Abstract
The estuarine copepod Acartia tonsa naturally carried diverse strains of bacteria on its body. The bacterial community composition (BCC) remained very conservative even when the copepod was fed different axenic algal species, indicating that the food per se did not much affect BCC associated with the copepod. In xenic algal treatments, however, copepod-associated BCC differed with each alga fed, even though the same bacterial source was used to inoculate the algae. In addition, starved copepods taken at the same location but at different times significantly differed in their BCC. Algal species composition and copepod life history therefore serve to regulate BCC associated with copepods, and spatial and temporal variations in algal species composition and copepod origin would alter bacteria–copepod interactions.
Keywords: denaturing gradient gel electrophoresis, bacteria, copepod, estuary
1. Introduction
Conventional ecological research views mesozooplankton and bacteria as two weakly and indirectly connected functional groups (e.g. Azam & Malfatti 2007), but in reality they are closely linked in occurrence and ecological functions (Harris 1993). For example, the exoskeleton and gut lining of a copepod provide favourable surfaces for bacterial attachment (Carman & Dobbs 1997). The equivalent bacterial abundance associated with copepods can be orders of magnitude higher than that in ambient water, indicating active bacterial colonization and growth in these microenvironments (Tang 2005). The observations that the copepod's body and the surrounding water share similar bacterial groups, but in different proportions, suggest an active exchange of bacteria between the compartments, but the different environments tend to favour different bacterial groups (Sochard et al. 1979; Delille & Razouls 1994). These earlier studies relied on culturing techniques or biochemical assays, and may have missed many bacterial phylotypes that were present. Modern molecular techniques allow for a more detailed phylogenetic investigation of these bacterial communities (e.g. Møller et al. 2007; Peter & Sommaruga 2008). In the natural environment, food particles are already colonized by bacteria (Simon et al. 2002); copepod feeding will therefore bring new bacteria to its gut. The food environment as exploited by the copepod, and subsequent interactions between the ingested bacteria and the gut environment, would determine the resultant bacterial community composition (BCC) associated with the copepod. Here we used the molecular fingerprint technique denaturing gradient gel electrophoresis (DGGE) followed by the sequencing of individual DGGE bands to study how different algal species, under both axenic and xenic conditions, affect the BCC associated with the copepod Acartia tonsa.
2. Material and methods
Axenic algal cultures (table 1) were maintained in f/2 medium at 20 psu; axenic status was confirmed by DAPI staining (Porter & Feig 1980) and DGGE using eubacterial primers. Xenic algal cultures were prepared by inoculating the algae with 5 μm filtered water from the York River estuary (VA, USA). Cell carbon content was estimated from cell size (Strathmann 1967), and the experimental food concentration was adjusted to approximately 350 ng C ml−1 to ensure maximum ingestion rate (Tang et al. 2001).
Table 1.
Axenic algal strains used in the present study. (Cell carbon content was estimated from cell size based on Strathmann (1967). All algal strains were obtained from Bigelow CCMP collection except for Dunaliella tertiolecta DE, which was obtained from NOAA-NMFS in Milford, CT.)
| axenic algal strain | taxonomic group | CCMP no. | carbon content (pg C cell−1) |
|---|---|---|---|
| Phaeodactylum tricornutum | Bacillariophyceae | 1327 | 11.6 |
| Thalassiosira weissflogiia | Bacillariophyceae | 1336 | 54 |
| Rhodomonas salinaa | Cryptophyceae | 1319 | 29.9 |
| Dunaliella tertiolectaa | Chlorophyceae | 1320 | 31.1 |
| Dunaliella tertiolecta DE | Chlorophyceae | n.a. | 31.1 |
Xenic algal food was prepared by inoculating the cultures with natural bacteria.
The calanoid copepod A. tonsa was collected from the York River estuary. Female copepods were first incubated in 0.2 μm filtered Instant Ocean artificial sea water (20 psu) for 24 hours to empty their gut contents. After starvation, a subsample of copepods were rinsed with sterile sea water and transferred to sterile Eppendorf vials as initial samples (5–7 copepods per vial; 3–4 replicates per treatment). The samples were preserved with 20 μl of 95 per cent ethanol (molecular biology grade) and stored at −20°C until DGGE analysis. Remaining starved copepods were transferred to incubation bottles with either axenic (15 copepods per 130 ml in triplicate) or xenic (13–15 animals per 130 ml in triplicate) algae. Because the axenic and xenic algae treatments were conducted at different times, the copepods were taken from different field populations for the experiments. The bottles were fastened onto a rotating wheel in an environmental room (19±1°C, 12 L : 12 D cycle) for 48 hours. The algae in the incubation bottles were renewed after 1 day; after 2 days of incubation, remaining live copepods were rinsed and preserved for DGGE analysis.
Bacterial DNA was extracted using phenol–chloroform–isoamylalcohol and zirconium beads (Zhou et al. 1996). Fragments of bacterial 16S rRNA genes were amplified using universal primers 341f-GC and 907r (Muyzer & Ramsing 1995). Approximately 500 ng of amplification product were loaded in each lane of a 7 per cent polyacrylamide gel with a denaturing gradient ranging from 40 to 70 per cent (urea/formamide). The gels were run for 20 hours, stained with SYBRGold (Molecular Probes) for 30 min, then destained with Milli-Q water for 10 min and illuminated on a UV table (Biometra). Cluster analysis of the banding patterns was done by GelCompare II, v. 3.5 (Applied Maths) using unweighted pair group method with arithmetic averages. A pairwise similarity matrix based on Dice correlation index was calculated. DNA from excised individual DGGE bands was re-amplified for sequencing using the primers 341f without GC-clamp and 907r (Muyzer & Ramsing 1995) and the PCR protocol of Grossart et al. (2005). Partial 16S rRNA gene sequences were deposited in GenBank with accession numbers EU675687–EU675713 and EU680798–EU680807. Phylogenetic trees were constructed using the ARB software package (www.arb-home.de) and a database of approximately 52 100 aligned sequences. Only sequences of more than 1400 nucleotides were used. Phylogenetic analyses were performed by the maximum-likelihood algorithm. The resulting tree was compared with trees calculated with the neighbour-joining or maximum-parsimony algorithms to test for stability. Partial sequences from DGGE bands (approx. 560 nucleotides) were added to the tree according to maximum-parsimony criteria and with the 50 per cent base frequency filter.
3. Results
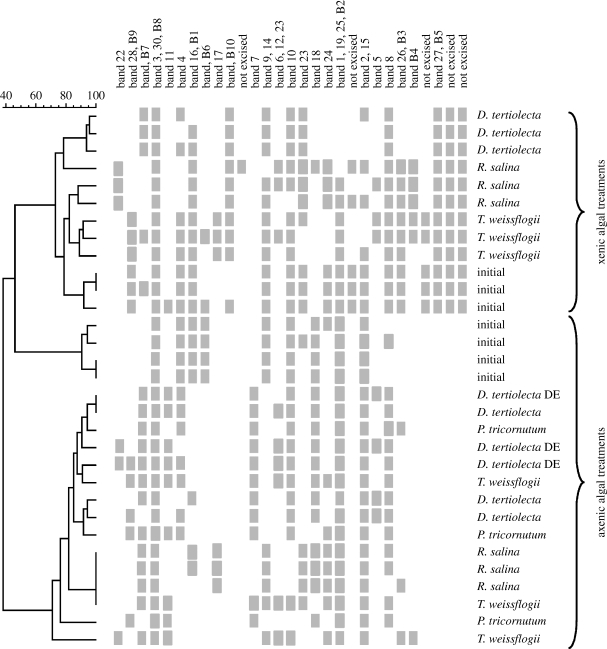
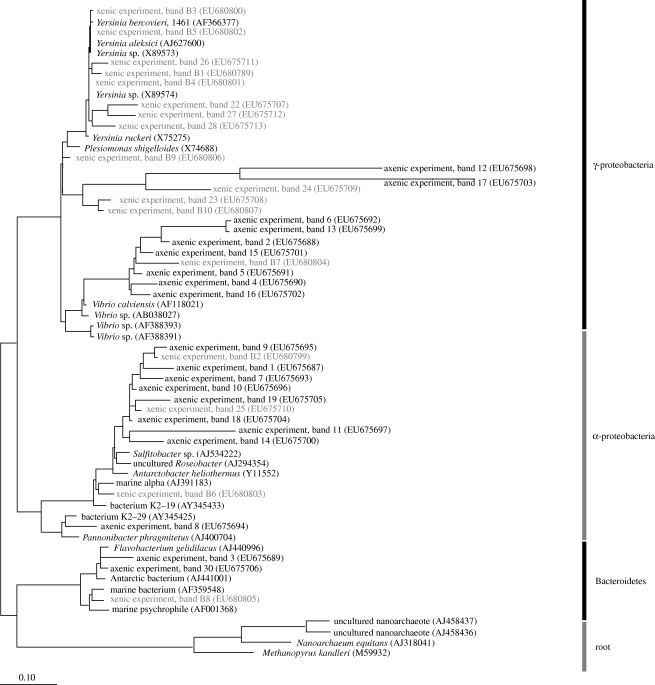
There was a large dissimilarity in the initial copepod-associated BCC between experiments (approx. 55%), reflecting differences in the experimental copepod populations (figure 1). In the axenic food experiment, the final copepod-associated BCC were very different from the initial samples (less than 40% similarity; figure 1), but were very similar among treatments: more than 80 per cent similarity except for one replicate of Phaeodactylum tricornutum (72%) and one replicate of Thalassiosira weissflogii (68%). This indicates that although the addition of food caused a change in copepod-associated BCC, the type of food per se had little effect on what bacterial community would establish on the copepods. Three bacterial classes were most common among the DGGE bands: α-proteobacteria; γ-proteobacteria; and Bacteroidetes (figure 2); all have members that commonly exist as symbionts or pathogens in marine organisms.
Figure 1.
Combined cluster diagram of DGGE banding patterns and the presence/absence table for specific bacteria associated with A. tonsa in axenic and xenic algal treatments. Full species name are given in table 1. Initial samples after starvation and prior to algal treatments are included for comparison. The dendrogram was calculated by Dice correlation index and UPGMA. Scale bar at the top indicates per cent similarity.
Figure 2.
Phylogenetic tree based on maximum likelihood including 16S rRNA sequences of all DGGE bands from feeding experiments with axenic (bold) and xenic (grey) algal food (see text for details). GenBank accession numbers are given in parentheses. Scale bar at the bottom represents relative phylogenetic distance.
Xenic food contributed new bacterial species to the copepod (seven bands that were not present in the initial samples; figure 1). To compare the two experiments, we normalized the number of bands in each sample to the total number of different bands in each experiment and then performed Welch's t-test. The result showed that the two experiments were significantly different from each other (p<0.001, t=−5.794, d.f.=13). In xenic treatments, the copepod-associated BCC clustered according to food type (except for one Rhodomonas salina sample), suggesting that different algal species delivered different bacterial assemblages to the grazers. The bacterial community mainly consisted of γ-proteobacteria, of which Yersinia of the Enterobacteriaceae group was frequently present (figure 2). In addition, the excised bands revealed several bacterial groups that are those commonly found on surfaces and in close association with marine organisms (Grossart et al. 2005): Pseudoalteromonas of the Alteromonadaceae group, Sulfitobacter and Roseobacter of the Rhodobacteraceae group and Halomonas species.
4. Discussion
Our experiments showed that an external source of bacteria (e.g. via food intake) was required to maintain a diverse bacterial community associated with A. tonsa. The dissimilarity between the two sets of initial samples suggests that the copepods were initially exposed to and retained very different BCC even after gut clearance, similar to the observations by Grossart et al. (2009). This implies that the life history of copepods has an influence on the occurrence of specific bacterial communities associated with them.
Members of Vibrionaceae frequently appeared among our 16S rRNA gene sequences. This group, including the pathogen Vibrio cholerae, is often found in close association with copepods (Heidelberg et al. 2002; Belkin & Colwell 2005). Vibrio species possess enzymes for chitin catabolism (Park et al. 2002) that make them well adapted to growing on the exoskeleton, gut lining and faecal pellets of copepods. Rhodobacteraceae are also commonly associated with copepods (Maran et al. 2007; Møller et al. 2007; this study), and form the second most abundant SSU rRNA gene cluster in marine plankton clone libraries (Giovannoni & Rappé 2000). The genus Yersinia is commonly found as gut flora (Brenner et al. 2005). Its frequent occurrence in our xenic treatments but near absence in the axenic treatments suggests that it relied on delivery via food intake to maintain a constant presence associated with the copepod. The absence of Pseudoalteromonas, Sulfitobacter and Roseobacter in the initial samples, but their frequent presence after xenic algal addition indicates that these species were also delivered via food intake, and were able to quickly establish a prominent presence associated with the copepod.
Although we made no distinction between bacteria attached to the exterior of the copepod and bacteria inside the copepod's gut, there is no a priori reason to expect that food intake would affect the composition of externally attached bacteria. By contrast, food intake is expected to affect bacterial dynamics inside the gut (Harris 1993). For example, when the copepod Calanus pacificus switched its diet from diatoms colonized by 3H-labelled bacteria to axenic diatoms, the copepod retained 31 per cent or less of the 3H signal after 43 hours (Lawrence et al. 1993). The authors interpreted it as the amount of ingested bacterial biomass assimilated into the copepod tissues. We, however, suggest that some of the residual 3H might represent ingested bacteria that became attached to the gut. Applying transmission electron microscopy (TEM) to paraffin sections, Peter & Sommaruga (2008) observed bacteria in the gut of starved Daphnia pulex (freshwater cladoceran), suggesting that some bacteria remained attached even after the zooplankton had cleared its gut content. They also reported the absence of bacteria in starved copepod Acanthodiaptomus denticornis. However, owing to the intrusive procedures of paraffin sectioning and TEM, even attached bacteria could have been lost, and negative results should be interpreted carefully. Using scanning electron microscopy, Nagasawa & Nemoto (1988) found gut bacteria in starved copepod Eucalanus bungii. Likewise, Hansen & Bech (1996) reported the presence of bacteria in the intestines of A. tonsa after the copepod had been starved and stripped of externally attached bacteria. In another study, when A. tonsa fed on axenic diatom, the abundance of attached bacteria exhibited a dome-shaped response to ingestion rate, which is consistent with the idea that bacterial abundance inside the gut was controlled by a balance between growth as stimulated by the copepod's feeding and loss due to defecation (Tang 2005). Conceptually, some researchers distinguish between ‘transient’ and ‘resident’ gut bacteria (Harris 1993). The former are the ones that are ingested but are either digested or released through defecation; the latter are the ones that permanently reside inside the gut. Such a distinction, however, may be difficult in practice. A copepod's gut must initially acquire bacteria from an external source, most probably from pre-colonized food particles. As shown in this study, the rather stable copepod-associated BCC among the axenic algal treatments suggests that certain bacterial groups may be preferentially retained by the copepod; on the other hand, ingestion of different algae that were pre-colonized by bacteria resulted in varying bacterial communities associated with the copepod. Other studies have shown that the bacterial communities attached to particles are more diverse than ambient free-living bacteria (Riemann & Winding 2001), and that different algal species harbour different bacterial communities (Grossart et al. 2006). Therefore, copepods exposed to different food environments will probably establish different resident bacterial communities inside their bodies. Some ingested bacteria will probably survive digestion and be incorporated into faecal matter (Lawrence et al. 1993). Although we did not examine the faecal matter, the dissimilarity in copepod-associated BCC among the xenic treatments in this study leads us to speculate that copepod feeding on different xenic food particles would produce faecal pellets that contain different bacteria, which may lead to different dissolution rate of the faecal materials, with important ramification for material fluxes in the ocean.
In this study, we showed that BCC associated with A. tonsa depended on several factors: life history of the copepods; the source of bacteria; and the food that delivered them. Extrapolating these laboratory results to the field, we hypothesize that the same copepod species exposed to different environments and food would establish different bacterial communities associated with its body. Conversely, copepod species of different feeding habits would acquire different bacterial communities via food intake, even if they share the same environment. Spatial and temporal variations in the food environment would therefore mediate changes in copepod–bacteria interactions.
Acknowledgments
This study was supported by the US NSF OCE-0352125, OCE-0814558, Jeffress Memorial Trust J-895 (USA), the German Science Foundation (DFG: GR 1540/11-1) and the Leibniz Foundation. The authors thank S. Brückner, S. L. Bickel and M. A. Lynch for their technical assistance. This is contribution no. 3009 of the Virginia Institute of Marine Science.
References
- Azam F., Malfatti F.2007Microbial structuring of marine ecosystems. Nat. Rev. Microbiol 5, 782–791doi:10.1038/nrmicro1747 [DOI] [PubMed] [Google Scholar]
- Belkin S., Colwell R.R.Ocean and health: pathogens in the marine environment. 2005New York, NY:Springer [Google Scholar]
- Brenner D.J., Krieg N.R., Staley J.T.vol. 22005New York, NY:Springer [Google Scholar]
- Carman K.R., Dobbs F.C.1997Epibiotic microorganisms ion copepods and other marine crustaceans. Microsc. Res. Tech 37, 116–135doi:10.1002/(SICI)1097-0029(19970415)37:2<116::AID-JEMT2>3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- Delille D., Razouls S.1994Community structures of heterotrohpic bacteria of copepod fecal pellets. J. Plankton Res 16, 603–615doi:10.1093/plankt/16.6.603 [Google Scholar]
- Giovannoni S., Rappé M.Evolution, diversity, and molecular ecology of marine prokaryotes. In Microbial ecology of the oceans ed. Kirchman D.L.2000. pp. 47–84 Chichester, UK:Wiley [Google Scholar]
- Grossart H.P., Levold F., Allgaier M., Simon M., Brinkhoff T.2005Marine diatom species harbour distinct bacterial communities. Environ. Microbiol 7, 860–873doi:10.1111/j.1462-2920.2005.00759.x [DOI] [PubMed] [Google Scholar]
- Grossart H.P., Kiørboe T., Tang K.W., Allgaier M., Yam E.M., Ploug H.2006Interactions between marine snow and heterotrophic bacteria: aggregate formation and microbial dynamics. Aquat. Microb. Ecol 42, 19–26doi:10.3354/ame042019 [Google Scholar]
- Grossart H.P., Dziallas C., Tang K.W.2009Bacterial diversity associated with freshwater zooplankton. Environ. Microbiol. Rep 1, 50–55doi:10.1111/j.1758-2229.2008.00003.x [DOI] [PubMed] [Google Scholar]
- Hansen B., Bech G.1996Bacteria associated with a marine planktonic copepod in culture. I. Bacterial genera in seawater, body surface, intestines and fecal pellets and succession during fecal pellet degradation. J. Plankton Res 18, 257–273doi:10.1093/plankt/18.2.257 [Google Scholar]
- Harris J.M.1993The presence, nature, and role of gut microflora in aquatic invertebrates: a synthesis. Microb. Ecol 25, 195–231doi:10.1007/BF00171889 [DOI] [PubMed] [Google Scholar]
- Heidelberg J.F., Heidelberg K.B., Colwell R.R.2002Bacteria of the gamma-subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl. Environ. Microb 68, 5498–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S.G., Ahmad A., Azam F.1993Fate of particle-bound bacteria ingested by Calanus pacificus. Mar. Ecol. Prog. Ser 97, 299–307doi:10.3354/meps097299 [Google Scholar]
- Maran B., et al. 2007Isolation and characterization of bacteria from the copepod Pseudocaligus fugu ectoparasitic on the panther puffer Takifugu pardalis with the emphasis on TTX. Toxicon 50, 779–790doi:10.1016/j.toxicon.2007.06.021 [DOI] [PubMed] [Google Scholar]
- Møller E.F., Riemann L., Søndergaard M.2007Bacteria associated with copepods: abundance, activity and community composition. Aquat. Microb. Ecol 47, 99–106doi:10.3354/ame047099 [Google Scholar]
- Muyzer G., Ramsing N.B.1995Molecular methods to study the organization of microbial communities. Water Sci. Technol 32, 1–9doi:10.1016/0273-1223(96)00001-7 [Google Scholar]
- Nagasawa S., Nemoto T.1988Presence of bacteria in guts of marine crustaceans and on their fecal pellets. J. Plankton Res 10, 559–564doi:10.1093/plankt/10.3.559 [Google Scholar]
- Park J.K., Wang L.-X., Roseman S.2002Isolation of a glucosamine-specific kinase, a unique enzyme of Vibrio cholerae. J. Biol. Chem 227, 15 573–15 578doi:10.1074/jbc.M107953200 [DOI] [PubMed] [Google Scholar]
- Peter H., Sommaruga R.2008An evaluation of methods to study the gut bacterial community composition of freshwater zooplankton. J. Plankton Res 30, 997–1006doi:10.1093/plankt/fbn061 [Google Scholar]
- Porter K.G., Feig Y.S.1980DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr 25, 943–948 [Google Scholar]
- Riemann L., Winding A.2001Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol 42, 274–282doi:10.1007/s00248-001-0018-8 [DOI] [PubMed] [Google Scholar]
- Simon M., Grossart H.-P., Schweitzer B., Ploug H.2002Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol 28, 175–211doi:10.3354/ame028175 [Google Scholar]
- Sochard M.R., Wilson D.F., Austin B., Colwell R.R.1979Bacteria associated with the surface and gut of marine copepods. Appl. Environ. Microbiol 37, 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann R.R.1967Estimating the organic carbon content of phytoplankton from cell volume or plasma volume. Limnol. Oceanogr 12, 411–418 [Google Scholar]
- Tang K.W.2005Copepods as microbial hotspots in the ocean: effects of host feeding activities on attached bacteria. Aquat. Microb. Ecol 38, 31–40doi:10.3354/ame038031 [Google Scholar]
- Tang K.W., Jakobsen H.H., Visser A.W.2001Phaeocystis globosa (Prymnesiophyceae) and the planktonic food web: feeding, growth and trophic interactions among grazers. Limnol. Oceanogr 46, 1860–1870 [Google Scholar]
- Zhou J., Bruns M.A., Tiedje J.M.1996DNA recovery from soils of diverse composition. Appl. Environ. Microbiol 62, 695–724 [DOI] [PMC free article] [PubMed] [Google Scholar]