Abstract
Epstein-Barr virus, a ubiquitous human herpesvirus, is associated with the development of carcinomas and lymphomas. We previously showed that transforming growth factor β1 (TGF-β1) mediated the virus to enter the lytic cycle, which is triggered by expression of Z Epstein-Barr virus replication activator (ZEBRA), through the ERK 1/2 MAPK signaling pathway. We report here that Akt, activated downstream from ERK 1/2, was required for TGF-β1-induced ZEBRA expression and enabled Smad3, a mediator of TGF-β1 signaling, to be acetylated by direct interaction with the co-activator CREB-binding protein and then to regulate TGF-β1-induced ZEBRA expression.
Epstein-Barr virus (EBV)3 is potentially oncogenic, being etiologically linked to a variety of malignancies, including nasopharyngeal carcinoma (1), endemic Burkitt's lymphoma (BL) (2), lymphomas of immunocompromised patients, and Hodgkin's disease (3). In vitro infection of B-lymphocytes usually results in viral latency, with only a small fraction of the viral genes being expressed: six nuclear proteins (EBNA 1, 2, 3A, 3B, 3C, and LP), three latent membrane proteins (LMP1, LMP2A, and LMP2B), and two small EBV-encoded RNAs (4). Entry into the viral lytic cycle is initiated by expression of the immediate-early EBV proteins, Z Epstein-Barr virus replication activator (ZEBRA) encoded by BZLF1 and BRLF1 (Rta) (5, 6). The two immediate-early proteins activate the viral early genes, resulting in a cascade of events leading to the progeny virions. The requirement of lytic gene expression for outgrowth of lymphoproliferations in a severe combined immunodeficiency (SCID) mouse model suggests the importance of reactivation for EBV pathogenesis (7).
EBV reactivation can be achieved in vitro by treatment of latent B cells with various activating agents including halogenated pyrimidines (8, 9), phorbol esters (10), anti-IgG antibodies (11, 12), sodium butyrate (13), and transforming growth factor (TGF) β1 (14). TGF-β1 regulates different physiological and pathological cellular processes including differentiation, immune response, inflammation, extracellular matrix synthesis, angiogenesis, and wound healing in humans (15). Moreover, TGF-β1 controls the expression of various genes and exerts its effects through a wide range of intracellular routes (for a review see Ref. 16). Binding of TGF-β1 to the TGF-β type II receptor triggers heterodimerization and transphosphorylation of the TGF-β type I receptor. The signal is then propagated through phosphorylation of receptor-regulated Smads (R-Smads), Smad2 and Smad3, which oligomerize with the common mediator (Co-Smad) Smad4 and then translocate to the nucleus, where they induce the expression of a large number of target genes via direct DNA binding or by association with other DNA-binding proteins (17–19). We have reported that Smad signaling alone is not sufficient in mediating TGF-β1 induction of ZEBRA (14). The co-activator CREB-binding protein (CBP) with acetyltransferase activity has been shown to associate and regulate Smad3-dependent transcription by acetylation (20, 21).
We show here that PI3-K/Akt pathway contributes to TGF-β1-induced ZEBRA expression by regulation of Smad3 association with CBP and acetylation by its acetyltransferase activity. Furthermore activation of PI3-K/Akt occurs downstream of a cascade, which includes ERK 1/2 MAPK signaling. These findings shed new light on the possible mechanisms underling the activation of EBV on BL cell lines by TGF-β1.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The Burkitt's lymphoma cell lines Mutu-I, Kem-I, Sav-I, and DG75 were maintained in RPMI 1640 medium (Sigma) supplemented with 2 mm glutamine (Invitrogen), 100 μg/ml primocin (Cayla, Toulouse, France), and 10% heat-inactivated fetal calf serum (Invitrogen). TGF-β1 stimulation was performed in this medium containing a reduced fetal calf serum concentration (0.5%). Purified recombinant TGF-β1 from R & D Systems (Minneapolis, MN) and from Strathmann Biotech AG (Hamburg, Germany) was added to a final concentration of 2 ng/ml. U0126 was from Promega (Madison, WI), wortmannin was from Alexis (San Diego, CA); Akt inhibitor X was from Calbiochem (VWR International); and protease inhibitor mixture and SB-431542 were purchased from Sigma. The acetyltransferase inhibition of CBP/p300 was performed with Lys-CoA-Tat (Ac-Lys[CoA]-YGRKKRRQRRR-OH) (22). Briefly, the histone acetyltransferase (HAT) inhibitor Lys-CoA (23) was linked with an 11-amino acid human immunodeficiency virus Tat transduction domain (24) to enable cellular permeability. As a control, Ac-DDDD-Tat (Ac-DDDD-YGRKKRRQRRR-OH) was synthesized on a Protein Technologies PS3 automated peptide synthesizer using the Fmoc strategy in combination with preloaded Fmoc-Arg(Pbf)-Wang resin. Couplings were preformed in triplicate using 4 equivalents of amino acid and 3.8 equivalents of the activating agent, N-[(dimethyamino)-1H-1,2,3-triazolo[4,5-b]pyridino-1-ylmethylene]-N-methylmethanaminium hexafluorophosphate N-oxide (HATU) (Applied Biosystems, Foster City, CA) for 1.5 h dissolved in a solution of 0.4 m of N-methylmorpholine in N,N-dimethylformamide. Fmoc deprotection involved exposure of the resin to a 20% (v/v) solution of piperidine in N,N-dimethylformamide (3 × 20 min.). Following amino acid couplings and N-terminal acetylation with acetic anhydride, the resin was washed (N,N-dimethylformamide, CH2Cl2, MeOH) and dried, and the peptide was cleaved from the resin using Reagent K (trifluoroacetic acid:phenol:H2O:thioanisole:1,2-ethanedithiol:triisopropylsilane, 81.5:5:5:5:2.5:1) for 4 h at room temperature. The cleaved peptide was precipitated with cold diethyl ether (−20 °C). The precipitate was collected by centrifugation, the supernatant was discarded, and the pellet was washed two times with cold diethyl ether. Precipitated peptide was dissolved in water, flash-frozen, lyophilized, and purified by preparative reverse-phase (C18) high pressure liquid chromatography (HPLC) using a gradient of H2O-CH3CN (each containing 0.05% (v/v) trifluoroacetic acid). The peptides were 95% pure by HPLC, and their structural identities were confirmed by mass spectrometry.
Phospho-protein Analysis
Mutu-I cells were treated with TGF-β1 for 17 h. The cells were then harvested, washed briefly with PBS, and resuspended in a buffer containing 20 mm MOPS, 2 mm EGTA, 5 mm EDTA, 30 mm sodium fluoride, 40 mm β-glycerophosphate, 20 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 3 mm benzamidine, 5 μm pepstatin A, 10 μm leupeptin, and 5% Nonidet P-40. Protein levels were quantified using the standard protocol of Bradford. Analysis of phospho-protein was performed by the Kinexus Bioinformatics Corporation using a KPSS1.3 KinetworksTM phospho-site screen (Kinexus Corporation).
RT-PCR
Total RNA from each uninduced and induced EBV-positive cells line was isolated using TRIzol reagent and RNase inhibitors (Invitrogen) according to the manufacturer's protocol. RNA concentration was determined spectrophotometrically. The template cDNA was synthesized by reverse transcription of the total RNA (3 μg) with Moloney murine leukemia virus reverse transcriptase (Promega) using an oligo(dT)15 primer (Promega). The primers used for PCR amplifications included: 5′-TTACACCTGACCCATACCAG-3′, 5′-ACATCTGCTTCAACAGGAGG-3′ for ZEBRA (25). cDNA of hypoxanthine-guanine phosphoribosyltransferase was used as an internal control as previously performed (26).
Antibodies
The anti-ZEBRA monoclonal antibody Z125 was obtained from E. Drouet (Faculté de Pharmacie, Grenoble, France). The antibodies against Akt, phospho-Akt (Ser473), p44/p42 MAPK, phospho-p44/p42 MAPK (Thr202/Tyr204), HP1G, Smad3, phospho-Smad3, acetyllysine, and CBP were purchased from Ozyme (St. Quentin-en-Yvelines, France). Human antiserum to EA (D and R) was kindly provided by Jean H. Joncas (Sainte Justine Hospital, Montreal, Canada). Human anti-VCA antibody was obtained from Ortho Diagnostic System, Inc. (Raritan, New Jersey). Anti-tubulin antibody was purchased from Sigma. Rabbit peroxidase-conjugated Ig, human peroxidase-conjugated IgG, and mouse peroxidase-conjugated IgG from Jackson ImmunoResearch Laboratories (West Grove, PA.) were used as secondary antibodies.
Immunoblot Analysis
Immunoblot analysis for the detection of ZEBRA, Akt, phospho-Akt (Ser473), EA-D, EA-R, total p42/p44, phospho-p44/p42, Smad3, phospho-Smad3 (Ser423/425), acetyllysine, and CBP was performed using 12% SDS-polyacrylamide gel. Equal amounts of protein were loaded in each lane. The proteins were transferred onto nitrocellulose (Sigma), blocked in PBS containing 5% nonfat dry milk, and incubated with primary antibody overnight at 4 °C. The membrane was washed in PBS, 0.1% Tween 20, incubated in secondary antibody for 1 h at room temperature (horseradish peroxidase-conjugated goat anti-mouse (1:3000), horseradish peroxidase-conjugated goat anti-rabbit (1:2000) or horseradish peroxidase-conjugated goat anti-human (1:2000) from Promega), and washed. Antibody binding was detected by enhanced chemiluminescence (West-pico or femto; Pierce). The images were captured using a DDC camera (LAS-1000; Fuji System).
Immunoprecipitation
The cells were extracted in radioimmunoprecipitation assay buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 0.1% protease inhibitor mixture). The cells were pelleted by centrifugation at 10,000 × g for 30 min at 4 °C. Protein concentrations were estimated in cleared cell lysates using a Micro BCATM protein assay kit from Pierce. Two μg of the indicated antibodies were added to 500 μl of the lysate and incubated overnight at 4 °C, followed by precipitation with 40 μl of protein A-Sepharose and incubation for 6 h at room temperature. The protein-antibody-Sepharose complexes were washed with radioimmunoprecipitation assay and heated at 100 °C for 5 min. The resulting immunoprecipitates were analyzed by immunoblotting as described in the figure legends.
Cell Fractionation
The cells were maintained in culture medium containing 0.5% fetal calf serum and were treated with TGF-β1 (2 ng/ml) and/or with indicated inhibitors. The cells were harvested at 4 °C, lysed, and cytoplasmic and nuclear extracts were prepared using a NE-PER nuclear and cytoplasmic extraction reagents kit (Pierce), as described by the manufacturer. Protease inhibitors were added (complete mini protease inhibitor mixture tablets, obtained from Roche Applied Science). Briefly, 10 mg of cells were resuspended in 50 μl of cytoplasmic extraction reagent I, mixed, and incubated on ice for 10 min. Then 2.75 μl of cytoplasmic extraction reagent II was added, followed by mixing and incubation on ice for 1 min. The intact nuclei were pelleted, and the supernatant cytoplasmic extract was collected. The nuclei were resuspended in 25 μl of ice-cold nuclear extraction reagent, incubated on ice repeatedly, and centrifuged to obtain the supernatant containing nuclear proteins. The protein concentrations were determined using a Micro BCATM protein assay kit from Pierce.
Determination of Cell Viability
The cells were treated as indicated under “Experimental Procedures” with different combination of TGF-β1, wortmannin, Akt inhibitor X, and U0126 for the indicated period of time and then tested for viability using a LIVE/DEAD reduced biohazard viability/cytotoxicity test (Molecular Probes, Invitrogen). Staining and analysis were performed as recommended by the manufacturer. The ability of this kit to detect cell death was controlled using a treatment with 0.4 mm of MnCl2 (27). The cell number was determined with a Kova counting chamber (Hycor, Penicuik, UK).
Lentivirus-mediated shRNA Silencing
shRNA-specific for human CBP (sc-29244-V) and a negative control (sc-108080) obtained from Santa Cruz (Santa Cruz Biotechnology) were used to infect Mutu-I, Kem-I, and Sav-I cells. Lentivirus particle was added to the cell culture in a 6-well dish and centrifuged for 1.5 h at 1200 × g, the cells were washed with PBS and cultured for 3 days before selection with 5 μg/ml puromycin (Sigma) for 10 days prior to cell analysis.
Plasmids, Transfection, and Reporter Assays
Plasmid −234Zp-CAT, (28) (generously provided by Henri Gruffat (ENS, Lyon, France), contains BZLF1 promoter bp −234 to +12, relative to the transcription initiation site, cloned upstream of the bacterial chloramphenicol acetyltransferase (CAT) reporter gene. Activity of the promoter was controlled by a 24-h treatment with phorbol 12-myristate 13-acetate as previously described (14). Plasmid pCMV-CBP (29) was kindly provided by Annick Harel-Bellan. Plasmids containing Smad3, coding sequences driven by the cytomegalovirus promoter were kindly provided by Peter ten Dijke (Ludwig Institute for Cancer Research, Uppsala, Sweden). The Smad3 sequence was mutated with the GenScript procedure (GenScript, Piscataway, NJ). Expression of Smad3 (wild type or mutated) as well as CBP was visualized in the transfected cells by Western blot (data not shown). Empty vectors, pcDNA3.1 (+), and pCMV2N3T, were used as controls for respectively Smad3 or CBP expression vectors. Transfection efficiency was monitored by using pRL-TK plasmid (Promega).
Plasmid DNA (5 μg) and pRL-TK vector (0.1 μg) was mixed with DG75 cells in 500 μl of RPMI 1640. The cells were exposed to a single pulse at 230V and 960 mF (Bio-Rad). The transfected cells were resuspended in 5 ml of complete culture medium. The cells were harvested 24 h later and washed with phosphate-buffered saline, and cell extract was subjected to the CAT enzyme-linked immunosorbent assay as recommended by the manufacturer (Roche Applied Science). Each transfection and reporter assay result shown was compiled from two independent experiments. Assay of Renilla luciferase was performed as recommended by the manufacturer (Promega).
RESULTS
TGF-β1-activated PI3-K/Akt Signaling Pathway Is Required for EBV Reactivation
Kinase pathways modulated by TGF-β1 in Mutu-I cells were investigated with the KPSS1.3 KinetworksTM phospho-site screen kit. As indicated in Table 1, TGF-β1 treatment stimulated a ∼6-fold increase in the level of Akt phosphorylation compared with the control, suggesting a strong activation of the PI3-K/Akt signaling pathway following TGF-β1 stimulation.
TABLE 1.
Effect of TGF-β1 on Akt-induced phosphorylation
Cell lysates of Mutu-I treated or not with TGF-β1 (2 ng/ml) for 17 h were analyzed using a KPSS1.3 KinetworksTM phospho-site screen from Kinexus Corporation. The level of Akt phosphorylation was estimated as a percentage of the control.
| No treatment | TGF-β1 | |
|---|---|---|
| P-Akt (Ser473) | 100 | 597 |
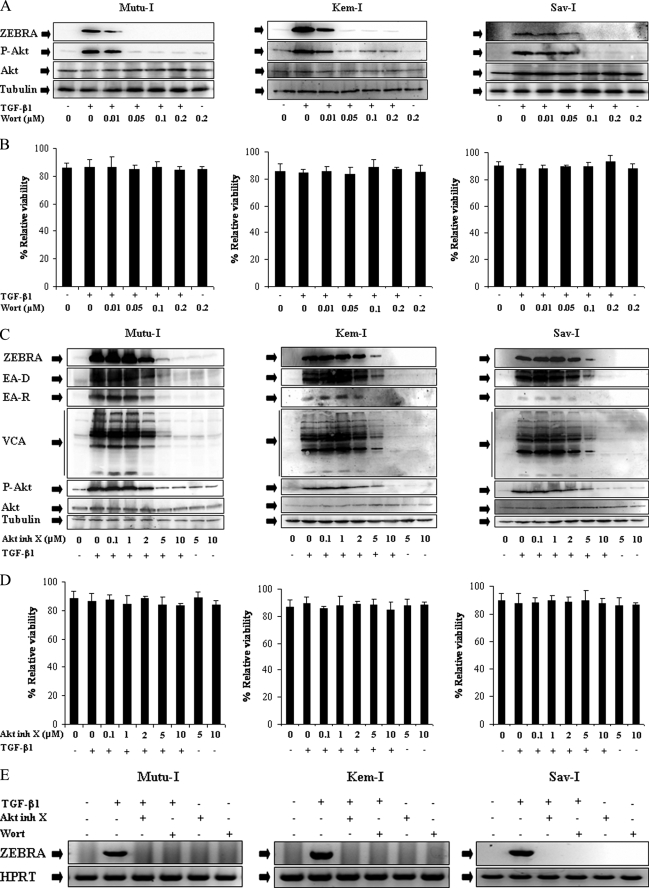
To examine the possible involvement of the PI3-K/Akt pathway in TGF-β1-induced EBV reactivation, the expression of EBV lytic proteins was analyzed in TGF-β1-stimulated Mutu-I, Kem-I, and Sav-I cells, pretreated with increasing concentrations of wortmannin, a selective inhibitor of PI3-K activity (30, 31), or Akt inhibitor X, a specific pharmacological inhibitor of Akt (32). As illustrated by Fig. 1A, treatment of cells with wortmannin resulted in a simultaneous inhibition of Akt phosphorylation and ZEBRA expression in a dose-dependent manner, with efficient inhibition being observed at a concentration of 0.05 μm for Mutu-I and Kem-I or 0.1 μm for Sav-I, without any significant effects on cell viability (Fig. 1B). Similar results were obtained using Akt inhibitor X (Fig. 1C), in the three cell lines, TGF-β1-stimulated expression of the EBV lytic proteins ZEBRA, EA-D, EA-R, and VCA was significantly inhibited at a concentration of 5 μm, and a complete inhibition was observed with 10 μm of Akt inhibitor X (Fig. 1C), without any significant effects on cell viability (Fig. 1D). These results indicated that the activated PI3-K/Akt pathway is critical for the TGF-β1-induced EBV lytic program in Mutu-I, Kem-I, and Sav-I cells.
FIGURE 1.
Inhibitors of PI3-K/Akt signaling pathway abolished TGF-β1-induced ZEBRA expression. A and C, Mutu-I, Kem-I, and Sav-I cells were treated with increasing concentration of wortmannin (Wort) (0.01, 0.05, 0.1, or 0.2 μm) (A) or Akt inhibitor X (Akt inh X) (0.1, 1, 2, 5, or 10 μm) (C) for 1 h, prior to incubation with TGF-β1 (2 ng/ml). Seventeen hours later, the cells were harvested and lysed. Equal amounts of protein were separated by SDS-PAGE and analyzed by Western blotting with antibodies to ZEBRA, phospho-Akt, Akt, and tubulin. B and D, viability assay was performed with the LIVE/DEAD reduced biohazard viability/cytotoxicity kit (Molecular Probes, Invitrogen). The ability of this kit to detect cell death was controlled using a treatment with 0.4 mm of MnCl2 (not shown). E, RT-PCR assay of ZEBRA was performed. 3 μg of total RNA from Mutu-I, Kem-I, and Sav-I cells pretreated, respectively, with 0.05, 0.05, and 0.1 μm of wortmannin or 10 μm of Akt inhibitor X for 1 h and then stimulated with TGF-β1 (2 ng/ml) for 17 h were reverse transcribed. cDNA coding for ZEBRA was then analyzed by PCR; cDNA of hypoxanthine-guanine phosphoribosyltransferase (HPRT) was used as an internal control.
RT-PCR analysis was performed to determine whether the observed ZEBRA induction in Mutu-I, Kem-I, or Sav-I cells resulted from a modulation of the transcription level. The result is shown in Fig. 1E; whereas the signal obtained when the cDNA encoding the ZEBRA protein showed a marked amplification after EBV reactivation, no signal was obtained when cells were pretreated with wortmannin or Akt inhibitor X, suggesting that the high level of ZEBRA protein expression observed upon EBV reactivation was due to an induction of transcription and that this induction was PI3-K/Akt-dependent.
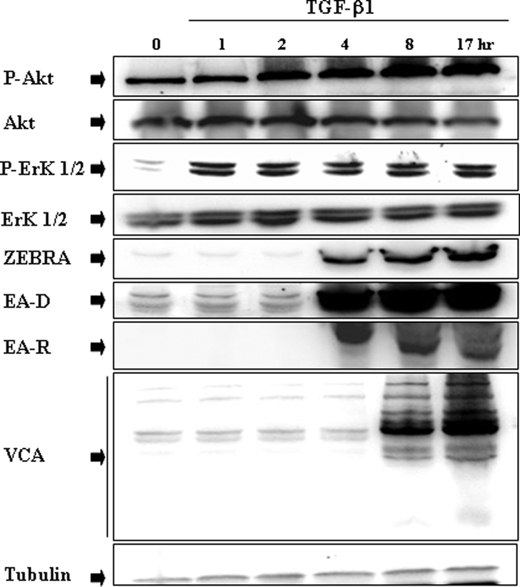
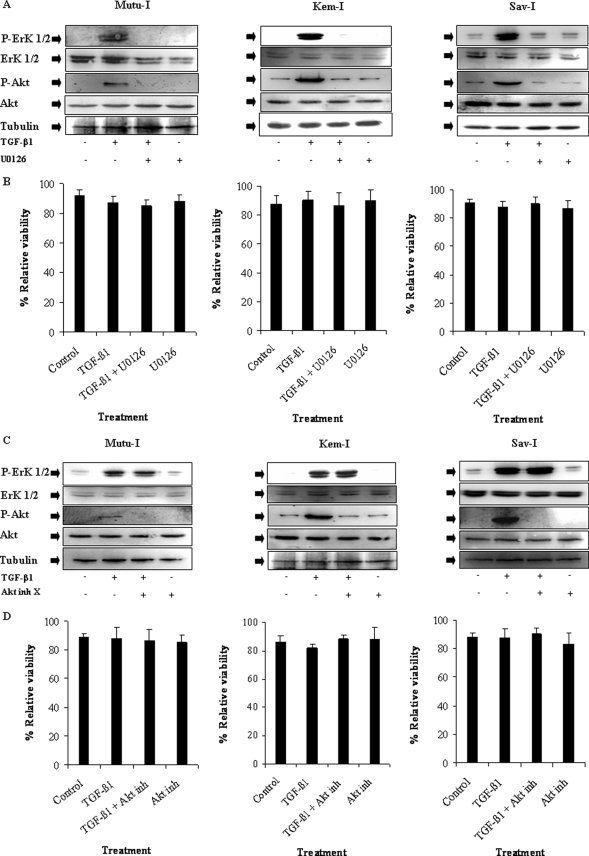
TGF-β1-induced PI3-K/Akt Pathway Activation Requires ERK 1/2 MAPK Signaling Pathway
We already established a key role of ERK 1/2 MAPK in the TGF-β1 signaling pathway, showing that U0126 a specific ERK 1/2 MAPK inhibitor completely abrogated TGF-β1-mediated ZEBRA induction in different BL cell lines and lymphoblastoid cell lines (14). A detailed time course study showing TGF-β1-induced signaling cascade was performed using immunoblot analysis. As shown in Fig. 2, phosphorylation of ERK 1/2 occurred before the increase of Akt phosphorylation and EBV lytic protein expression were observed, suggesting that activation of ERK 1/2 MAPK pathway in response to TGF-β1 occurred at a step upstream from PI3-K/Akt. The possible role of ERK 1/2 MAPK in TGF-β1-induced PI3-K/Akt signaling activation was investigated in Mutu-I, Kem-I, and Sav-I cells. Lysates of those BL cells treated with 2 ng/ml of TGF-β1 for 17 h, with or without a pretreatment of U0126 (20 μm), were analyzed by Western blotting against the phosphorylated form of Akt. Our data indicated that although the total Akt remained unchanged, U0126 completely inhibited TGF-β1-mediated phosphorylation of Akt (Fig. 3A), without altering cell viability (Fig. 3B).
FIGURE 2.
Time course of TGF-β1-induced phosphorylation of Akt and ERK 1/2, and EBV lytic gene expression in Mutu-I cells. Mutu-I cells were incubated in the presence of TGF-β1 (2 ng/ml) for various periods of time. The cells were lysed, and equal amounts of proteins were separated by SDS-PAGE. Phosphorylated Akt and Akt were analyzed, respectively, with anti-phospho-Akt and Akt antibodies by Western blotting. The membrane was then reprobed separately with a panel of specific antibodies directed against phospho-ERK 1/2, ERK 1/2, ZEBRA, EA-D, EA-R, and VCA. The amounts of protein loaded were assayed by reprobing the membrane with anti-tubulin antibody.
FIGURE 3.
TGF-β1-induced Akt phosphorylation in an ERK 1/2-dependent manner. A, Mutu-I, Kem-I, and Sav-I cells were incubated in the presence of 20 μm of U0126 for 1 h and then treated by TGF-β1 (2 ng/ml) for 17 h. The cells were harvested, washed, and lysed. Equal amounts of protein were analyzed by Western blotting using specific antibodies against phospho-Akt, Akt, phospho-ERK 1/2, and ERK 1/2. The amounts of protein loaded were assayed by reprobing the membrane with anti-tubulin antibody. C, Mutu-I, Kem-I, and Sav-I cells were treated with 10 μm of Akt inhibitor X (Akt inh X), prior to TGF-β1 (2 ng/ml) stimulation. Seventeen hours later, the cells were harvested and resuspended in Laemmli sample buffer. Equals amounts of protein were separated by SDS-PAGE and analyzed by Western blotting using phospho-Akt, Akt, phospho-ERK 1/2, ERK 1/2, and tubulin antibodies. B and D, viability assay was performed with the LIVE/DEAD reduced biohazard viability/cytotoxicity kit (Molecular Probes, Invitrogen). The ability of this kit to detect cell death was controlled using a treatment with 0.4 mm of MnCl2 (not shown).
Moreover, pretreatment with Akt inhibitor X affected neither cell viability (Fig. 3D) nor TGF-β1-mediated ERK 1/2 phosphorylation (Fig. 3C), showing that TGF-β1-mediated ERK 1/2 phosphorylation is Akt-independent. Together theses results show that the TGF-β1-induced signaling cascade, which included ERK 1/2 phosphorylation, is required for the PI3-K/Akt signaling pathway activation that precedes expression of ZEBRA, EA, and VCA proteins.
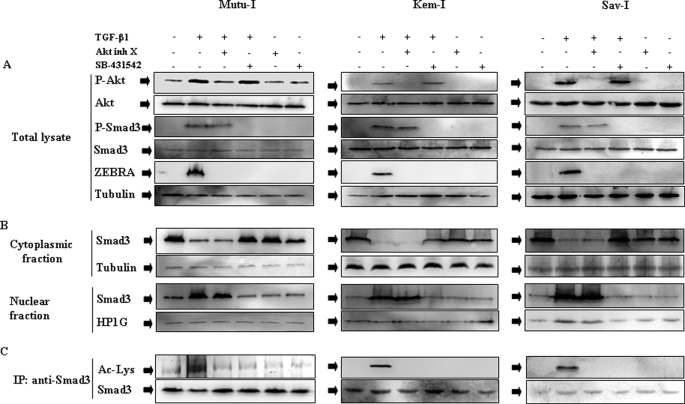
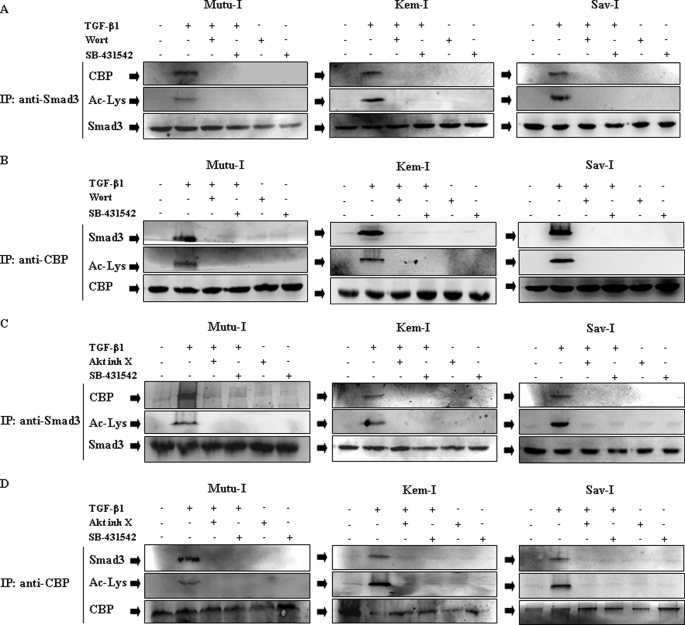
The involvement of ALK5 receptor to trigger the TGF-β1-mediated activation of the PI3-K/Akt pathway was examined. Mutu-I, Kem-I, and Sav-I cells were pretreated with SB-431542, which has been shown to inhibit specifically activin receptor-like kinase ALK5 (33, 34). This inhibitor did not alter TGF-β1 induced phosphorylation of Akt, whereas the control Akt inhibitor X completely abolished the increase in phosphorylation of Akt (see Fig. 5A). This result indicates that a noncanonical signaling pathway, independent of ALK5, was used to activate PI3-K/Akt pathway.
FIGURE 5.
PI3-K/Akt pathway regulates TGF-β1-induced Smad3 acetylation without any effect on Smad3 phosphorylation, and translocation. A, cells were treated or not with 10 μm of Akt inhibitor X (Akt inh X) or 10 μm SB-431542 for 1 h and then simulated with TGF-β1 (2 ng/ml) for 6 h. The cells were harvested, washed, and resuspended in Laemmli sample buffer. Cell extracts were analyzed by Western blotting against phospho-Akt, Akt, phospho-Smad3, Smad3, ZEBRA, and tubulin. B, nuclear and cytosolic extracts were prepared as described under “Experimental Procedures.” Equal amounts of each extracts were fractioned by SDS-PAGE, and the content of Smad3 was determined by Western blotting using anti-Smad3 antibody. Loading of nuclear and cytosolic fractions was assayed by blotting with antibodies respectively to HP1G (for nuclear fraction) and tubulin (for cytosolic fraction). C, cell lysates were immunoprecipitated (IP) with anti-Smad3 antibody, and acetylated Smad3 was detected by immunoblot with anti-acetyllysine antibody.
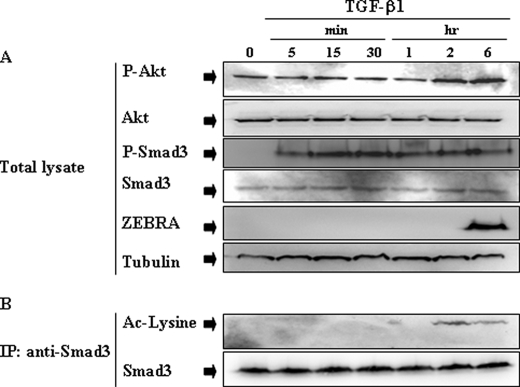
Time Course of TGF-β1-induced Smad3 Modifications in BL Cells
Because Smads are the primary mediators of TGF-β1 signaling, the TGF-β1-induced Smad3 modifications were checked. Mutu-I cells were stimulated with TGF-β1 (2 ng/ml) for various periods of time (between 5 min and 6 h) and then examined for Smad3 phosphorylation level using antibody against the phosphorylated form of Smad3 (Ser423/425). As seen in Fig. 4A, TGF-β1 treatment induced Smad3 phosphorylation beginning at 5 min and sustained for 6 h. The acetylation level of Smad3 was estimated after immunoprecipitation from lysates of TGF-β1-stimulated Mutu-I cells, followed by immunoblotting with anti-acetyllysine antibody. As illustrated by Fig. 4B, in Mutu-I cells, TGF-β1 increased acetylation of Smad3, beginning at 2 h and sustained for 6 h. As expected, TGF-β1-induced modifications of Smad3 depend upon the activity of ALK5 receptor, because treatment of the different BL cells with the inhibitor of ALK5, SB-431542, completely inhibited the phosphorylation of Smad3 (Fig. 5A), its translocation into nucleus in response TGF-β1 treatment (Fig. 5B) as well as its acetylation (Fig. 5C). This was observed for Mutu-I, Kem-I, and Sav-I cells.
FIGURE 4.
Time course of TGF-β-induced Smad3 phosphorylation and acetylation. A, Mutu-I cells were treated with TGF-β1 (2 ng/ml) for various periods of time. At the indicated time points, the cells were harvested, and lysed. Equal amounts of protein were separated by SDS-PAGE and analyzed by Western blotting with antibodies to phospho-Akt, Akt, phospho-Smad3, Smad3, ZEBRA, and tubulin. B, simultaneously, cell lysates were immunoprecipitated (IP) with anti-Smad3 antibody, and acetylated Smad3 was detected by immunoblot with anti-acetylated lysine antibody.
Role of PI3-K/Akt Signaling in TGF-β1-induced Smad3 Pathway Activation
The effect of Akt on Smad3 phosphorylation and its translocation into the nucleus was explored. Mutu-I, Kem-I, and Sav-I cells were pretreated or not with Akt inhibitor X and then stimulated by TGF-β1. The Western blot revealing phospho-Smad3 showed no effect on TGF-β1-induced Smad3 phosphorylation (Fig. 5A). Fractionation of the different BL cells into cytosol and nuclear fractions were analyzed by immunoblot with anti-Smad3 antibodies. Fig. 5B showed that TGF-β1 stimulation of Mutu-I, Kem-I, and Sav-I cells increased translocation of Smad3 into the nucleus. Incubation of the cells with Akt inhibitor X did not affect TGF-β1-induced Smad3 translocation (Fig. 5B). Taken together, these data clearly imply that Akt regulated TGF-β1-induced ZEBRA expression in BL cells, without any effect on Smad3 phosphorylation or translocation into the nucleus.
The possible role of Akt on TGF-β1-mediated Smad3 acetylation in BL cell lines was examined. Smad3 was immunoprecipitated from lysates of TGF-β1-stimulated Mutu-I, Kem-I, and Sav-I cells, followed by immunoblotting with anti-acetyllysine antibody. Inhibition of Akt by Akt inhibitor X completely abrogated TGF-β1-induced Smad3 acetylation (Fig. 5C). These results suggest that acetylation of Smad 3 occurs after signaling downstream of Akt.
PI3-K/Akt Signaling Pathway Regulates Smad3-CBP Association
Several groups have shown that the co-activator CBP regulates Smad3-dependent transcription by acetylation (21, 35). The fact that Akt inhibition impaired TGF-β1-mediated Smad3 acetylation suggested that association of CBP with Smad3 might be Akt-dependent. Therefore, the effect of PI3-K/Akt inhibition on Smad3-CBP association was examined in Mutu-I, Kem-I, and Sav-I cells. Smad3 immunoprecipitates from TGF-β1-stimulated cell lysates were immunoblotted with CBP antibody. The results indicated that association of CBP with Smad3 increased dramatically following TGF-β1 stimulation (Fig. 6). Incubation of BL cells with wortmannin or Akt inhibitor X (Fig. 6, A and C, respectively) impaired this interaction between Smad3 and CBP. To confirm theses results, reciprocal immunoprecipitation was performed and showed association of CBP with Smad3 in response to TGF-β. Blockade of PI3-K/Akt by wortmannin or Akt inhibitor X (Fig. 6, B and D, respectively) abolished TGF-β1-induced association of CBP with Smad3. As shown above, pretreatment of cells with SB-431542 completely inhibited phosphorylated Smad3 translocation into the nucleus where Smad3 and CBP association occurred. Thus, no Smad3-CBP interaction was observed in the presence of SB-431542 (Fig. 6). These results conclusively indicate that PI3-K/Akt signaling pathway integrates TGF-β1-induced Smad3-dependent ZEBRA expression in Mutu-I, Kem-I, or Sav-I BL cells by regulating the interaction between CBP and Smad3 and regulating Smad3 acetylation.
FIGURE 6.
Both Akt inhibitor X and wortmannin impair TGF-β-induced Smad3 association with CBP. Mutu-I, Kem-I, and Sav-I cells were treated, respectively, with 0.05, 0.05, and 0.1 μm of wortmannin (Wort) (A and B), 10 μm of Akt inhibitor X (Akt inh X) (C and D), or 10 μm of SB-431542 (A–D) for 1 h, prior to incubation with 2 ng/ml of TGF-β1. The cell lysates were immunoprecipitated (IP) with Smad3 (A and C) or CBP (B and D) antibodies, respectively. The immunoprecipitates were immunoblotted with anti-Smad3 or anti-CBP antibodies as indicated. The membranes were then reprobed with anti-acetyllysine antibody.
Contribution of PI3-K/Akt-mediated Smad3 Acetylation to TGF-β1-mediated EBV Reactivation
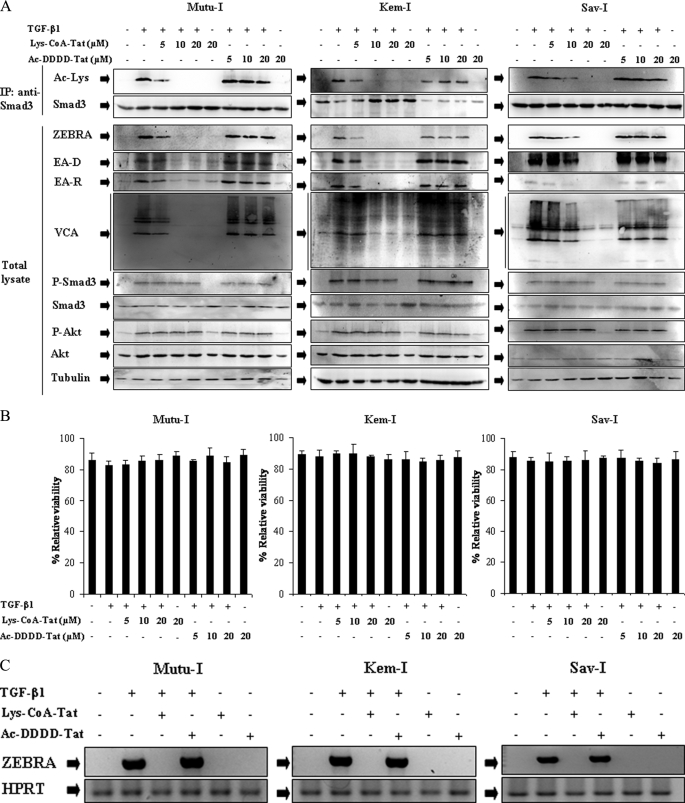
To address the question of the requirement of CBP acetyltransferase activity for Smad3 modification and for TGF-β1-mediated EBV-lytic expression, we used Lys-CoA-Tat, a chemical inhibitor of CBP HAT function (22). Ac-DDDD-Tat was used as a control. Mutu-I, Kem-I, and Sav-I cells were pretreated with increasing concentrations of Lys-CoA-Tat before induction for 17 h with TGF-β1. Smad3 was immunoprecipitated from lysates of TGF-β1-stimulated BL cells, followed by immunoblotting with anti-acetyllysine antibody. As illustrated by Fig. 7A, whereas phosphorylation of Smad3 was not affected by the inhibitor, treatment with Lys-CoA-Tat abolished Smad3 acetylation. Moreover, in Mutu-I and Kem-I cells, TGF-β1-stimulated cells, expression of the EBV lytic proteins ZEBRA, EA-D, EA-R, and VCA was significantly inhibited at a concentration of 5 μm of Lys-CoA-Tat, and a complete inhibition was observed with 10 μm, whereas 10 and 20 μm of Lys-CoA-Tat was necessary for, respectively, partial or total inhibition of EBV reactivation in Sav-I cells (Fig. 7A). The TGF-β1-mediated Akt phosphorylation was not affected by Lys-CoA-Tat, suggesting that its activation did not depend on Smad3 acetylation. Moreover, pretreatment of the BL cells with a similar concentration of the control Ac-DDDD-Tat did not affect ZEBRA, EA, and VCA expression. No significant effect on cell viability of Lys-CoA-Tat or Ac-DDDD-Tat was observed (Fig. 7B). These results indicated that acetylation of Smad3 is critical for TGF-β1-induced EBV lytic program in Mutu-I, Kem-I, and Sav-I cells.
FIGURE 7.
Lys-CoA-Tat inhibits TGF-β1-induced Smad3 acetylation and EBV-lytic cycle. A, Mutu-I, Kem-I, and Sav-I cells pretreated, respectively, with 10, 10, and 20 μm of Lys-CoA-Tat or 20 μm of Ac-DDDD-Tat for 1 h were stimulated with TGF-β1 (2 ng/ml) for 17 h. The cell lysates were immunoprecipitated (IP) with anti-Smad3 antibody, and acetylated Smad3 was detected by immunoblot with anti-acetyllysine antibody. Equal amounts of protein were separated by SDS-PAGE and analyzed by Western blotting with antibodies to ZEBRA, EA, VCA, phosphoSmad3, Smad3, phospho-Akt, Akt, and tubulin. B, viability assay was performed with the LIVE/DEAD reduced biohazard viability/cytotoxicity kit (Molecular Probes, Invitrogen). The ability of this kit to detect cell death was controlled using a treatment with 0.4 mm of MnCl2 (not shown). C, 3 μg of total RNA from Mutu-I, Kem-I, and Sav-I cells pretreated, respectively, with 10, 10, and 20 μm of Lys-CoA-Tat or 20 μm of Ac-DDDD-Tat for 1 h and then stimulated with TGF-β1 (2 ng/ml) for 17 h, were reverse transcribed. cDNA coding for ZEBRA was then analyzed by PCR; cDNA of hypoxanthine-guanine phosphoribosyltransferase (HPRT) was used as an internal control.
RT-PCR analysis was performed to determine whether the observed inhibition of ZEBRA expression in Mutu-I, Kem-I, or Sav-I cells by Lys-CoA-Tat resulted from an inhibition at the transcription level. The result is shown in Fig. 7C; although the signal of the cDNA encoding the ZEBRA protein showed a marked amplification after TGF-β1-mediated EBV reactivation, no signal was obtained when cells were pretreated with Lys-CoA-Tat, suggesting that the inhibition of ZEBRA protein expression observed upon the treatment with the inhibitor was due to inhibition of transcription and that this inhibition was dependent on inhibition of Smad3 acetylation.
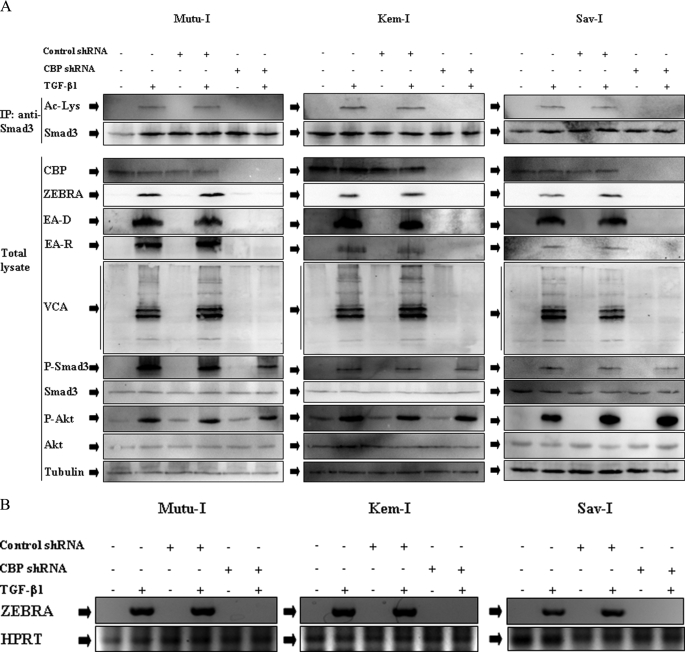
To further address the question of the requirement of CBP for Smad3 acetylation, Mutu-I, Kem-I, and Sav-I cells were infected with human CBP shRNA lentiviral particles or control particles harboring the gene conferring resistance to puromycin. After infection of Mutu-I, Sav-I, and Kem-I cells and selection with puromycin, CBP expression was completely abolished (Fig. 8A). TGF-β1-stimulated expression of the EBV lytic proteins ZEBRA, EA-D, EA-R, and VCA was assayed. Infection of the BL cells with CBP shRNA not only inhibited expression of CBP but also Smad3 acetylation and EBV lytic proteins expression (Fig. 8A) or BZLF1 transcription (Fig. 8B). No effect of CBP shRNA on phosphorylation of Smad3 and Akt was observed, indicating that phosphorylation of Akt occurred before Smad3 acetylation. Control shRNA had no effect either on CBP or lytic EBV protein levels and did not affect Smad3 acetylation. This result demonstrated that CBP is required for Smad3 acetylation, which is required for TGF-β1-mediated EBV lytic cycle expression.
FIGURE 8.
CBP shRNA inhibits TGF-β1-induced Smad3 acetylation and EBV-lytic cycle. A, Mutu-I, Kem-I, and Sav-I cells infected with CBP shRNA lentiviral particles or control particles were stimulated or not with TGF-β1 (2 ng/ml) for 17 h. The cell lysates were immunoprecipitated (IP) with anti-Smad3 antibody, and acetylated Smad3 was detected by immunoblot with anti-acetyllysine antibody. Simultaneously, equal amounts of protein from total lysate were separated by SDS-PAGE and analyzed by Western blotting with antibodies to ZEBRA, EA, VCA, phospho-Smad3, Smad3, phospho-Akt, Akt, and tubulin. B, 3 μg of total RNA from Mutu-I, Kem-I, and Sav-I cells infected with CBP shRNA lentiviral particles or control particles stimulated or not with TGF-β1 (2 ng/ml) for 17 h were reverse transcribed. cDNA coding for ZEBRA was then analyzed by PCR; cDNA of hypoxanthine-guanine phosphoribosyltransferase (HPRT) was used as an internal control.
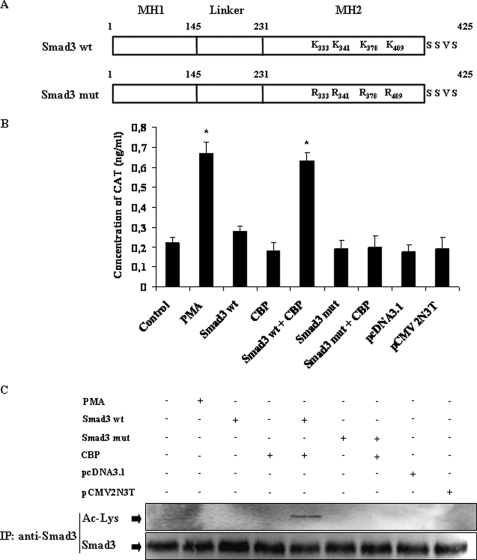
To ascertain the contribution of acetylated Smad3 for the activity of the BZLF1 promoter, Zp, DG75 cells were transfected with the Zp-CAT together with the pCMV-Smad3 vector alone or with pCMV-CBP. A robust CAT activity was detected when Smad3 and CBP were co-transfected with Zp, whereas no increase above the basal level was observed when Smad3 or CBP alone was used in the transient expression assay (Fig. 9B). In those cells Smad3 was acetylated as shown by the immunoblot probed with the anti-acetyllysine antibody (Fig. 9C). It was shown that the lysine residues located at positions 333, 341, 378, and 409 of the Smad3 protein were crucial for its CBP-mediated acetylation (21). No stimulation of the CAT activity was obtained when a Smad3 mutated in the four lysines of its MH2 domain were expressed with the CBP sequence; as expected, no acetylation of Smad3 was observed (Fig. 9C). This result showed that acetylation of Smad3 regulates its transcriptional activity on the Zp promoter.
FIGURE 9.
Acetylation of Smad3 participates in TGF-β1-induced Zp activation. A, schematic representation of the Smad3 protein and its mutant used in transient transfection experiments. B, the DG75 cells were transiently transfected with reporter gene construct containing the wild type Zp (−234 to +12) inserted upstream of the CAT gene (Control), or with a combination of expression plasmids for wild type of Smad3 (Smad3 wt) or its mutant (Smad3 mut), CBP, or empty vectors (pcDNA3.1 and pCMV2N3T) as indicated. The positive control consisted of Zp (−234 to +12) transfected DG75 cells and treated by 20 ng/ml of phorbol 12-myristate 13-acetate (PMA) immediately following transfection. Twenty-four hours later, the cells were harvested and lysed, and CAT activity was quantified as described under “Experimental Procedures.” The error bars represent the standard deviation. Significant differences between control and the other samples were determined by Student's t test; *, p < 0.05. C, cell lysates were immunoprecipitated (IP) with anti-Smad3 antibody, and equal amounts of immunoprecipitated Smad3 were analyzed by Western blotting with anti-acetyllysine antibodies.
DISCUSSION
The focus of this study was to gain a better understanding of the molecular mechanisms by which TGF-β1 induces disruption of EBV latency. We previously reported that TGF-β1 induces ZEBRA expression through the ERK 1/2 MAPK signaling pathway and involves more than Smad phosphorylation (14), and here we provide evidence for the involvement of the PI3-K/Akt signaling pathway as a key effector in TGF-β1-mediated ZEBRA expression in EBV-positive BL cell lines. Using phospho-protein and immunoblotting analysis, we showed that TGF-β1 treatment rapidly increased the PI3-K/Akt pathway activity, as judged by phosphorylation of Akt. Pharmacological inhibitors both of PI3-K and of Akt dramatically impaired TGF-β1-induced ZEBRA expression, suggesting that PI3-K/Akt signal is a major effector mediating the progression of the signal from TGF-β1 to the induction of ZEBRA expression. These data are in accordance with previous observations showing that the PI3-K/Akt pathway is required for disruption of viral latency in anti-Ig-treated Akata cells (12, 30). Our data are also consistent with reports demonstrating that TGF-β1 activates the PI3-K/Akt signaling pathway in different types of cell including BL cell lines (36, 37). Moreover, by several lines of evidence, our data suggest that the ERK 1/2 MAPK pathway is involved in TGF-β1-induced PI3-K/Akt signaling pathway activation in Mutu-I, Kem-I, and Sav-I cells, because incubation with U0126 completely inhibits Akt phosphorylation, in response to TGF-β1 stimulation (Fig. 3A), whereas Akt inhibition had no effect on TGF-β1-induced ERK 1/2 phosphorylation (Fig. 3C). Furthermore, a detailed time course study showing the kinetics of ERK 1/2 MAPK, PI3-K/Akt pathway activation, and EBV lytic gene expression in response TGF-β1 stimulation indicates that activation of ERK 1/2 occurs before detection of the increase in phosphorylation of Akt (Fig. 2). However, TGF-β1-mediated phosphorylation of Akt occurred in the presence of the ALK5 inhibitor, suggesting that a noncanonical signaling pathway was used to activate Akt kinases.
Nevertheless, the canonical TGF-β1 pathway also took place with the modifications of Smad3. Smad3 phosphorylation and translocation occurred downstream from ALK5 independently of PI3-K/Akt (Fig. 5). Conversely, Akt inhibitor X completely abolishes TGF-β1-mediated Smad3 acetylation (Fig. 5) by abolishing interactions of Smad3 with the transcriptional co-activator CBP (Fig. 6). Indeed, the ability of Smad to modulate transcription in response to TGF-β1 can result from functional cooperativity with general transcription co-activators such as CBP (21). We show here that acetylation of Smad3 was required to induce ZEBRA expression and EBV reactivation because the treatment with Lys-CoA-Tat, a potent HAT activity inhibitor of CBP, abolishes Smad3 acetylation and ZEBRA expression (Fig. 7). This result indicates an essential function for HAT in EBV reactivation. We further demonstrate that the HAT function was catalyzed by CBP because abolition of CBP expression by a specific shRNA completely suppressed Smad3 acetylation and TGF β1-mediated EBV reactivation (Fig. 8).
Smads mediated TGF-β1-induced BZLF1 expression through binding to its promoter Zp (38). Smad3 acetylation was crucial for transcriptional activity of this factor on Zp promoter because mutation of the four lysine residues abolished the activation of the promoter by the overexpression of the transcription factor together with the CBP co-activator. Thus, we show here for the first time that CBP has a crucial role in EBV reactivation through the HAT function necessary for the activity of Smad protein. This finding may account for the well established reactivation of EBV by the known histone deacetylase inhibitor sodium butyrate (13).
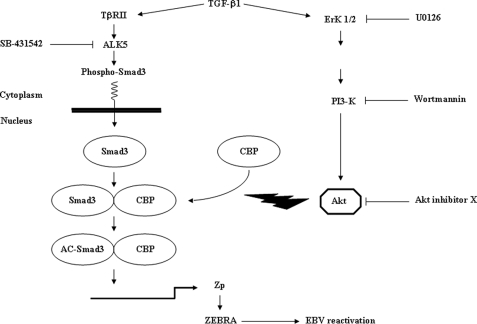
In conclusion (schematized in Fig. 10), canonical (implicating Smad3) as well as noncanonical (implicating ERK 1/2 and PI3-K/Akt) pathways, in concert, contribute to TGF-β1-mediated EBV reactivation. The PI3-K/Akt signaling pathway is required for TGF-β1-induced ZEBRA expression by regulating the interaction of the transcriptional co-activator CBP with Smad3, allowing acetylation of this protein. Our findings shed new light on the possible mechanisms underling disruption of EBV latency on Mutu-I, Kem-I, and Sav-I BL cell lines by the physiological factor TGF-β1.
FIGURE 10.
Proposed mechanism of TGF-β1-mediated ZEBRA expression. Canonical (implicating Smad3) as well as noncanonical (implicating PI3-K/Akt) pathways, in concert, contribute to TGF-β1-mediated ZEBRA expression. TGF-β1 mediates its activity through binding to its receptor II which recruits and phosphorylates its receptor I/ALK5; Smad3 phosphorylation takes place and translocation to the nucleus occurs. However, activated Akt, the non-Smad mediator of TGF-β1 directed CBP-Smad3 interaction and Smad3 acetylation, which enables the increase of ZEBRA expression.
Acknowledgments
We thank Emmanuel Drouet for the anti-ZEBRA monoclonal antibodies, Henri Gruffat for the Zp plasmid, and Nicolas Bidère and Damien Arnoult for helpful discussion. We are grateful to Antoine Durrbach and Bernard Charpentier for constant support.
This work was supported by the French network Herpesviruses and Cancer, Association pour la Recherche sur le Cancer Grant 3572, Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), Tunisian Ministry of Higher Education, Research and Technology, INSERM, Institut Francilien de Recherche en Néphrologie et Transplantation, Association Nouvelles Recherches Biomédicales, Association pour l'Utilisation du Rein Artificiel, and Naturalia and Biologia.
- EBV
- Epstein-Barr virus
- TGF
- transforming growth factor
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- ZEBRA
- Z Epstein-Barr virus replication activator
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- BL
- Burkitt's lymphoma
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- HPLC
- high pressure liquid chromatography
- PBS
- phosphate-buffered saline
- MOPS
- 4-morpholinepropanesulfonic acid
- RT
- reverse transcription
- shRNA
- small hairpin RNA
- CAT
- chloramphenicol acetyltransferase
- PI3-K
- phosphatidylinositol 3-kinase
- EA
- early antigen
- VCA
- viral capsid antigen
- HAT
- histone acetyltransferase.
REFERENCES
- 1.Desgranges C., de-The G., Wolf H., zur Hausen H. (1975) IARC Sci. Publ. 11, 191–193 [PubMed] [Google Scholar]
- 2.Henle W., Henle G., Niederman J. C., Klemola E., Haltia K. (1971) J. Infect. Dis. 124, 58–67 [DOI] [PubMed] [Google Scholar]
- 3.Pallesen G., Hamilton-Dutoit S. J., Rowe M., Young L. S. (1991) Lancet 337, 320–322 [DOI] [PubMed] [Google Scholar]
- 4.Kieff (2007) Epstein-Barr Virus and Its Replication, Lippincott William & Wilkins, Philadelphia, PA [Google Scholar]
- 5.Ragoczy T., Miller G. (1999) J. Virol. 73, 9858–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zalani S., Holley-Guthrie E., Kenney S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9194–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong G. K., Gulley M. L., Feng W. H., Delecluse H. J., Holley-Guthrie E., Kenney S. C. (2005) J. Virol. 79, 13993–14003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber P. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 83–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampar B., Derge J. G., Martos L. M., Walker J. L. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.zur Hausen H., O'Neill F. J., Freese U. K., Hecker E. (1978) Nature 272, 373–375 [DOI] [PubMed] [Google Scholar]
- 11.Tovey M. G., Lenoir G., Begon-Lours J. (1978) Nature 276, 270–272 [DOI] [PubMed] [Google Scholar]
- 12.Takada K. (1984) Int. J. Cancer 33, 27–32 [DOI] [PubMed] [Google Scholar]
- 13.Luka J., Kallin B., Klein G. (1979) Virology 94, 228–231 [DOI] [PubMed] [Google Scholar]
- 14.Fahmi H., Cochet C., Hmama Z., Opolon P., Joab I. (2000) J. Virol. 74, 5810–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence D. A. (1996) Eur. Cytokine Netw 7, 363–374 [PubMed] [Google Scholar]
- 16.Bierie B., Moses H. L. (2006) Nat. Rev. Cancer 6, 506–520 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Feng X., We R., Derynck R. (1996) Nature 383, 168–172 [DOI] [PubMed] [Google Scholar]
- 18.Massague J., Wotton D. (2000) EMBO J. 19, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y., Massague J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 20.Shen X., Hu P. P., Liberati N. T., Datto M. B., Frederick J. P., Wang X. F. (1998) Mol. Biol. Cell 9, 3309–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue Y., Itoh Y., Abe K., Okamoto T., Daitoku H., Fukamizu A., Onozaki K., Hayashi H. (2007) Oncogene 26, 500–508 [DOI] [PubMed] [Google Scholar]
- 22.Guidez F., Howell L., Isalan M., Cebrat M., Alani R. M., Ivins S., Hormaeche I., McConnell M. J., Pierce S., Cole P. A., Licht J., Zelent A. (2005) Mol. Cell Biol. 25, 5552–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau O. D., Kundu T. K., Soccio R. E., Ait-Si-Ali S., Khalil E. M., Vassilev A., Wolffe A. P., Nakatani Y., Roeder R. G., Cole P. A. (2000) Mol. Cell 5, 589–595 [DOI] [PubMed] [Google Scholar]
- 24.Nagahara H., Vocero-Akbani A. M., Snyder E. L., Ho A., Latham D. G., Lissy N. A., Becker-Hapak M., Ezhevsky S. A., Dowdy S. F. (1998) Nat. Med. 4, 1449–1452 [DOI] [PubMed] [Google Scholar]
- 25.Arbach H., Viglasky V., Lefeu F., Guinebretiere J. M., Ramirez V., Bride N., Boualaga N., Bauchet T., Peyrat J. P., Mathieu M. C., Mourah S., Podgorniak M. P., Seignerin J. M., Takada K., Joab I. (2006) J. Virol. 80, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touitou R., Arbach H., Cochet C., Feuillard J., Martin A., Raphael M., Joab I. (2003) J. Gen. Virol. 84, 949–957 [DOI] [PubMed] [Google Scholar]
- 27.El Mchichi B., Hadji A., Vazquez A., Leca G. (2007) Cell Death Differ. 14, 1826–1836 [DOI] [PubMed] [Google Scholar]
- 28.Gruffat H., Manet E., Sergeant A. (2002) EMBO Rep. 3, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez S., Ait-Si-Ali S., Robin P., Trouche D., Harel-Bellan A. (1997) J. Biol. Chem. 272, 31016–31021 [DOI] [PubMed] [Google Scholar]
- 30.Iwakiri D., Takada K. (2004) J. Immunol. 172, 1561–1566 [DOI] [PubMed] [Google Scholar]
- 31.Fukagawa Y., Nishikawa J., Kuramitsu Y., Iwakiri D., Takada K., Imai S., Satake M., Okamoto T., Fujimoto M., Okita K., Nakamura K., Sakaida I. (2008) Electrophoresis 29, 3192–3200 [DOI] [PubMed] [Google Scholar]
- 32.Thimmaiah K. N., Easton J. B., Germain G. S., Morton C. L., Kamath S., Buolamwini J. K., Houghton P. J. (2005) J. Biol. Chem. 280, 31924–31935 [DOI] [PubMed] [Google Scholar]
- 33.Callahan J. F., Burgess J. L., Fornwald J. A., Gaster L. M., Harling J. D., Harrington F. P., Heer J., Kwon C., Lehr R., Mathur A., Olson B. A., Weinstock J., Laping N. J. (2002) J. Med. Chem. 45, 999–1001 [DOI] [PubMed] [Google Scholar]
- 34.Inman G. J., Nicolas F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., Hill C. S. (2002) Mol. Pharmacol. 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 35.Das F., Ghosh-Choudhury N., Venkatesan B., Li X., Mahimainathan L., Choudhury G. G. (2008) J. Cell Physiol. 214, 513–527 [DOI] [PubMed] [Google Scholar]
- 36.Ghosh Choudhury G., Abboud H. E. (2004) Cell Signal 16, 31–41 [DOI] [PubMed] [Google Scholar]
- 37.Wilkes M. C., Mitchell H., Penheiter S. G., Dore J. J., Suzuki K., Edens M., Sharma D. K., Pagano R. E., Leof E. B. (2005) Cancer Res. 65, 10431–10440 [DOI] [PubMed] [Google Scholar]
- 38.Liang C. L., Chen J. L., Hsu Y. P., Ou J. T., Chang Y. S. (2002) J. Biol. Chem. 277, 23345–23357 [DOI] [PubMed] [Google Scholar]