Abstract
Admixture between wild and captive populations is an increasing concern in conservation biology. Understanding the extent of admixture and the processes involved requires identification of admixed and non-admixed individuals. This can be achieved by statistical methods employing Bayesian clustering, but resolution is low if genetic differentiation is weak. Here, we analyse stocked brown trout populations represented by historical (1943–1956) and contemporary (2000s) samples, where genetic differentiation between wild populations and stocked trout is weak (pairwise FST of 0.047 and 0.053). By analysing a high number of microsatellite DNA markers (50) and making use of linkage map information, we achieve clear identification of admixed and non-admixed trout. Moreover, despite strong population-level admixture by hatchery strain trout in one of the populations (70.8%), non-admixed individuals nevertheless persist (7 out of 53 individuals). These remnants of the indigenous population are characterized by later spawning time than the majority of the admixed individuals. We hypothesize that isolation by time mediated by spawning time differences between wild and hatchery strain trout is a major factor rescuing a part of the indigenous population from introgression.
Keywords: admixture analysis, historical DNA, isolation by time, microsatellite DNA, Salmo trutta, stocking
1. Introduction
Admixture between wild indigenous populations and translocated or escaped captive conspecifics is an increasing problem in conservation biology (Allendorf et al. 2001; Randi 2008). Captive breeding may lead to selection for domestication, and interbreeding between wild and captive populations may decrease the mean fitness of the wild populations (Lynch & O'Hely 2001). Salmonid fishes are particularly affected by leakage from captive populations, either due to fish farm escapees or deliberate stocking of wild populations with exogenous farmed strains (Hutchings & Fraser 2008). Several studies have demonstrated decreased fitness of farmed fish relative to wild indigenous populations (Araki et al. 2008), but significant admixture may nevertheless occur depending on the immigration rate due to farmed fish and local environmental conditions (Hansen et al. in press). However, it is not known whether all individuals in strongly introgressed populations become admixed or whether partial reproductive isolation mechanisms could delay admixture. Addressing this issue requires reliable identification of admixed versus non-admixed individuals.
This is in itself a challenge as genetic differentiation (FST) between farmed and wild populations is typically low (Vähä & Primmer 2006). An assessment of individual assignment methods suggested that more than 24 microsatellite loci are required for reliably distinguishing admixed versus non-admixed individuals at FST of 12 per cent (Vähä & Primmer 2006). However, developments in PCR multiplexing and automated genotyping now allow for analysing high numbers of microsatellite loci. Moreover, linkage maps developed for many organisms enable selection of markers such as to optimize individual assignment, both in terms of covering as many linkage groups as possible and by making use of information from weakly linked loci (Falush et al. 2003; Randi 2008).
We analysed two brown trout (Salmo trutta) populations that have previously been shown to be admixed with stocked hatchery strain trout, i.e. the Skjern (SKJ) and Storaa (STO) river populations (Hansen et al. in press). The populations were represented by pre- (1940–1950s) and post-stocking (2000s) samples. We furthermore analysed samples from the hatchery strain used for stocking. We made use of a linkage map (Gharbi et al. 2006) and analysed 50 microsatellite loci with known linkage relationships. We asked the following questions: (i) do non-admixed individuals persist even in strongly introgressed populations? (ii) If so, can mechanisms be suggested that may prevent complete admixture?
2. Material and methods
The geographical locations of the STO and SKJ rivers in western Jutland, Denmark, are shown in figure 1, along with information on sample sizes and sampling years. From 1977 to 1992, SKJ was stocked with 945 000 trout from the Hårkær hatchery strain (HAT). STO was stocked with the same strain. Exact details are not available, but the number of stocked trout probably exceeded 100 000. Contemporary samples consisted of adipose fin clips of spawning anadromous trout and historical samples of dried scales from anadromous trout caught during the spawning run. HAT samples consisted of muscle tissue from juveniles.
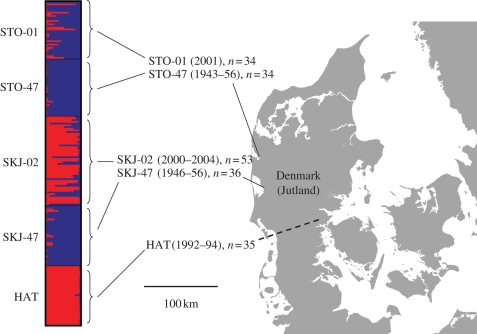
Figure 1.
Map showing the location of populations, information on sample sizes and year of sampling. Individual admixture proportions, estimated using Structure 2.2 and assuming k = 2 are shown as vertical bars, with clusters denoted by red and blue colours.
Details on DNA extraction are provided in Hansen et al. (in press). Fifty microsatellite loci from the brown trout linkage map (Gharbi et al. 2006) were analysed. One linkage group was represented by three loci, whereas 12 linkage groups were represented each by two loci. The remaining 23 loci were from different linkage groups (see electronic supplementary material, table S1 for details). Mapping distances between linked loci varied between 5.6 and 49.9 cM in females; males show much lower recombination rates (Gharbi et al. 2006). In four linkage groups, loci were tightly linked in males, but appeared unlinked in females and were treated as such in the statistical analyses.
Genetic variation was quantified by estimated observed (Ho) and expected (He) heterozygosity and by allelic richness (El Mousadik & Petit 1996). We specifically tested for deviations from Hardy–Weinberg equilibrium owing to heterozygote deficits caused by Wahlund effects or technical artefacts such as null alleles. This was achieved by permutation tests measuring if FIS was significantly higher than 0. Genetic differentiation was estimated with θST (Weir & Cockerham 1984), with confidence intervals determined by bootstrapping 10 000 times over loci. These analyses were conducted using Fstat 2.9.1 (Goudet 1995). The Bayesian simulation-based test by Foll & Gaggiotti (2008) implemented in the software Bayescan 1.0 was used for identifying loci potentially under diversifying selection. The analysis involved all five samples and was based on 10 pilot runs each consisting of 5 × 103 iterations followed by 105 iterations.
Bayesian clustering implemented in Structure 2.2 (Falush et al. 2003) was used for estimating the number of groups (k) represented by all sampled individuals and for estimating individual admixture proportions. We used the ‘linkage model’, based on female mapping distances. Although recombination rates differ between sexes, we assume that the relative mapping distances (used by Structure) are roughly the same. However, as a control, we also conducted analyses based on average mapping distances for both sexes and achieved very similar results (not shown). For estimating k, we included all samples and assumed k from 1 to 6 with no prior information about the population of origin of individuals. Burn-in was 105 iterations, followed by 5 × 105 iterations. Ten replicates were conducted for each k. We plotted the probability of the data ((p(D)) and the ad hoc statistic Δk (Evanno et al. 2005) for determining the most likely k. Subsequently, we estimated individual admixture proportions for SKJ and STO separately, involving historical and contemporary samples from the focal population along with the HAT sample.
3. Results
Eighteen tests showed FIS values significantly higher than 0 after false discovery rate correction (Benjamini & Yekutieli 2001) (table 1; electronic supplementary material, table S2). There was a tendency towards more significant tests in historical samples, but the relative similarity of He and Ho values suggests that technical artefacts have not had a major influence (table 1).
Table 1.
Summary statistics for microsatellite data. (Mean observed (Ho) and expected (He) heterozygosity and allelic richness across loci within samples, and number of tests where FIS is significantly higher than 0; significant at the 5 per cent level after false discovery rate correction.)
| sample | Ho (s.d.) | He (s.d.) | average allelic richness (s.d.) | number of tests with FIS significantly higher than 0 |
|---|---|---|---|---|
| SKJ-02 | 0.754 (0.173) | 0.765 (0.173) | 9.9 (4.8) | 2 |
| SKJ-47 | 0.753 (0.172) | 0.775 (0.181) | 10.7 (5.6) | 5 |
| STO-01 | 0.754 (0.196) | 0.779 (0.177) | 10.8 (5.5) | 3 |
| STO-47 | 0.725 (0.184) | 0.772 (0.177) | 10.5 (5.3) | 7 |
| HAT | 0.702 (0.193) | 0.710 (0.176) | 7.8 (3.8) | 1 |
Pairwise θST values were low, ranging from 0.005 between the historical samples SKJ-47 and STO-47 to 0.053 between HAT and STO-47 (table 2). The Bayescan analysis did not provide strong support for any of the loci being under diversifying selection (Bayesian p > 0.01 for all loci), and we assume that most or all of the loci are neutral.
Table 2.
Genetic differentiation (θST) between samples. (Pairwise θST estimates between samples with 95 per cent confidence intervals determined by bootstrapping over loci.)
| SKJ-02 | SKJ-47 | STO-01 | STO-47 | |
|---|---|---|---|---|
| SKJ-47 | 0.025 (0.019–0.032) | |||
| STO-01 | 0.016 (0.012–0.021) | 0.008 (0.005–0.012) | ||
| STO-47 | 0.029 (0.024–0.035) | 0.005 (0.002–0.008) | 0.010 (0.006–0.015) | |
| HAT | 0.025 (0.018–0.033) | 0.047 (0.038–0.057) | 0.040 (0.032–0.049) | 0.053 (0.042–0.064) |
The Structure analysis suggested that the number of clusters (k) was 2 (see details in electronic supplementary material, figure S1). Inspection of individual admixture proportions (figure 1) showed that one cluster corresponded to HAT and the other cluster to the historical samples STO-47 and SKJ-47, which could not be separated because of low genetic differentiation. Admixture between wild and hatchery trout was evident in the contemporary samples STO-01 and SKJ-02.
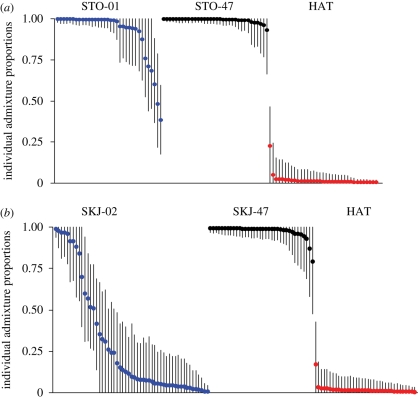
Separate analysis of STO suggested an average HAT admixture proportion of 0.091. The majority of individuals were non-admixed, but 7 out of 34 individuals showed HAT individual admixture coefficients of more than 10 per cent, with 90 per cent credible intervals in three individuals not including 0 or 1, thereby providing strong evidence of admixture (figure 2a). In SKJ-02, the average HAT admixture proportion was very high (70.8%). Fourty-six out of 53 individuals showed HAT individual admixture proportions exceeding 10 per cent (figure 2b). Nevertheless, seven individuals showed HAT admixture proportions of less than 10 per cent, providing evidence for the presence of non-admixed individuals.
Figure 2.
Individual admixture proportions and 90 per cent credible intervals, estimated using Structure 2.2. A value of 1 corresponds to a ‘pure’ indigenous trout and 0 to a ‘pure’ hatchery trout. Contemporary samples are denoted by blue, historical samples by black and hatchery trout by red. (a) Analysis of the STO population and (b) analysis of the SKJ population.
4. Discussion
The results demonstrate that even in the case of low genetic differentiation (θST around 0.05), increasing the number of loci enables clear identification of admixed and non-admixed individuals. This can be illustrated by comparing individual admixture proportions in SKJ-02 (figure 2b) with individual admixture proportions from the same population using only eight microsatellite loci (fig. 3f in Hansen 2002). In the latter case, credible intervals were very wide, precluding unambiguous identification of admixed and non-admixed individuals.
The question remains whether the increased resolution results only from the high number of loci, or whether linkage information has also increased the information content. An analysis of linkage disequilibrium reported in table S3 in the electronic supplementary material shows higher than average correlation between alleles at some linked loci, but there is considerable variation between linkage groups and samples, and the conclusion is not clear-cut. In any case, information from linkage maps allows for covering as many known linkage groups as possible, thereby providing more representative estimates of admixture at the genome level.
The presence of non-admixed trout in the strongly admixed SKJ population is particularly noteworthy. Stocking commenced in 1977 and interbreeding has taken place for more than 20 years (ca six generations), which would be expected to obliterate any remnants of ‘pure’ indigenous individuals. This suggests partial reproductive isolation between wild and hatchery trout. One such mechanism could be spatial isolation; a remnant of the indigenous population may spawn in a part of the river system with few hatchery trout. However, a previous study of SKJ trout points to another mechanism; using eight microsatellite markers, Hansen et al. (2006) assigned trout to two groups, i.e. hatchery strain trout and F1 hybrids versus a composite group of non-admixed indigenous trout and hybrid-indigenous back-crosses. Among all spawning trout, 65 per cent were ripe in November–December, but individuals assigned to the indigenous-backcross group were not ripe until January–February. The individuals in the present study exhibiting more than 90 per cent indigenous SKJ admixture proportions were all late spawners. Hence, our results suggest a link between non-admixed individuals and spawning time.
Reproductive isolation through isolation by time (IBT) has recently attracted considerable interest (Hendry & Day 2005). IBT may be common in hatchery–wild salmonid interactions, as genetic components of spawning time differences have been demonstrated in at least one species (Sakamoto et al. 1999), hatchery managers often select broodstock for early spawning (Hansen et al. 2006) and temporal segregation of spawning has been observed in sympatric wild and hatchery strain salmonids (Shields et al. 2005). We hypothesize that IBT has rescued a small proportion of the indigenous SKJ population from full introgression. However, as there is a continuous distribution from early- to late-spawning trout, the last remnants of indigenous trout may become admixed over time, unless precluded by natural selection or targeted management measures.
In conclusion, our results demonstrate the increased resolution of individual assignment and admixture analysis that can be achieved by genotyping higher numbers of loci and using linkage map information. Further, our findings caution against assuming that no ‘pure’ indigenous individuals can be found in strongly admixed populations. Partial reproductive isolation mechanisms can delay introgression and there may still be components of the indigenous populations left worthy of conservation measures.
Acknowledgements
The fin clips from the fish were taken under license from the Ministry of Food, Fisheries and Agriculture.
We acknowledge financial support from the Danish Natural Science Research Council (272-05-0202).
References
- Allendorf F. W., Leary R. F., Spruell P., Wenburg J. K.2001The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16, 613–622 (doi:10.1016/S0169-5347(01)02290-X) [Google Scholar]
- Araki H., Berejikian B. A., Ford M. J., Blouin M. S.2008Fitness of hatchery-reared salmonids in the wild. Evol. Appl. 1, 342–355 (doi:10.1111/j.1752-4571.2008.00026.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D.2001The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 [Google Scholar]
- El Mousadik A., Petit R. J.1996High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theor. Appl. Genet. 92, 832–839 (doi:10.1007/BF00221895) [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J.2005Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611–2620 (doi:10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K.2003Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Gaggiotti O.2008A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993 (doi:10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi K., et al. 2006A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172, 2405–2419 (doi:10.1534/genetics.105.048330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J.1995Fstat (version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486 [Google Scholar]
- Hansen M. M.2002Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol. Ecol. 11, 1003–1015 (doi:10.1046/j.1365-294X.2002.01495.x) [DOI] [PubMed] [Google Scholar]
- Hansen M. M., Bekkevold D., Jensen L. F., Mensberg K. L. D., Nielsen E. E.2006Genetic restoration of a stocked brown trout Salmo trutta population using microsatellite DNA analysis of historical and contemporary samples. J. Appl. Ecol. 43, 669–679 (doi:10.1111/j.1365-2664.2006.01185.x) [Google Scholar]
- Hansen M. M., Fraser D. J., Meier K., Mensberg L. D.In press Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol. Ecol. (doi:10.1111/j.1365-294X.2009.04198.x) [DOI] [PubMed] [Google Scholar]
- Hendry A. P., Day T.2005Population structure attributable to reproductive time: isolation by time and adaptation by time. Mol. Ecol. 14, 901–916 (doi:10.1111/j.1365-294X.2005.02480.x) [DOI] [PubMed] [Google Scholar]
- Hutchings J. A., Fraser D. J.2008The nature of fisheries- and farming-induced evolution. Mol. Ecol. 17, 294–313 (doi:10.1111/j.1365-294X.2007.03485.x) [DOI] [PubMed] [Google Scholar]
- Lynch M., O'Hely M.2001Captive breeding and the genetic fitness of natural populations. Conserv. Genet. 2, 363–378 (doi:10.1023/A:1012550620717) [Google Scholar]
- Randi E.2008Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 17, 285–293 (doi:10.1111/j.1365-294X.2007.03417.x) [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Danzmann R. G., Okamoto N., Ferguson M. M., Ihssen P. E.1999Linkage analysis of quantitative trait loci associated with spawning time in rainbow trout (Oncorhynchus mykiss). Aquaculture 173, 33–43 (doi:10.1016/S0044-8486(98)00463-3) [Google Scholar]
- Shields B. A., Stubbing D. N., Summers D. W., Giles N.2005Temporal and spatial segregation of spawning by wild and farm-reared brown trout, Salmo trutta L., in the River Avon, Wiltshire, UK. Fish. Manag. Ecol. 12, 77–79 (doi:10.1111/j.1365-2400.2004.00429.x) [Google Scholar]
- Vähä J. P., Primmer C. R.2006Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol. Ecol. 15, 63–72 (doi:10.1111/j.1365-294X.2005.02773.x) [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C.1984Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (doi:10.2307/2408641) [DOI] [PubMed] [Google Scholar]