v. 6.0 (www.treefam.org) (Li et al. 2006; Ruan et al. 2008). The trees have been accessed between 15 December 2008 and 15 January 2009.
(a). Opsins
Several recent reviews have focused on the evolution of opsins both within the vertebrates themselves (Lamb et al. 2007; Bowmaker 2008) and across the animal kingdom (Oakley 2003; Terakita 2005). As is well known, rhodopsin (RHO) is expressed in rods and the colour vision opsins are typically specific for distinct types of cones. However, some overlap of expression may occur; mice express the two cone opsins called S (ancestral UV, OPN1SW) and L (OPN1LW) in the same cones in a large part of the retina (Applebury et al. 2000). Note that the long-wavelength opsin was called M in the cited reference. Rabbits and guinea pigs also co-express two opsins (S and L) in their cones, but not as extensively as shown for the mouse. Co-expression of these opsins is not observed in adult rats and gerbils, but transiently during development (Szel et al. 2000).
Our initial analyses indicated that human RHO (RH1), OPN1SW (blue = ancestral ultraviolet), and OPN1LW (red) occupy corresponding positions on the three chromosomes Hsa3, Hsa7 and HsaX, suggesting that they might have arisen from a common ancestral opsin by chromosome duplication (Nordström et al. 2004). However, this interpretation was too simplistic because analyses from a broad range of vertebrates have shown that the ancestral gnathostome had five visual opsin genes: RHO, RH2 (ancestral vertebrate green), OPN1SW (ancestral ultraviolet), OPN1SW2 (blue) and OPN1LW (red) (Bowmaker 2008). Furthermore, chromosomal data in teleost fishes show that OPN1SW2 and OPN1LW are syntenic (i.e. they are located in the same chromosome region). Because phylogenetic sequence analyses show that the earliest branching in the visual opsin tree involves these two genes (Bowmaker 2008), the ancestral chromosome probably contained both of these genes before the chromosome quadruplication took place. If so, all duplicates of OPN1LW must have been lost as shown in the hypothetical chromosome duplication scheme shown in figure 3. However, the chromosomal locations of the visual opsin genes in teleost fishes seem to add further complexity to their evolutionary history, implying that even the duplication scheme proposed in figure 3 is too simplistic.
Figure 3.
Chromosomal locations of vertebrate visual opsin gene family members in the human genome (Hsa for Homo sapiens) and postulated positions for the lost ancestral blue and green opsins. The proposed position for the ancient green opsin (RHO2) is confirmed by the present location of the orthologue in the chicken genome in a region with conserved synteny with Hsa1. Because phylogenetic trees for vertebrate visual opsins have OPN1LW and OPN1SW as the most basal divergence (Bowmaker 2008), we postulate that there was an ancestral gene pair before the chromosome quadruplication. However, other scenarios are possible. This hypothetical chromosome duplication scenario needs to be investigated further by studies in genomes of representatives from additional vertebrate classes. The transducin α subunit (GNAT) involved in phototransduction is located in the same paralogon, with GNAT1 expressed in rods, and GNAT2 in cones.
Additionally, some of the adjacent gene families in these chromosome regions need to be revised. As recently noted by Kuraku et al. (2009), the PLXN gene family is not a single series of paralogues, but in fact involves an ancestral gene pair with the ancestor of the PLXNA family and the ancestor of the PLXNB family. After considering this as well as additional information, the chromosome duplication scenario actually receives even more support from adjacent gene families as shown in figure 3 than it did in our previous study (Nordström et al. 2004).
The evolution of vertebrate visual opsins is further complicated by the fact that even though the cephalochordate amphioxus has five ‘ciliary’ opsins, all these are related to the encephalopsin/TMT-opsin subfamily, and no orthologues of the vertebrate visual opsins (including RHO) were identified (Holland et al. 2008) in a recent analysis of its genome. Opsins similar to the vertebrate ancient and RPE-retinal G-protein receptor (RGR) opsins have been identified in Ciona intestinalis (Kusakabe et al. 2001; Horie et al. 2008). To resolve this, chromosomal information is needed for tunicates, amphioxus and the cyclostome groups of hagfishes and lampreys, as well as the earliest gnathostome branch comprised of the cartilaginous fishes.
(b). Transducins α, β and γ
Transducins are trimeric G proteins found in photoreceptor cells. Each G protein consists of unrelated α, β and γ subunits and each of these has multiple members in vertebrate genomes. In mammals 16 α (GNA) subunit genes (Birnbaumer 2007) have been identified that are sorted into subfamilies based on sequence phylogeny (Nordström et al. 2004) and functional coupling (Birnbaumer 2007). Two transducin α genes (GNAT) are expressed in the retina of mammals where they stimulate the activity of phosphodiesterase, with GNAT1 expressed in rods and GNAT2 in cones (Downes & Gautam 1999). A third member, GNAT3 (gustducin), is expressed in taste cells. The two transducins and gustducin are almost equally closely related to each other (Downes & Gautam 1999). Interestingly, the gustducin gene has been found to be involved in the phototransduction cascade of the lizard parietal eye, whereas the transducins were not (Su et al. 2006). Each of the three genes is located close to a Giα (G-protein inhibitory α subunit; GNAI) subunit gene and these are also equally distantly related to each other (Downes & Gautam 1999). The GNAT and GNAI genes have the same exon–intron organization (Peng et al. 1992). This led us to propose that an ancestral gene pair consisting of one GNAT gene and one GNAI gene was duplicated twice to give the three pairs on Hsa1p, 3p and 7q (see Nordström et al. 2004). These chromosome arms, together with Hsa12p, form a paralogon containing numerous other gene families.
Early work suggested that the α subunit was responsible for the signalling of each G protein, but recently the roles of the β and γ subunits have received intense interest (for a recent review, see Birnbaumer 2007). The G-protein β subunit gene family consists of seven human members (Downes & Gautam 1999). Members GNB1 to GNB4 are approximately equally closely related to each other (figure 4), whereas GNB5 is more distantly related and GNB1L and GNB2L1 even more remote (Nordström et al. 2004). GNB1–4 show broad tissue distribution with differential expression in retinal cell types. Rods and amacrine cells express GNB1 while cones and bipolar cells express GNB3. GNB1–4 diverged from each other after the vertebrate lineage split from the urochordate lineage represented by two tunicates in figure 4 (see also Nordström et al. 2004). GNB1–4 (figure 5) are located on Hsa 1p, 3q, 7q and 12p and are flanked by many other gene families in this paralogon. Also GNB5 is expressed in rods as well as in the brain (Watson et al. 1996). Its role in phototransduction is little known but it has been reported to contribute to rod adaptation because knock-out mice show reduced G-protein deactivation (Krispel et al. 2003). GNB5 seems to have no close relatives resulting from 2R.
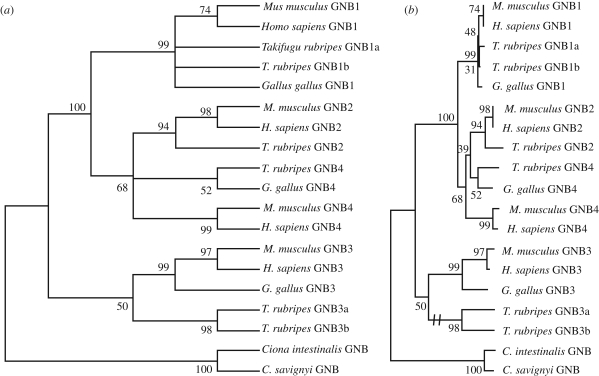
Figure 4.
Phylogenetic trees for the G-protein β subunit family GNB1–4. The species included are two mammals (human and mouse), one bird (chicken) and a pufferfish (Takifugu rubripes). As outgroup the GNB gene of the tunicates Ciona intestinalis and Ciona savignyi were used. The trees were calculated using the neighbour-joining method as implemented in Mega 4.0 (Tamura et al. 2007) with default settings. Panel (a) shows the obtained tree collapsed for nodes with less than 50 per cent bootstrap support. Panel (b) shows the same tree with branch lengths representing evolutionary distance. The two T. rubripes genes GNB3a and GNB3b have evolved at a much higher rate than all of the other sequences and their common branch has been shortened as shown by the double dash in panel (b).
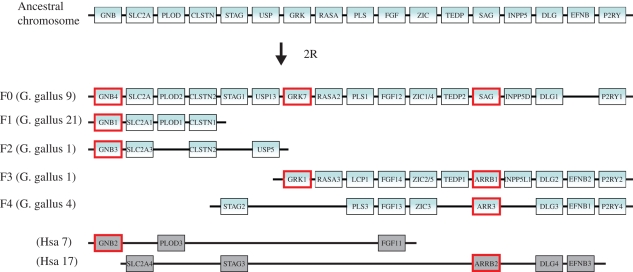
Figure 5.
Chromosomal locations of vertebrate phototransduction gene families GNB1/2/3/4, GRK1/7 and Arrestins/SAG together with several adjacent gene families in the chicken genome (Gga for Gallus gallus) and in the human genome (Hsa for Homo sapiens) in paralogon F as named by (Nakatani et al. 2007). The phototransduction genes are highlighted by red boxes. Rods express GNB1, GRK1 and SAG, and cones express GNB3, GRK7 and ARR3. The top chromosome shows the deduced ancestral chromosome that gave rise to the duplicated chromosomal segments in gnathostomes, represented here by the chicken. The five chicken chromosomes together account for three copies of the ancestral chromosome (further analyses in additional species are required to say whether segment F1 shall be combined with F3 or F4). As the GNB2 gene and the ARRB2 gene are missing in the chicken genome database, the human orthologues and their chromosomal regions are shown instead. Numerous rearrangements have occurred after the chromosome quadruplication. Note that the gene order has been re-shuffled to highlight the similarity between the chromosomes.
The G-protein γ subunit family consists of 12 mammalian members (Serb & Oakley 2005; Birnbaumer 2007) and many of these are expressed in the brain. Distinct γ subunit genes are used in rods and cones: GNGT1 and GNGT2. Whereas mammalian GNGT1 is expressed exclusively in the eye, the zebrafish orthologue is expressed both in the pineal gland and retina, but driven by different transcriptions factors (Ferguson et al. 1998). The human GNGT1 and GNGT2 genes are located on human chromosomes 7q and 17q, respectively, thereby belonging to the extended Hox paralogon (Nordström et al. 2004; Sundström et al. 2008). GNGT1 shows a low level of identity (<80%) compared to that observed for other γ subtypes that typically show greater than 90 per cent amino acid identity between teleosts and mammals (Leung et al. 2006). This lower level of identity may reflect adaptation to different environmental conditions. Alternatively, more rapid divergence of mammalian GNGT1 may have been facilitated by a duplication that generated a gene adjacently on the same chromosome (Hsa7q) called GNG11 (Kaupp & Seifert 2002). GNG11 probably arose in a mammalian ancestor and is highly expressed in a variety of tissues such as the pineal gland, the heart, lung and skeletal muscle (Balcueva et al. 2000; Chen et al. 2007).
(c). Phosphodiesterases
The phosphodiesterases (PDEs) form a superfamily with 21 members in mammals that are classified into 11 families (Omori & Kotera 2007). Some of these enzymes hydrolyze cGMP to GMP, whereas others hydrolyze cAMP to AMP. The retina expresses the three members of a PDE family called PDE6A–C, all of which act on cGMP. Many of the PDEs function as dimers, including the PDE6 family. The PDE6A and PDE6B genes encode the subunits PDE6α and PDE6β, respectively, that form a PDEαβ heterodimer in rods. The PDE6C gene gives rise to the PDE6α′ subunit which is expressed in cones and forms a homodimer, PDE6α′α′ (see Cote 2004 for review).
We previously reported (Nordström et al. 2004) that the three PDE6 protein sequences are more closely related to each other than to all other members of the PDE superfamily (see also Omori & Kotera 2007). The duplications that gave rise to the three copies took place in early vertebrate evolution as supported by sequence-based phylogeny, taxonomic distribution in vertebrate classes, and chromosomal positions of the three genes in the large paralogon described (see fig. 3 in Lundin et al. 2003), consisting largely of human chromosomes 4, 5, 2/8 and 10. This corresponds to paralogon C in Nakatani et al. (2007) and part of this paralogon has been described in detail in several species in connection with the NPY receptor family (Larsson et al. 2008).
The enzymatically active PDE6 dimer is inhibited by two copies of a γ subunit. Activation of PDE6 takes place when two transducin α subunits bind and remove the γ subunits. The rod γ subunit is encoded by a gene called PDE6G, whereas cones have a distinct γ subunit encoded by the PDE6H gene (Cote 2004). Sequence-based phylogeny suggests that the two genes resulted from a duplication in early vertebrate evolution. The genes are located on Hsa12 and Hsa17, the same chromosomes as two of the Hox clusters, albeit some distance away from the Hox clusters. However, the PDE6 γ genes are closer to the Hox-cluster genes in the dog genome (data not shown), suggesting that they may be part of the same chromosome duplication events, but more detailed studies involving additional species are necessary to confirm this.
In addition to cones and rods, PDE has been detected in bipolar cells, namely PDE1C in ON bipolars (Dhingra et al. 2008). However, because PDE inhibitors do not affect the response of ON bipolars (Nawy 1999), it has been suggested that PDE has a modulatory role in these cells and that their glutamate receptor mGluR6 activates Gαo to regulate ion channels (Dhingra et al. 2008).
(d). Cyclic nucleotide-gated ion channels
There are six members of the CNG gene family in mammalian genomes. Four of these encode α subunits (CNGA1–CNGA4) and two encode β subunits (CNGB1 and CNGB3) (Kaupp & Seifert 2002). Functional ion channels are heterotetramers, in rods and cones consisting of three CNGA subunits and one CNGB subunit (Zheng et al. 2002). Rods and cones use distinct subunits with rods having CNGA1 and CNGB1, whereas cone channels consist of CNGA3 and CNGB3 (Paillart et al. 2006). Olfactory neurons in contrast use three distinct subunits: CNGA2, CNGA4 and CNGB1b with two copies of the first-mentioned (Zheng et al. 2002). The CNG channels are non-selective cation channels, thus allowing influx of both Na+ and Ca2+.
Our previous analyses showed that the duplication that led to the α and β lineage took place before the protostome–deuterostome divergence (Nordström et al. 2004). The phylogenetic trees showed expansion of both lineages in early vertebrate evolution and this receives further support from data from additional vertebrate genomes in the TreeFam database (see also Paillart et al. 2006). Although quadruplicate resemblance could be detected for the four CNGA genes, they are not located in any of the large paralogons described in detail (Nordström et al. 2004). Our subsequent analyses suggest that some of the CNGA genes are located close to a few other gene families that seem to have been translocated after the chromosome duplications (data not shown), thereby obscuring the paralogon relationship. The two CNGB genes are the only survivors of the chromosome quadruplications that involved segments that are located on human chromosomes 8, 16, 18 and 20. Again, translocations make analyses complicated but it seems like these chromosome regions share an evolutionary history with the paralogon containing the four opioid receptor genes and several other gene families on Hsa8 and 20 (the CNGB genes are located close to these clusters), also involving Hsa1 and 6 (Dreborg et al. 2008), presumably due to chromosomal rearrangements.
Some teleost fishes have duplicates of both CNGA3 and CNGB3 (see TreeFam) and it will be interesting to see if these duplicates have become specialized by restriction of their expression to distinct cone types in the teleosts that have more colour opsins than mammals.
(e). Guanylyl cyclases
Many cell types possess the ability to produce cGMP via GC, either by soluble cytosolic GCs or by membrane GCs. The membrane-penetrating GC proteins (mGC) can be further subdivided into two major categories, those that serve as receptors for extracellular ligands and those that are activated by intracellular proteins (Sharma 2002). The last-mentioned type harbours three members, two of which are expressed in the retina. Initially named ROS-GC1 and ROS-GC2 for rod outer segment-GCs, these two enzymes have also been given other names: ROS-GC1 is also called GC-D or GUCY2D, whereas ROS-GC2 is also called GC-F or GUCY2F in human and several other mammals. A third membrane-bound GC was discovered in rat olfactory sensory neurons and was, unfortunately, named D, and the rodent orthologue of human GUCYD is called GUCY2E. The proposed human orthologue of rodent GC-D is called GC-E (Yamagami & Suzuki 2005). Further confusion arises because the third form in chicken has been named F. These non-receptor mGCs function as homodimers (Sharma 2002).
Both GUCY2D and GUCY2F are expressed in both rods and cones in the mammals that have been investigated. Form D predominates and in bovine rods it has been found to be expressed at 25-fold higher level than form F (Helten et al. 2007). The GC mutations that have been identified in human are in the GUCY2D gene (Kitiratschky et al. 2008) and affect both rods and cones. GUCY2D (ROS-GC1) is also expressed in some non-retinal tissues, namely the pineal gland (Venkataraman et al. 2000), which is of course related to the retina by expression of several other phototransduction proteins, and the mitral cells of the olfactory bulb (Duda et al. 2001). On the other hand, expression of GUCY2F (ROS-GC2) has, to our knowledge, not been detected outside of rods and cones .
Phylogenetic analyses suggest that the three GUCY2 genes arose in early vertebrate evolution, possibly in 2R. The human genes D and F are located on chromosomes Hsa17p and Xq, respectively. The human gene GUCY2E is on Hsa11q. These regions are not immediately obvious to belong to a paralogon resulting from 2R. More detailed studies of neighbouring genes are required to explore this possibility. The GUCY2D gene seems to be duplicated in four of the five fish genomes that are available and these duplicates display a phylogeny compatible with origin in 3R. This needs to be confirmed by chromosomal analyses. Also GUCY2F is duplicated in fish genomes (Imanishi et al. 2004).
Finally, it should be noted that also soluble GCs are expressed in the retina. In the rat retina expression was detected in ganglion cells and some bipolar and amacrine cells, but not in rods and cones (Ding & Weinberg 2007).
(f). G-protein-coupled receptor kinases
GRKs are Ser/Thr kinases that phosphorylate cytoplasmic parts of G-protein-coupled receptors and reduce their signalling to G proteins. The human genome contains seven GRK genes with GRK1 (previously called rhodopsin kinase, RHOK) predominantly expressed in rods and GRK7 (previously GPRK7) in cones, albeit with some interesting species differences. GRK1 is expressed mainly in rods but also in cones in humans (Zhao et al. 1998), as in monkey (Sears et al. 2000; Weiss et al. 2001) and mouse (Lyubarsky et al. 2000) retinas, but not in the cones of dog and pig retinas (Weiss et al. 2001). GRK1 is expressed exclusively in rods of the medaka fish (Hisatomi et al. 1998). In contrast, GRK7 is concentrated in cones in both cone- and rod-dominant mammals (Weiss et al. 2001) and in fish (Hisatomi et al. 1998). An intriguing exception is that the expression of GRK7 is not detectable in mouse and rat retinas, nor is the GRK7 sequence present in mouse or rat cDNA or mouse genomic libraries and deposited genomic sequences (Weiss et al. 2001), although it is present in the guinea pig and rabbit genomes. GRK1 and GRK7 are attached to the membrane by an isoprenyl moiety (Inglese et al. 1992; Zhang et al. 2004).
In our earlier phylogenetic analyses, these two retinal GRKs did not branch together although their chromosomal positions suggested that they arose from a common ancestral gene in 2R (Nordström et al. 2004). More extensive data from various vertebrate genomes now add further support for their origin in early vertebrate evolution, as shown by their presence in both teleosts and tetrapods (Wada et al. 2006). Furthermore, their chromosomal locations can now be assigned to agree with paralogon F (Nakatani et al. 2007) with extensive regions of similarity (i.e. representatives from the same gene families) on Hsa3 (GRK7) and Hsa13 (GRK1) as well as HsaX (Nordström et al. 2004) and Hsa 1, 2 and 11 (Nakatani et al. 2007; see figure 5). These chromosome regions form a paralogon that also contains the GNB and arrestin gene families (figure 5). Teleost fishes have duplicates of both GRK1 and GRK7 that seem to agree with 3R. Furthermore, there is a third member forming a basal branch for this GRK family in teleost fishes (Wada et al. 2006; Imanishi et al. 2007) that may have been lost in mammals but appears to be present in chicken (Imanishi et al. 2007) and which appears not to be duplicated in teleost-specific tetraploidization 3R. More detailed analyses of chromosomal location in different teleost genomes and in the chicken genome are required to determine if this gene is a third product of 2R in addition to GRK1 and GRK7. If so, the names of some of the members of this gene family will probably require revision to comply with orthology.
The remaining five GRKs in the human genome belong to two other paralogons. GRKs 4, 5 and 6 (Nordström et al. 2004) are located in the NPY receptor paralogon involving Hsa4, 5, 2/8 and 10 (Larsson et al. 2008), i.e. paralogon C in Nakatani et al. (2007). GRK4 was previously called GPRK2L. GRKs 2 and 3 (formerly called β-adrenergic receptor kinases 1 and 2, abbreviated ADRBK1 and 2) are located on Hsa11 and 22, respectively, and may belong to paralogon H (Nakatani et al. 2007). Both GRK2 and GRK3 are present in the genomes of mammals and chicken. However, more extensive analyses are required to confirm their duplication before the radiation of gnathostomes.
(g). Arrestins (ARR)
Arrestins bind to phosphorylated G-protein-coupled receptors to reduce or arrest their signalling and may also signal internalization of cell-surface receptors as well as induce other signal transduction pathways than the G-protein-mediated signalling (Lefkowitz & Shenoy 2005). Arrestin undergoes translocation to the membrane with which it seems to interact by means of an amphipathic helix (Han et al. 2001). Mammalian genomes encode four arrestins (Gurevich & Gurevich 2006). One is called SAG (S antigen) and is expressed in rods where it terminates RHO signalling. ARR3 is expressed in cones and inhibits cone opsins. ARRB1 and ARRB2 are expressed in other organs. The phylogenetic tree for the arrestins suggested quadruplication in early vertebrate evolution (Nordström et al. 2004) as supported by the location of most of the family members as well as several adjacent gene family members in a paralogon. As shown in figure 5, SAG and ARR3 belong to gene regions with extensive similarity between chromosomes also in chicken and human, in line with 2R.
Both SAG and ARR3, as well as ARRB2, have been duplicated in teleost fishes, presumably in 3R (see TreeFam), thereby perhaps allowing for further subfunctionalization, particularly for ARR3 in different cone types as discussed above for CNGA and CNGB.
(h). Recoverin (RCVRN)
Recoverin is a negative regulator of GRK1, i.e. RHOK (Chen 2002; Makino et al. 2004). It contains a Ca2+-binding domain with similarity to calmodulin (Ames & Ikura 2002) and is attached to membrane by a myristoyl group upon Ca2+ binding (Desmeules et al. 2002). The number of members of the RCVRN family depends on what criteria are used to define a family. One classification encompasses recoverin itself plus the five human hippocalcin (HPCA)-like proteins, one of which is neurocalcin delta (NCALD), see the TreeFam database. Recoverin is expressed only in the retina (De Raad et al. 1995). Also NCALD is expressed in the retina in amacrine and ganglion cells, but not in rods and cones (Krishnan et al. 2004). NCALD is also expressed in many other organs (Krishnan et al. 2004). NCALD is also activated by a Ca2+-myristoyl switch (Krishnan et al. 2004) that allows it to stimulate GC.
In our previous analyses we proposed that there was a local gene duplication of the ancestral hippocalcin gene before 2R, whereupon the tetraploidizations gave rise to three additional copies (Nordström et al. 2004). Recoverin, on the other hand, had no duplicates in any tetrapod in our earlier studies. The phylogenetic tree of representative sequences (fig. 9 in Nordström et al. 2004) indicated a duplication before the actinopterygian–sarcopterygian divergence, but none of the tetrapods included in the analysis possessed two gene copies. Only the pufferfish Takifugu rubripes displayed multiple gene copies but we considered this insufficient to propose a duplication in 2R. More recent information from additional teleost species now indeed confirms a duplication preceding the split of the lineages leading to tetrapods and teleost fishes because both the frog Xenopus tropicalis and several teleosts have both of these genes (called RCVRN and RCV1); see TreeFam. (Note that RCV1 is sometimes used in the literature for recoverin itself; here we use RCV1 for the X. tropicalis protein SwissProt P35243). Unfortunately, the X. tropicalis genome has not yet been assembled to large linkage groups, let alone chromosomes, so it is still too early to say whether the chromosomal locations confirm duplication in 2R. Hopefully, the teleost genomes may shed light on this (analyses in progress). Interestingly, also the platypus has the second recoverin gene RCV1, but it lacks an orthologue of RCVRN itself (Warren et al. 2008). Thus, it seems like placental mammals have lost the early vertebrate duplicate of the recoverin gene quite recently. In addition, the early recoverin duplicate RCV1 has been duplicated in the teleosts, possibly as a result of 3R. Naturally, it will be highly interesting to see which retinal cells express the RCV1 gene(s) and if there has been sub- or neo-functionalization relative to the RCVRN gene.
(i). Guanylyl cyclase-activating proteins
More distant relatives of the recoverin superfamily are the GUCAs (also called GCAP), which can be considered a separate family. We used these remote relatives to root the phylogenetic tree in our previous analysis of the recoverin superfamily (Nordström et al. 2004). The GUCA family members possess Ca2+-binding domains like other relatives of the calmodulin superfamily and perform the function that their name describes, i.e. they activate GCs. Three GUCA genes have been identified in mammalian genomes. GUCA1A (GCAP1) and GUCA1B (GCAP2) activate the two GCs present in rods and cones (Palczewski et al. 2004). The third member, GUCA1C (GCAP3), was discovered in human retina (Haeseleer et al. 1999) but is a pseudogene in mouse (Imanishi et al. 2002) although it is present in platypus, chicken, the frog X. tropicalis and in duplicate in teleosts (see TreeFam). The GUCA1C gene is expressed in cones in human and zebrafish (Imanishi et al. 2002). The loss of a functional GUCA1C in mouse is compatible with their reduced dependence on cone-based (colour) vision. The GUCA1A and GUCA1B genes are located adjacent to each other on Hsa6 and the GUCA1C gene is on Hsa3 and is more closely related to GUCA1A according to phylogenetic analyses (Imanishi et al. 2004), suggesting that the local duplication on Hsa6 took place before GUCA1C arose from GUCA1A. Although the phylogenies suggest duplications in early vertebrate evolution, it is not apparent that these regions of human chromosomes 3 and 6 were duplicated in 2R.
Additional GUCA gene copies exist in teleost fishes with a total of six in zebrafish and seven and eight in the pufferfishes, T. rubripes and Tetraodon nigroviridis, respectively. Some of these duplicates are likely to have arisen in 3R, but more detailed analyses of their chromosomal locations as well as sequence phylogenies are required to resolve their evolutionary relationships.
3. Discussion
The retina is thought to have undergone rapid evolution at the dawn of the vertebrates with dramatic increase in the number of visual ciliary opsins from one to five (Bowmaker 2008). Our previous simplistic model for opsin gene duplications by chromosome duplication is elaborated here to comply with data from several additional species representing the various vertebrate classes. The scheme proposed in figure 3 is speculative and involves several gene losses, but is compatible both with the phylogenetic tree for vertebrate opsins and the chromosomal locations of the opsin genes in mammals and chicken. Note also that the transducin α subunit genes (GNAT) are located in these chromosome regions together with the GNAI genes, suggesting that GNAI and GNAT were generated from a common ancestral GNA gene by a local duplication prior to the chromosome duplications. Because a GNA that inhibits adenylyl cyclase is present also in invertebrates, it seems that GNAI has retained the ancestral properties after the gene duplication, whereas GNAT evolved coupling to PDE.
For a gene family like the opsins, which is subjected to strong selective forces, it should be expected that the evolutionary rate may be somewhat uneven and therefore give rise to phylogenetic trees that do not necessarily reflect the actual order of events for the gene duplications. Chromosomal location may therefore be a useful complement to disentangle the evolutionary history. However, data from more species, particularly actinopterygian (ray-finned) fishes and cartilaginous fishes, as well as the various pre-gnathostomes, is required before more definitive conclusions can be drawn. Indeed, preliminary analyses of teleosts indicate that the scheme proposed in figure 3 perhaps does not give the whole story.
The duplications of the opsin genes seem to have been reasonably concomitant with the diversification of retinal cell types, not only the cones and rods which express the various opsins but also many other retinal cell types. Our analyses reported previously (Nordström et al. 2004) and expanded here show that a large number of gene families besides the opsins received additional copies in the two tetraploidizations preceding the origin and radiation of the gnathostomes.
Of the 13 gene families in the phototransduction cascade investigated here, no less than 10 seem to have expanded in 2R, namely the opsins, G-protein (transducin) α, β and γ subunits (GNAT, GNBT and GNGT), PDE6 α/β/α′ subunits (PDE6A, B and C) and γ subunits (PDE6G and H), CNG α and β subunits, G-protein-coupled receptor kinases (GRK1 and 7) and arrestins (ARR3 and SAG). Note that the CNG α and β subunits can be considered as two separate gene families because both existed before the origin of the gnathostomes (in our previous study (Nordström et al. 2004) we regarded them as a single family). The remaining three gene families, GCs (GUCY), GC-activating proteins (GUCA) and recoverin (RCVRN and RCV1) also received additional members during this phase of (pre)vertebrate evolution but the gene duplicates either do not seem to be located in any of the more well-characterized paralogons (GUCY and GUCA) or have not yet been investigated in detail (RCVRN–RCV1). However, it cannot be ruled out that also these gene families expanded as a result of 2R. As an extension of the present analysis, also the GNB5 gene may be considered although its role in cones and rods may be minor relative to its functions elsewhere as indicated by its wide distribution in the brain as well as in other organs in human and mouse (Jones et al. 1998). GNB5 is distantly related to GNB1–4 and does not seem to have any surviving 2R duplicates.
Interestingly, the only retinal gene that did not seem to have been duplicated in our previous study (Nordström et al. 2004), namely recoverin, has indeed also been duplicated, but the duplicate RCV1 has been lost in placental mammals. It would be interesting to try to deduce if the loss of RCV1 coincides with loss of any cell types in the mammalian retina.
Most, if not all, of these gene families have also expanded in the teleost fish lineage through the teleost-specific tetraploidization, 3R. Because many teleosts have retained more of the original retinal complexity of the gnathostome ancestor when compared with the degenerate mammals (that have lost two of the ancestral four colour vision opsins), they may have been able to benefit from the additional 3R duplicates by evolving additional functions or abilities by either neo- or sub-functionalization. It should perhaps be expected that all these additional genes might have led to great divergence of the retinas in the various teleost fish lineage. More detailed comparative studies of, for instance, cell type-specific gene expression using the various gene family members and multiple fish species will therefore be highly interesting.
An intriguing finding was that the rhodopsin gene expressed in the rods of ray-finned fishes lacks introns, whereas an intron-containing rhodopsin gene is expressed in the pineal gland of teleost fishes. It was proposed that the intron-less rhodopsin gene arose early in ray-finned fish evolution by retrotransposition (Bellingham et al. 2003). More recent results suggest that at least the zebrafish has one additional intron-containing rhodopsin gene whose expression pattern remains to be investigated.
Some of the gene families seem to have undergone no obvious subfunctionalization of the daughter genes. The two GCs GUCY2D and GUCY2F are expressed both in cones and rods. However, the level of expression differs considerably, with form D being expressed at 25-fold higher level than F in bovine rods (Helten et al. 2007). It will be interesting to see if other vertebrate taxa have different proportions between the two forms. Another pair of 2R daughter genes shows variable regulation across mammals, namely GRK1 and GRK7, as detailed above, indicating that changes in the regulation of expression have taken place long after the duplication.
In our previous study, we noted that some of the retinal gene families might have been located in the same paralogons. However, some of the gene families seemed to have been quite far apart in their respective chromosome regions making coordinated gene regulation unlikely. While some of these expanded paralogons may have been prematurely assigned due to lack of information from species other than human, our present analyses have identified another expanded paralogon harbouring three distinct retinal gene families, namely GNB, GRK and ARR as shown in figure 5. It still seems far-fetched to assume coordinated gene regulation, particularly in consideration of the numerous intra-chromosomal rearrangements that have scrambled the order of the gene families. On the other hand, duplication by tetraploidization has been proposed to allow equilibrium to be maintained in biochemical pathways and may subsequently be followed by more gradual changes in the levels, sites or time points of expression that may result in specialization.
One intriguing possibility for specialization involves the pineal gland. Several of the genes investigated are expressed both in the retina and the pineal gland, namely GNAT1, arrestin, recoverin, RHOK and GUCY2D (Takano et al. 2003). However, it is a bit surprising to see no example, to our knowledge, of a gene family with one 2R product exclusively expressed in the pineal gland and one or more specifically expressed in the retina. The pineal opsin expressed in the chicken pineal gland, pinopsin (Downes & Gautam 1999), is the result of a duplication preceding 2R. This is compatible with the idea that the origin of the pineal gland precedes 2R because amphioxus has a putative pineal homologue, the dorsal lamellar body (Lacalli 2008). The mammalian G-protein γ subunit GNG11 is expressed in the pineal and elsewhere, but not in the retina (Balcueva et al. 2000), but this is a much more recent duplicate than 2R.
4. Conclusions
These extended analyses of the previously described phototransduction gene families corroborate their expansion as a result of the basal vertebrate tetraploidizations, 2R. Furthermore, we have described here investigations of additional phototransduction gene families that follow the same pattern. Thus, the 2R events seem to have paved the way for the evolution of elaborate vision in the vertebrates by offering an expanded gene repertoire that could be selected for novel or more highly specialized functions. Presumably, the 3R tetraploidization in the teleost fish ancestor contributed additional genetic raw material for functional specialization in this lineage. These observations suggest that the basal vertebrate tetraploidizations may have contributed to functional specialization also in other organs.
Acknowledgements
We thank Susanne Dreborg for tentative analyses of some of the gene families and Görel Sundström and Ola Gustafsson for analyses of the opsins and their paralogon. We are grateful to Lars G. Lundin for critical reading of the manuscript. This work was supported by grants from the Swedish Research Council and the Australian Research Council (DP0880983).
Footnotes
One contribution of 13 to a Theme Issue ‘The evolution of phototransduction and eyes’.
References
- Abi-Rached L., Gilles A., Shiina T., Pontarotti P., Inoko H.2002Evidence of en bloc duplication in vertebrate genomes. Nat. Genet. 31, 100–105 (doi:10.1038/ng855) [DOI] [PubMed] [Google Scholar]
- Ames J. B., Ikura M.2002Structure and membrane-targeting mechanism of retinal Ca2+-binding proteins, recoverin and GCAP-2. Adv. Exp. Med. Biol. 514, 333–348 [DOI] [PubMed] [Google Scholar]
- Applebury M. L., et al. 2000The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27, 513–523 (doi:10.1016/S0896-6273(00)00062-3) [DOI] [PubMed] [Google Scholar]
- Arendt D.2008The evolution of cell types in animals: emerging principles from molecular studies. Nat. Rev. Genet. 9, 868–882 (doi:10.1038/nrg2416) [DOI] [PubMed] [Google Scholar]
- Balcueva E. A., Wang Q., Hughes H., Kunsch C., Yu Z., Robishaw J. D.2000Human G protein gamma(11) and gamma(14) subtypes define a new functional subclass. Exp. Cell Res. 257, 310–319 (doi:10.1006/excr.2000.4893) [DOI] [PubMed] [Google Scholar]
- Bellingham J., Tarttelin E. E., Foster R. G., Wells D. J.2003Structure and evolution of the teleost extraretinal rod-like opsin (errlo) and ocular rod opsin (rho) genes: is teleost rho a retrogene? J. Exp. Zool. B Mol. Dev. Evol. 297, 1–10 [DOI] [PubMed] [Google Scholar]
- Birnbaumer L.2007Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 alpha subunits plus betagamma dimers. Biochim. Biophys. Acta 1768, 772–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker J. K.2008Evolution of vertebrate visual pigments. Vision Res. 48, 2022–2041 (doi:10.1016/j.visres.2008.03.025) [DOI] [PubMed] [Google Scholar]
- Chen C. K.2002Recoverin and rhodopsin kinase. Adv. Exp. Med. Biol. 514, 101–107 [DOI] [PubMed] [Google Scholar]
- Chen H., Leung T., Giger K. E., Stauffer A. M., Humbert J. E., Sinha S., Horstick E. J., Hansen C. A., Robishaw J. D.2007Expression of the G protein gammaT1 subunit during zebrafish development. Gene Expr. Patterns 7, 574–583 (doi:10.1016/j.modgep.2007.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A., Koh E. G., Chia J. M., Brenner S., Aparicio S., Venkatesh B.2004Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21, 1146–1151 (doi:10.1093/molbev/msh114) [DOI] [PubMed] [Google Scholar]
- Cote R. H.2004Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int. J. Impot. Res. 16(Suppl. 1), S28–S33 [DOI] [PubMed] [Google Scholar]
- Coulier F., Popovici C., Villet R., Birnbaum D.2000MetaHOX gene clusters. J. Exp. Zool. 288, 345–351 (doi:10.1002/1097-010X(20001215)288:4<345::AID-JEZ7>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- De Raad S., Comte M., Nef P., Lenz S. E., Gundelfinger E. D., Cox J. A.1995Distribution pattern of three neural calcium-binding proteins (NCS-1, VILIP and recoverin) in chicken, bovine and rat retina. Histochem. J. 27, 524–535 [DOI] [PubMed] [Google Scholar]
- Dehal P., Boore J. L.2005Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314 (doi:10.1371/journal.pbio.0030314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H.2006Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (doi:10.1038/nature04336) [DOI] [PubMed] [Google Scholar]
- Desmeules P., Grandbois M., Bondarenko V. A., Yamazaki A., Salesse C.2002Measurement of membrane binding between recoverin, a calcium-myristoyl switch protein, and lipid bilayers by AFM-based force spectroscopy. Biophys. J. 82, 3343–3350 (doi:10.1016/S0006-3495(02)75674-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A., Sulaiman P., Xu Y., Fina M. E., Veh R. W., Vardi N.2008Probing neurochemical structure and function of retinal ON bipolar cells with a transgenic mouse. J. Comp. Neurol. 510, 484–496 (doi:10.1002/cne.21807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. D., Weinberg R. J.2007Distribution of soluble guanylyl cyclase in rat retina. J. Comp. Neurol. 502, 734–745 [PubMed] [Google Scholar]
- Dowling J. E., Werblin F. S.1969Organization of retina of the mudpuppy, Necturus maculosus. I. Synaptic structure. J. Neurophysiol. 32, 315–338 [DOI] [PubMed] [Google Scholar]
- Downes G. B., Gautam N.1999The G protein subunit gene families. Genomics 62, 544–552 (doi:10.1006/geno.1999.5992) [DOI] [PubMed] [Google Scholar]
- Dreborg S., Sundström G., Larsson T. A., Larhammar D.2008Evolution of vertebrate opioid receptors. Proc. Natl Acad. Sci. USA 105, 15 487–15 492 (doi:10.1073/pnas.0805590105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T., Venkataraman V., Krishnan A., Nagele R. G., Sharma R. K.2001Negatively calcium-modulated membrane guanylate cyclase signaling system in the rat olfactory bulb. Biochemistry 40, 4654–4662 (doi:10.1021/bi0027985) [DOI] [PubMed] [Google Scholar]
- Dyer M. A., Cepko C. L.2001Regulating proliferation during retinal development. Nat. Rev. Neurosci. 2, 333–342 (doi:10.1038/35072555) [DOI] [PubMed] [Google Scholar]
- Ferguson S. S., Zhang J., Barak L. S., Caron M. G.1998Molecular mechanisms of G protein-coupled receptor desensitization and resensitization. Life Sci. 62, 1561–1565 (doi:10.1016/S0024-3205(98)00107-6) [DOI] [PubMed] [Google Scholar]
- Fu Y., Yau K. W.2007Phototransduction in mouse rods and cones. Pflugers Arch. 454, 805–819 (doi:10.1007/s00424-006-0194-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong R. F., Holland P. W. H.2002Were vertebrates octoploid? Phil. Trans. R. Soc. Lond. B 357, 531–544 (doi:10.1098/rstb.2001.1035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernàndez J., Holland P. W. H.1994Archetypal organization of the amphioxus Hox gene cluster. Nature 370, 563–566 (doi:10.1038/370563a0) [DOI] [PubMed] [Google Scholar]
- Gurevich V. V., Gurevich E. V.2006The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 110, 465–502 (doi:10.1016/j.pharmthera.2005.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F., Sokal I., Li N., Pettenati M., Rao N., Bronson D., Wechter R., Baehr W., Palczewski K.1999Molecular characterization of a third member of the guanylyl cyclase-activating protein subfamily. J. Biol. Chem. 274, 6526–6535 (doi:10.1074/jbc.274.10.6526) [DOI] [PubMed] [Google Scholar]
- Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C.2001Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880 (doi:10.1016/S0969-2126(01)00644-X) [DOI] [PubMed] [Google Scholar]
- Hartline H. K.1938The response of single optic nerve fibers of the vertebrate eye to illumination of the retina. Am. J. Physiol. 121, 400–415 [Google Scholar]
- Helten A., Saftel W., Koch K. W.2007Expression level and activity profile of membrane bound guanylate cyclase type 2 in rod outer segments. J. Neurochem. 103, 1439–1446 (doi:10.1111/j.1471-4159.2007.04923.x) [DOI] [PubMed] [Google Scholar]
- Hisatomi O., Matsuda S., Satoh T., Kotaka S., Imanishi Y., Tokunaga F.1998A novel subtype of G-protein-coupled receptor kinase, GRK7, in teleost cone photoreceptors. FEBS Lett. 424, 159–164 (doi:10.1016/S0014-5793(98)00162-8) [DOI] [PubMed] [Google Scholar]
- Hoegg S., Meyer A.2005Hox clusters as models for vertebrate genome evolution. Trend Genet. 21, 421–424 (doi:10.1016/j.tig.2005.06.004) [DOI] [PubMed] [Google Scholar]
- Holland P. W. H., Garcia-Fernàndez J., Williams N. A., Sidow A.1994Gene duplications and the origins of vertebrate development. Development Supplement, 125–133 [PubMed] [Google Scholar]
- Holland L. Z., et al. 2008The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111 (doi:10.1101/gr.073676.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Sakurai D., Ohtsuki H., Terakita A., Shichida Y., Usukura J., Kusakabe T., Tsuda M.2008Pigmented and nonpigmented ocelli in the brain vesicle of the ascidian larva. J. Comp. Neurol. 509, 88–102 (doi:10.1002/cne.21733) [DOI] [PubMed] [Google Scholar]
- Huang L., Max M., Margolskee R. F., Su H., Masland R. H., Euler T.2003G protein subunit G gamma 13 is coexpressed with G alpha o, G beta 3, and G beta 4 in retinal ON bipolar cells. J. Comp. Neurol. 455, 1–10 (doi:10.1002/cne.10396) [DOI] [PubMed] [Google Scholar]
- Imanishi Y., Li N., Sokal I., Sowa M. E., Lichtarge O., Wensel T. G., Saperstein D. A., Baehr W., Palczewski K.2002Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur. J. Neurosci. 15, 63–78 (doi:10.1046/j.0953-816x.2001.01835.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi Y., Yang L., Sokal I., Filipek S., Palczewski K., Baehr W.2004Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1–8) in pufferfish (Fugu rubripes). J. Mol. Evol. 59, 204–217 (doi:10.1007/s00239-004-2614-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi Y., Hisatomi O., Yamamoto S., Satoh T., Kotaka S., Kobayashi Y., Tokunaga F.2007A third photoreceptor-specific GRK found in the retina of Oryzias latipes (Japanese killifish). Zool. Sci. 24, 87–93 (doi:10.2108/zsj.24.87) [DOI] [PubMed] [Google Scholar]
- Inglese J., Koch W. J., Caron M. G., Lefkowitz R. J.1992Isoprenylation in regulation of signal transduction by G-protein-coupled receptor kinases. Nature 359, 147–150 (doi:10.1038/359147a0) [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. 2004Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957 (doi:10.1038/nature03025) [DOI] [PubMed] [Google Scholar]
- Jones P. G., Lombardi S. J., Cockett M. I.1998Cloning and tissue distribution of the human G protein beta 5 cDNA. Biochim. Biophys. Acta 1402, 288–291 [DOI] [PubMed] [Google Scholar]
- Kaupp U. B., Seifert R.2002Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 [DOI] [PubMed] [Google Scholar]
- Kitiratschky V. B., Wilke R., Renner A. B., Kellner U., Vadala M., Birch D. G., Wissinger B., Zrenner E., Kohl S.2008Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest. Ophthalmol. Vis. Sci. 49, 5015–5023 (doi:10.1167/iovs.08-1901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Venkataraman V., Fik-Rymarkiewicz E., Duda T., Sharma R. K.2004Structural, biochemical, and functional characterization of the calcium sensor neurocalcin delta in the inner retinal neurons and its linkage with the rod outer segment membrane guanylate cyclase transduction system. Biochemistry 43, 2708–2723 (doi:10.1021/bi035631v) [DOI] [PubMed] [Google Scholar]
- Krispel C. M., Chen C. K., Simon M. I., Burns M. E.2003Prolonged photoresponses and defective adaptation in rods of Gbeta5−/− mice. J. Neurosci. 23, 6965–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S., Meyer A., Kuratani S.2009Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol. Biol. Evol. 26, 47–59 (doi:10.1093/molbev/msn222) [DOI] [PubMed] [Google Scholar]
- Kusakabe T., Kusakabe R., Kawakami I., Satou Y., Satoh N., Tsuda M.2001Ci-opsin1, a vertebrate-type opsin gene, expressed in the larval ocellus of the ascidian Ciona intestinalis. FEBS Lett. 506, 69–72 (doi:10.1016/S0014-5793(01)02877-0) [DOI] [PubMed] [Google Scholar]
- Lacalli T. C.2008Basic features of the ancestral chordate brain: a protochordate perspective. Brain Res. Bull. 75, 319–323 (doi:10.1016/j.brainresbull.2007.10.038) [DOI] [PubMed] [Google Scholar]
- Lamb T. D., Collin S. P., Pugh E. N., Jr2007Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci. 8, 960–976 (doi:10.1038/nrn2283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar D., Lundin L.-G., Hallböök F.2002The human Hox-bearing chromosome regions did arise by block or chromosome (or even genome) duplications. Genome Res. 12, 1910–1920 (doi:10.1101/gr.445702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson T. A., Olsson F., Sundström G., Lundin L. G., Brenner S., Venkatesh B., Larhammar D.2008Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol. Biol. 8, 184 (doi:10.1186/1471-2148-8-184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz R. J., Shenoy S. K.2005Transduction of receptor signals by beta-arrestins. Science 308, 512–517 (doi:10.1126/science.1109237) [DOI] [PubMed] [Google Scholar]
- Leung T., Chen H., Stauffer A. M., Giger K. E., Sinha S., Horstick E. J., Humbert J. E., Hansen C. A., Robishaw J. D.2006Zebrafish G protein gamma2 is required for VEGF signaling during angiogenesis. Blood 108, 160–166 (doi:10.1182/blood-2005-09-3706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. 2006TreeFam: a curated database of phylogenetic trees of animal gene families. Nucl. Acids Res. 34, D572–D580 (doi:10.1093/nar/gkj118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin L. G.1993Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16, 1–19 (doi:10.1006/geno.1993.1133) [DOI] [PubMed] [Google Scholar]
- Lundin L. G., Hallböök F., Larhammar D.2003Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J. Struct. Funct. Genom. 3, 53–63 (doi:10.1023/A:1022600813840) [PubMed] [Google Scholar]
- Lyubarsky A. L., Chen C., Simon M. I., Pugh E. N., Jr2000Mice lacking G-protein receptor kinase 1 have profoundly slowed recovery of cone-driven retinal responses. J. Neurosci. 20, 2209–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino C. L., Dodd R. L., Chen J., Burns M. E., Roca A., Simon M. I., Baylor D. A.2004Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 123, 729–741 (doi:10.1085/jgp.200308994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T., Gruss P.2002Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 25, 32–38 (doi:10.1016/S0166-2236(00)02028-2) [DOI] [PubMed] [Google Scholar]
- Miyata T., Suga H.2001Divergence pattern of animal gene families and relationship with the Cambrian explosion. BioEssays 23, 1018–1027 (doi:10.1002/bies.1147) [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Takeda H., Kohara Y., Morishita S.2007Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17, 1254–1265 (doi:10.1101/gr.6316407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy S.1999The metabotropic receptor mGluR6 may signal through G(o), but not phosphodiesterase, in retinal bipolar cells. J. Neurosci. 19, 2938–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Larsson T. A., Larhammar D.2004Extensive duplications of phototransduction genes in early vertebrate evolution correlate with block (chromosome) duplications. Genomics 83, 852–872 (doi:10.1016/j.ygeno.2003.11.008) [DOI] [PubMed] [Google Scholar]
- Oakley T. H.2003The eye as a replicating and diverging, modular developmental unit. Trends Ecol. Evol. 18, 623–627 (doi:10.1016/j.tree.2003.09.005) [Google Scholar]
- Ohno S.1970Evolution by gene duplication Berlin, Germany: Springer-Verlag [Google Scholar]
- Omori K., Kotera J.2007Overview of PDEs and their regulation. Circ. Res. 100, 309–327 (doi:10.1161/01.RES.0000256354.95791.f1) [DOI] [PubMed] [Google Scholar]
- Paillart C., Zhang K., Rebrik T. I., Baehr W., Korenbrot J. I.2006Cloning and molecular characterization of cGMP-gated ion channels from rod and cone photoreceptors of striped bass (M. saxatilis) retina. Vis. Neurosci. 23, 99–113 [DOI] [PubMed] [Google Scholar]
- Palczewski K., Sokal I., Baehr W.2004Guanylate cyclase-activating proteins: structure, function, and diversity. Biochem. Biophys. Res. Commun. 322, 1123–1130 (doi:10.1016/j.bbrc.2004.07.122) [DOI] [PubMed] [Google Scholar]
- Panopoulou G., Poustka A. J.2005Timing and mechanism of ancient vertebrate genome duplications—the adventure of a hypothesis. Trends Genet. 21, 559–567 (doi:10.1016/j.tig.2005.08.004) [DOI] [PubMed] [Google Scholar]
- Peng Y. W., Robishaw J. D., Levine M. A., Yau K. W.1992Retinal rods and cones have distinct G protein beta and gamma subunits. Proc. Natl Acad. Sci. USA 89, 10 882–10 886 (doi:10.1073/pnas.89.22.10882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki D. C., Oakley T. H.2007Key transitions during the evolution of animal phototransduction: novelty, ‘tree-thinking’, co-option, and co-duplication. Integr. Comp. Biol. 47, 759–769 (doi:10.1093/icb/icm050) [DOI] [PubMed] [Google Scholar]
- Postlethwait J. H., Woods I. G., Ngo-Hazelett P., Yan Y.-L., Kelly P. D., Chu F., Hill-Force A., Walbot W. S.2000Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10, 1890–1902 (doi:10.1101/gr.164800) [DOI] [PubMed] [Google Scholar]
- Putnam N. H., et al. 2008The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (doi:10.1038/nature06967) [DOI] [PubMed] [Google Scholar]
- Ruan J., et al. 2008TreeFam: 2008 Update. Nucl. Acids Res. 36, D735–D740 (doi:10.1093/nar/gkm1005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears S., Erickson A., Hendrickson A.2000The spatial and temporal expression of outer segment proteins during development of Macaca monkey cones. Invest. Ophthalmol. Vis. Sci. 41, 971–979 [PubMed] [Google Scholar]
- Serb J. M., Oakley T. H.2005Hierarchical phylogenetics as a quantitative analytical framework for evolutionary developmental biology. Bioessays 27, 1158–1166 (doi:10.1002/bies.20291) [DOI] [PubMed] [Google Scholar]
- Sharma R. K.2002Evolution of the membrane guanylate cyclase transduction system. Mol. Cell. Biochem. 230, 3–30 (doi:10.1023/A:1014280410459) [PubMed] [Google Scholar]
- Stadler P. F., Fried C., Prohaska S. J., Bailey W. J., Misof B. Y., Ruddle F. H., Wagner G. P.2004Evidence for independent Hox gene duplications in the hagfish lineage: a PCR-based gene inventory of Eptatretus stoutii. Mol. Phylogenet. Evol. 32, 686–694 (doi:10.1016/j.ympev.2004.03.015) [DOI] [PubMed] [Google Scholar]
- Su C. Y., Luo D. G., Terakita A., Shichida Y., Liao H. W., Kazmi M. A., Sakmar T. P., Yau K. W.2006Parietal-eye phototransduction components and their potential evolutionary implications. Science 311, 1617–1621 (doi:10.1126/science.1123802) [DOI] [PubMed] [Google Scholar]
- Sundström G., Larsson T. A., Larhammar D.2008Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol. Biol. 8, 254 (doi:10.1186/1471-2148-8-254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szel A., Lukats A., Fekete T., Szepessy Z., Rohlich P.2000Photoreceptor distribution in the retinas of subprimate mammals. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 17, 568–579 (doi:10.1364/JOSAA.17.000568) [DOI] [PubMed] [Google Scholar]
- Takano Y., Ohguro H., Ohguro I., Yamazaki H., Mamiya K., Ishikawa F., Nakazawa M.2003Low expression of rhodopsin kinase in pineal gland in Royal College of Surgeons rat. Curr. Eye Res. 27, 95–102 (doi:10.1076/ceyr.27.2.95.15953) [DOI] [PubMed] [Google Scholar]
- Takezaki N., Figueroa F., Zaleska-Rutczynska Z., Klein J.2003Molecular phylogeny of early vertebrates: monophyly of the agnathans as revealed by sequences of 35 genes. Mol. Biol. Evol. 20, 287–292 (doi:10.1093/molbev/msg040) [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Terakita A.2005The opsins. Genome Biol. 6, 213 (doi:10.1186/gb-2005-6-3-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., De Vos W., Taylor J. S., Meyer A., Van de Peer Y.2004Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl Acad. Sci. USA 101, 1638–1643 (doi:10.1073/pnas.0307968100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman V., Nagele R., Duda T., Sharma R. K.2000Rod outer segment membrane guanylate cyclase type 1-linked stimulatory and inhibitory calcium signaling systems in the pineal gland: biochemical, molecular, and immunohistochemical evidence. Biochemistry 39, 6042–6052 (doi:10.1021/bi9929960) [DOI] [PubMed] [Google Scholar]
- Venkatesh B., et al. 2007Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 5, e101 (doi:10.1371/journal.pbio.0050101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienne A., Rasmussen J., Abi-Rached L., Pontarotti P., Gilles A.2003Systematic phylogenomic evidence of en bloc duplication of the ancestral 8p11.21-8p21.3-like region. Mol. Biol. Evol. 20, 1290–1298 (doi:10.1093/molbev/msg127) [DOI] [PubMed] [Google Scholar]
- Wada Y., Sugiyama J., Okano T., Fukada Y.2006GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the zebrafish rods and cones. J. Neurochem. 98, 824–837 (doi:10.1111/j.1471-4159.2006.03920.x) [DOI] [PubMed] [Google Scholar]
- Warren W. C., et al. 2008Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175–183 (doi:10.1038/nature06936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A. J., Aragay A. M., Slepak V. Z., Simon M. I.1996A novel form of the G protein beta subunit Gbeta5 is specifically expressed in the vertebrate retina. J. Biol. Chem. 271, 28 154–28 160 [DOI] [PubMed] [Google Scholar]
- Weiss E. R., Ducceschi M. H., Horner T. J., Li A., Craft C. M., Osawa S.2001Species-specific differences in expression of G-protein-coupled receptor kinase (GRK) 7 and GRK1 in mammalian cone photoreceptor cells: implications for cone cell phototransduction. J. Neurosci. 21, 9175–9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel T. G.2008Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 48, 2052–2061 (doi:10.1016/j.visres.2008.03.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami S., Suzuki N.2005Diverse forms of guanylyl cyclases in medaka fish—their genomic structure and phylogenetic relationships to those in vertebrates and invertebrates. Zool. Sci. 22, 819–835 (doi:10.2108/zsj.22.819) [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu X. H., Zhang K., Chen C. K., Frederick J. M., Prestwich G. D., Baehr W.2004Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J. Biol. Chem. 279, 407–413 (doi:10.1074/jbc.M306559200) [DOI] [PubMed] [Google Scholar]
- Zhao X., Huang J., Khani S. C., Palczewski K.1998Molecular forms of human rhodopsin kinase (GRK1). J. Biol. Chem. 273, 5124–5131 (doi:10.1074/jbc.273.9.5124) [DOI] [PubMed] [Google Scholar]
- Zheng J., Trudeau M. C., Zagotta W. N.2002Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 36, 891–896 (doi:10.1016/S0896-6273(02)01099-1) [DOI] [PubMed] [Google Scholar]