Abstract
Members of the Family Bufonidae, true toads, are famous for their endogenously synthesized cardioactive steroids that serve as defensive toxins. Evolution of resistance to these toxins is not understood. We sequenced a key region of the toxin's binding site in the Na+/K+ ATPase for relevant taxa representing Hyloidea (including bufonids), Ranoidea and Archaeobatrachia and tested for positive selection in a phylogenetic context. Bufonidae were distinct from other Hyloidea at 4–6 of 12 sites and, with one exception, had a homologous amino acid sequence. Melanophryniscus stelzneri had a distinct sequence, consistent with other independent evidence for a differentiated toxin. Tests within Bufonidae detected positive selection within the binding region, providing, to our knowledge, the first evidence of this type for positive selection within Amphibia. There was no evidence for positive selection on Bufonidae or M. stelzneri lineages. Sequence change in Leptodactylus ocellatus, a leptodactylid predator of Bufonidae, provides a molecular basis for predator resistance possibly associated with gene duplication.
Keywords: Bufonidae, positive selection, cardioactive steroids, Na+/K+ ATPase
1. Introduction
Finding robust evidence for adaptive evolution of functional traits is among the foremost challenges of modern genetics. A promising approach is to use molecular and functional data to develop robust hypotheses of positive selection testable in a phylogenetic context (Jost et al. 2008). We apply this approach to test for positive selection for the resistance of true toads (Anura: Bufonidae) to defensive toxins.
Endogenous cardioactive steroids that inhibit activity of the Na+/K+ ATPase (sodium pump) are synthesized by all bufonids. Skin secretions of bufonid frogs contain levels of endogenous cardioactive steroids 25–40 000-fold higher than other anurans (Flier et al. 1980), providing an effective defence against most predators. With the exception of the genera Melanophryniscus and Dendrophryniscus, cardiotonic steroids produced are bufadienolides (Flier et al. 1980; Mebs et al. 2007). Most other anurans are not tolerant to cardioactive steroids, but the hylid Scinax ruber and leptodactylid Leptodactylus macrosternum (Leptodactylus ocellatus species group) are natural predators of bufonids in South America (Chen & Chen 1933; Crossland & Azevedo-Ramos 1999).
Amino acid sites that determine tolerance to cardioactive steroids have been identified in the sodium pump polymer. The initial extracellular loop of the α1 subunit of the sodium pump is a primary binding site for cardiotonic steroids. Changes within and around this region have been identified as facilitating resistance in bufonids as well as in some insects and rodents (Jaisser et al. 1992; Croyle et al. 1997).
This study sequenced the initial extracellular loop, and part of the surrounding transmembrane regions of the sodium pump, from representative anurans in order to determine whether resistance, conferred by changes in this region, is restricted to and conserved within Bufonidae. Specifically, we tested the hypothesis that positive selection has driven change in the binding region of the sodium pump along lineages ancestral to, or within, Bufonidae.
2. Material and methods
DNA was extracted from ethanol-preserved tissue using Puregene DNA Purification Kit (Gentra). Amplification of the initial extracellular loop including parts of the M1 and M2 transmembrane regions was performed using the primers ATP1_178Fwd (WGARATCCTGGCACGAGATG), or ATP1_222Fwd2a (ATGGGATACGGGGCCGGA), with ATP1_178Rvs (GAGGMACCATGTTCTTGAAGG), with an annealing temperature of 56°C for 30 cycles. PCR products were cleaned using ExoSapIT or Qiaquick PCR purification kit (Qiagen) after gel electrophoresis.
Products were sequenced using ATP1_178_Fwd and ATP1_178_Rvs with BigDye Terminator v. 3.1, with an annealing temperature of 53°C for 25 cycles, or subcloned using Invitrogen Topo protocol with PCR@4 vectors and Top10 chemically competent cells, purified using Purelink Quick Plasmid Miniprep kit (Invitrogen) and sequenced with M13 forward and M13 reverse primers. Reactions were cleaned using sodium acetate/EDTA/ethanol and sequenced with an ABI Prism 3100 Sequencer. Sequence traces were aligned and edited using Sequencer 3.1.
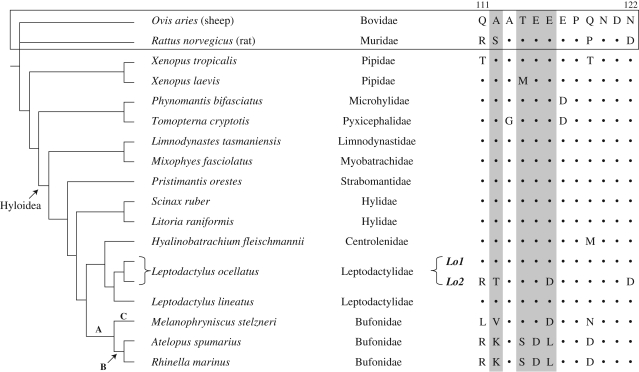
The phylogeny published by Frost et al. (2006) was used for analysis. Tests for positive selection were performed on three lineages (figure 1) using the improved branch-sites model A implemented in PAML (Yang 1997; Zhang et al. 2005). Significance was calculated by log likelihood comparison and Bayes empirical Bayes probabilities. Amino acid naming is consistent with Croyle et al. (1997). The binding region spans amino acids 111–122.
Figure 1.
Phylogeny and sequences of the binding site. Sequences of sheep and rat are included for comparison. Lineages tested for positive selection are A: Bufonidae, B: Bufonidae excluding Melanophryniscus and C: Melanophryniscus. Sites detected are highlighted in grey.
3. Results
One hundred and eighty-six base pairs of exon sequence were generated for 27 hyloid samples, 19 Bufonidae, one Centrolenidae, one Strabomantidae, two Hylidae, two Leptodactylidae, one Limnodynastidae and one Myobatrachidae, and two ranoid outgroups, one Microhylidae and one Pyxicephalidae (GenBank accession numbers FJ976618–FJ976647). Rhinella marinus (Z11798.2), Xenopus laevis (NM_001090595.1) and Xenopus tropicalis (NM_204076.1) were obtained from GenBank.
Most non-bufonid hyloid frogs had a typical non-resistant amino acid sequence at the binding site, homologous to sheep (Ovis aries; figure 1). Exceptions were Hyalinobatrachium fleischmanni (Centrolenidae), with an M at site 119, and L. ocellatus (Leptodactylidae). Direct sequencing of L. ocellatus found an unusual density of ‘heterozygotic sites’ in the M1–M2 extracellular loop. Cloning subsequently showed that there were two distinct conformations of the binding site. The nucleotide sequence outside the binding site was identical between the two sequences, however, within the binding site, one had a typical non-resistant sequence and the other had four non-synonymous changes: Q111R, A112T, E116D and N122D (figure 1). With the exception of Melanophryniscus stelzneri, the amino acid sequence of the binding site of all bufonids studied was homologous to R. marinus. Melanophryniscus stelzneri was differentiated both from other Bufonidae and non-resistant hyloid frogs (figure 1). Both ranoid outgroups had a D at site 117, and Tomopterna cryptotis had a G at site 113. The archaeobatrachians differed from the O. aries sequence; X. tropicalis had a T at sites 111 and 119 and X. laevis had an M at site 114.
Owing to conservation within Bufonidae, only sequences for R. marinus, Atelopus spumarius and M. stelzneri were included in tests of positive selection. A model allowing positively selected sites on lineage B performed significantly better than the null model (table 1, p < 0.01), but not for either Bufonidae or Melanophryniscus lineages. Four changes within the extracellular loop, A112K, T114S, E115D and E116L, were detected as being subject to positive selection on lineage B (p < 0.05).
Table 1.
Results of branch-site tests in PAML. *p < 0.05, **p < 0.01.
| lineage | Lln A | Lln null | p (χ2) | sites |
|---|---|---|---|---|
| A | −1091.17 | −1091.17 | 1 | |
| B | −1081.82 | −1088.12 | 0.002** | 112A 0.964*, 114T 0.965* |
| 115E 0.986*, 116E 0.992** | ||||
| C | −1090.91 | −1090.91 | 1 |
4. Discussion
Our results demonstrate that amino acid changes in the M1–M2 binding site associated with cardioactive steroid resistance evolved subsequent to divergence of Bufonidae from other hyloid anurans; the molecular basis for cardioactive steroid resistance of M. stelzneri is distinct from that of other bufonids studied and positive selection on at least four sites in the M1–M2 extracellular loop has contributed to the evolution of cardioactive steroid resistance. Evidence that resistance in a member of Leptodactylidae has evolved independently of resistance in Bufonidae is also presented.
Resistance is not ancestral in hyloid frogs. Bufonidae were highly differentiated from other hyloid frogs at 4–6 of 12 amino acids in the binding site. Binding site amino acid sequence of non-bufonid hyloid frogs studied, except H. fleischmanni (Centrolenidae) and resistant L. ocellatus (Leptodactylidae) (figure 1), was homologous to the non-resistant sheep sequence, clearly demonstrating the typically conserved nature of this site. The 119M difference of H. fleischmanni was not shared by other taxa. The L. ocellatus sequence Lo2 had 112T and 122D not shared by other taxa, but also shared 111R with lineage B and 116D with M. stelzneri. Despite these shared changes, robust phylogenetic separation of L. ocellatus from Bufonidae provides strong evidence for independent origins of resistance (figure 1). If resistance is not ancestral to Hyloidea, it must have arisen early in bufonid evolution.
Differentiation of both toxin and resistance between lineage B and Melanophryniscus suggests divergence early in the evolution of these traits. The split between Melanophryniscus and lineage B represents the oldest sampled split within Bufonidae (Pramuk et al. 2008) and, unlike bufadieonolides produced by other bufonids, cardioactive steroids in skin secretions of M. stelzneri are polar (Flier et al. 1980; Mebs et al. 2007). The cardioactive steroid binding site of M. stelzneri is distinct from other bufonids studied (figure 1) and indicative of resistance, as 111L reduces cardioactive steroid binding, and the change 108H (data not shown) occurs at a transmembrane site that influences resistance (Croyle et al. 1997). Sites 112V, 116D and 119N were also different. Selection for cardioactive steroid resistance may have acted on lineage B and Melanophryniscus before and after divergence.
Tests detected positive selection at four sites on lineage B, but not on the Bufonidae (A) or M. stelzneri (C) lineages. Sites subject to positive selection, 112K, 114S, 115D and 116L probably contribute to the 650-fold resistance of the lineage B sodium pump and to maintaining its function (Jaisser et al. 1992). Two of these sites, 112 and 116, were also modified in both L. ocellatus (Lo2) and M. stelzneri, confirming that the presence of cardioactive steroids has exerted selective pressure on these sites. Low power of tests makes it impossible to conclude that positive selection did not also occur on the M. stelzneri or Bufonidae lineages, or on other sites, such as 111R, which increases resistance 12.5-fold (Croyle et al. 1997). This is the first time that positive selection with a functional basis has been detected in amphibians, using phylogenetic methods.
A molecular basis for independently evolved resistance was discovered in L. ocellatus, a predator of bufonids. Two sequences, Lo1 and Lo2, cloned from L. ocellatus were differentiated within the 36 bp binding region. These were identical outside the binding site, but different from the closely related, non-resistant L. lineatus. The binding site amino acid sequence of Lo1 was homologous to non-resistant hyloids, while Lo2 had changes Q111R and N122D, a combination known to confer 1250-fold resistance to cardiotonic steroids (Price & Lingrel 1988). This suggests strong selection and recent divergence of sequences, possibly the consequence of hybridization between resistant and non-resistant races, or gene duplication associated with positive selection (Zhang 2003). This provides further evidence of the role of toad toxins as a selection pressure driving sequence change in the extracellular cardiotonic steroid binding site.
This study clearly shows that resistance to cardioactive steroids is not ancestral to hyloid frogs and that positive selection has driven sequence change for toxin resistance. Further research could include the complete α1 gene, or other α isoforms that interact with cardioactive steroids. Combined with protein structure modelling and simulated mutagenesis, this provides exciting opportunities to improve our understanding of positive selection in a functional context. How the unique, resistant sequence structure of M. stelzneri interacts with its yet uncharacterized cardioactive steroid, and the role of gene duplication and parallel selection in the evolution of resistance in L. ocellatus, would also be worthy of further investigation.
Acknowledgements
The Australian Government's Department of Environment and Water Resources provided funding through the National Heritage Trust. The Museum of Vertebrate Zoology (UC Berkeley), Louisiana Museum of Natural Science (LSU), University of Kansas National History Museum, Field Museum of Natural History (Chicago), National Museum of Natural History (Smithsonian, Washington, DC), Port Elizabeth Museum (South Africa) and the West Australian Museum (Perth) provided tissue grants. We also thank John Trueman, Lindell Bromham and Thayalini Shanmuganathan for discussion.
Appendix A
Specimen voucher numbers: WAM156571 Anaxyrus americanus; WAM109046 Duttaphrynus melanostictus; WAM109355 Ingerophrynus biporcatus; USNM534123 Rhaebo haematiticus; USNM331340 Incilius nebulifer; USNM303015 Rhinella crucifer; USNM302395 Rhinella granulosus; USNM320100 Anaxyrus cognatus; PEM A7511 P. bifasciatus; PEM A7515-6 Tomopterna cryptotis; CSIROVPC_Mfas01 Mixophyes fasciolatus; CSIROVPC_Ltas01 Limnodynastes tasmaniensis; CSIROVPC_Lran01 Litoria raniformis; KU_JES2472 Incilius coccifer; KU_WED59183 S. ruber; KU_JES1789 L. ocellatus; KU_JJW300 Pristimantis orestes; USNM253720 M. stelzneri; USNM_FS188295 Leptodactylus lineatus; USNM563019 H. fleischmanni; MVZHerp247525 Rhinella margaritifer; MVZHerp177905 Bufo bufo; MVZHerp241202 Peltophryne peltocephala; MVZHerp150267 Rhinella macrorhina; MVZHerp142938 Anaxyrus exsul; MVZHerp223373 Amietophrynus steindachneri; FMNH_HKV65808 Phrynoides aspera; FMNH255319 Ingerophrynus macrotis; LSU_H-15310 Atelopus spumarius.
References
- Chen K. K., Chen A. L.1933Relative susceptibility of the nebulous toad (Bufo valliceps) and the leopard frog (Rana pipiens) to different substances. J. Pharmacol. Exp. Ther. 47, 295–306 [Google Scholar]
- Crossland M. R., Azevedo-Ramos C.1999Effects of Bufo (Anura: Bufonidae) toxins on tadpoles from native and exotic Bufo habitats. Herpetologica 55, 192–199 [Google Scholar]
- Croyle M. L., Woo A. L., Lingrel J. B.1997Extensive random mutagenesis analysis of the Na+/K+-ATPase α subunit identifies known and previously unidentified amino acid residues that alter ouabain sensitivity: implications for ouabain binding. Eur. J. Biochem. 248, 488–495 (doi:10.1111/j.1432-1033.1997.00488.x) [DOI] [PubMed] [Google Scholar]
- Flier J., Edwards M. W., Daly J. W., Myers C. W.1980Widespread occurrence in frogs and toads of skin compounds interacting with the ouabain site of Na, K-ATPase. Science 208, 503–505 (doi:10.1126/science.6245447) [DOI] [PubMed] [Google Scholar]
- Frost D. R., et al. 2006The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 297, 1–291 (doi:10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2) [Google Scholar]
- Jaisser F., Canessa C. M., Horisberger J. D., Rossier B. C.1992Primary sequence and functional expression of a novel ouabain-resistant Na+,K+-ATPase. The beta subunit modulates potassium activation of the Na,K-pump. J. Biol. Chem. 267, 16 895–16 903 [PubMed] [Google Scholar]
- Jost M. C., Hillis D. M., Lu Y., Kyle J. W., Fozzard H. A., Zakon H. H.2008Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 25, 1016–1024 (doi:10.1093/molbev/msn025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebs D., Wagner M., Pogoda W., Maneyro R., Kwet A., Kauert G.2007Lack of bufadienolides in the skin secretion of red bellied toads, Melanophryniscus spp. (Anura, Bufonidae), from Uruguay. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 144, 398–402 (doi:10.1016/j.cbpc.2006.11.009) [DOI] [PubMed] [Google Scholar]
- Pramuk J. B., Robertson T., Sites J. W., Jr, Noonan B. P.2008Around the world in 10 million years: biogeography of the nearly cosmopolitan true toads (Anura: Bufonidae). Global Ecol. Biogeogr. 17, 72–83 [Google Scholar]
- Price E. M., Lingrel J. B.1988Structure–function relationships in the Na,K-ATPase α subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry 27, 8400–8408 (doi:10.1021/bi00422a016) [DOI] [PubMed] [Google Scholar]
- Yang Z.1997PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 [DOI] [PubMed] [Google Scholar]
- Zhang J.2003Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298 (doi:10.1016/S0169-5347(03)00033-8) [Google Scholar]
- Zhang J., Nielsen R., Yang Z.2005Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479 (doi:10.1093/molbev/msi237) [DOI] [PubMed] [Google Scholar]