Abstract
Previous studies showed that cytoplasmic and mitochondrial forms of yeast valyl-tRNA synthetase (ValRS) are specified by the VAS1 gene through alternative initiation of translation. Sequence comparison suggests that the yeast cytoplasmic (or mature mitochondrial) ValRS contains an N-terminal appendage that acts in cis as a nonspecific tRNA-binding domain (TRBD) and is absent from its bacterial relatives. We show here that Escherichia coli ValRS can substitute for the mitochondrial and cytoplasmic functions of VAS1 by fusion of a mitochondrial targeting signal and a TRBD, respectively. In addition, the bacterial ValRS gene can be converted into a dual functional yeast gene encoding both cytoplasmic and mitochondrial activities by fusion of a DNA sequence specifying both the mitochondrial targeting signal and TRBD. In vitro assays suggested that fusion of a nonspecific TRBD to the bacterial enzyme significantly enhanced its yeast tRNA-binding and aminoacylation activities. These results not only underscore the necessity of retaining a TRBD for functioning of a tRNA synthetase in yeast cytoplasm, but also provide insights into the evolution of tRNA synthetase genes.
Aminoacyl-tRNA synthetases (aaRSs)2 are a group of ancient enzymes responsible for protein translation, each of which catalyzes the attachment of a specific amino acid to its cognate tRNAs, forming aminoacyl-tRNAs. These charged tRNAs are then delivered by elongation factor-1 to ribosomes for protein translation. In prokaryotes, there are typically 20 aminoacyl-tRNA synthetases, one for each amino acid (1–4). In eukaryotes, protein synthesis occurs not only in the cytoplasm, but also in organelles, such as mitochondria and chloroplasts (5). Thus, eukaryotes, such as yeast, commonly have two genes that encode distinct sets of proteins for each aminoacylation activity, one localized in the cytoplasm and the other in the mitochondria. Each set aminoacylates the isoaccepting tRNAs within its respective cell compartment and is sequestered from the isoacceptors confined in other compartments. In most cases, cytoplasmic and mitochondrial synthetase activities are encoded by two distinct nuclear genes, regardless of the cell compartments to which they are confined. However, in some cases, cytoplasmic and mitochondrial forms of a tRNA synthetase with a given amino acid specificity are encoded by the same nuclear gene through alternative initiation of translation, examples of which include ALA1 (coding for alanyl-tRNA synthetase) (6, 7), GRS1 (coding for glycyl-tRNA synthetase) (8), HTS1 (coding for histidyl-tRNA synthetase) (9), and VAS1 (coding for valyl-tRNA synthetase (ValRS)) (10). Because the isozymes are targeted to different compartments, the two isoforms of ValRS, for example, cannot be substituted for each other in vivo. A similar scenario has been observed for genes encoding mitochondrial and cytoplasmic forms of Arabidopsis thaliana alanyl-tRNA synthetase, threonyl-tRNA synthetase, and ValRS (11).
Many yeast cytoplasmic tRNA synthetases contain an N- or C-terminal polypeptide extension that is absent from their bacterial homologs (12). A well studied example is the appended domain (Ad) of yeast glutaminyl-tRNA synthetase (GlnRS), which binds crude yeast tRNAs, single-stranded RNA, and pseudoknot RNA with comparable affinities; the Kd values are ∼0.6 μm (13, 14). Similar examples have been reported in yeast ValRS (15) and tRNA synthetases of higher eukaryotes, such as the EMAPII-like domain of plant methionyl-tRNA synthetase (16), the repeat domain of human methionyl-tRNA synthetase (17), and the N-terminal domain of mammalian lysyl-tRNA synthetase (18, 19). In addition to serving as a cis-acting tRNA-binding domain (TRBD), the Ads of some yeast tRNA synthetases were found to participate in protein-protein interactions, such as those of yeast glutamyl-, methionyl- (20), and seryl-tRNA synthetases (21). These interactions were shown to enhance their tRNA binding and aminoacylation (20, 21).
Interestingly, many of the Ads of yeast tRNA synthetases contain one or several canonical nuclear localization signals (22) that are thought to play a role in the nuclear import of these otherwise “cytoplasmic” proteins. Nuclear aminoacylation of tRNAs is thought to serve as a functional checkpoint for the maturity of tRNAs before they are exported from the nucleus (23, 24). By contrast, in higher eukaryotes, nine aminoacyl-tRNA synthetases and three auxiliary proteins (p43, p38, and p18) form a multienzyme complex through interactions of their hydrophobic Ads (25). In addition, ValRS from mammalian cells is exclusively isolated as a high molecular mass complex with the elongation factor EF-1H (26–28). Recently, several tRNA synthetases from prokaryotes and eukaryotes have been shown to take part in functions beyond aminoacylation, including roles in mitochondrial RNA splicing, transcriptional and translational regulation, cytokine-like activity, and amino acid biosynthesis (29, 30).
In this report, we focused on the cross-species and cross-compartmental complementation activities of a bacterial tRNA synthetase in an attempt to understand further the evolutionary pathway that has converted a bacterial tRNA synthetase gene into a dual functional yeast gene possessing both cytoplasmic and mitochondrial activities. Our results show that although Escherichia coli ValRS per se cannot substitute for the cytoplasmic activity of yeast VAS1, fusion of a nonspecific TRBD (such as Arc1p or the Ads of yeast GlnRS and ValRS) to the bacterial enzyme enabled the otherwise nonfunctional enzyme to rescue the growth defect of a VAS1 knock-out strain on 5-fluoroorotic acid (5-FOA). In contrast, the E. coli enzyme, when targeted to mitochondria, could substitute for the mitochondrial activity of VAS1 without the assistance of a cis-acting nonspecific TRBD. These results, together with others, suggest that acquiring a cis- or trans-acting TRBD might be necessary and sufficient for functioning by a yeast cytoplasmic tRNA synthetase, which might explain why so many yeast cytoplasmic aaRSs contain an N- or C-terminal Ad (12). In contrast, obtaining a cis- or trans-acting TRBD does not appear to be necessary for functioning of most yeast mitochondrial aaRSs.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
Cloning of the wild-type yeast VAS1 gene into the high copy number yeast shuttle vector pADH was previously described (31). A short DNA duplex coding for a FLAG (for Western blotting) or His6 tag (for protein purification) was inserted in-frame at the 3′ end of the VAS1 gene. To clone the E. coli valS gene, the open reading frame of this gene was first amplified by PCR using bacterial genomic DNA as the template. The forward (with an inserted SpeI site, underlined) and reverse (with an inserted XbaI site, underlined) primers used contained sequences complementary to base pairs −15 to +18 (5′-AACCTGGAAACTAGTATGGAAAAGACATATAAC-3′) and +2842 to +2871 (5′-ATCACTGTGTTTTCTAGACAGCGCGGCGAT-3′) of E. coli valS, respectively. After the PCR, the amplified DNA products were digested with the restriction enzymes SpeI and XbaI and then cloned into the appropriate sites of pADH for expression. Cloning of the Bacillus subtilis valS gene into pADH followed a similar protocol.
For fusion of the Ad of yeast ValRS (residues 1–97), the Ad of yeast GlnRS (residues 1–228), or Arc1p to E. coli ValRS, DNA sequence coding for the respective protein domain was PCR-amplified as an SpeI-SpeI fragment and inserted in-frame into the SpeI site at the 5′ end of the open reading frame of E. coli valS, yielding Ad(ScValRS)-EcValRS, Ad(ScGlnRS)-EcValRS, and Arc1p-EcValRS. For fusion of a mitochondrial targeting signal (MTS) to B. subtilis and E. coli ValRSs, DNA sequence coding for the MTS of the mitochondrial precursor form of yeast ValRS (residues 1–46) was PCR-amplified as an SpeI-SpeI fragment and inserted in-frame into the SpeI site at the 5′ end of the open reading frames of the B. subtilis and E. coli valS genes. The orientation of the SpeI-SpeI fragment was subsequently verified by DNA sequencing. Expressions of these constructs were under the control of a constitutive ADH promoter. Fusion of the DNA segment containing the promoter and sequence coding for the MTS and Ad of yeast ValRS to E. coli valS followed a similar protocol, except that the vector used for cloning and expression was pRS425 in this case.
Complementation Assays for Cytoplasmic Function
The yeast VAS1 knock-out strain, CW1, was described previously (31). This strain is maintained by a plasmid containing the wild-type VAS1 gene and a URA3 marker. Complementation assays for cytoplasmic ValRS activity were carried out by introducing a test plasmid carrying the gene of interest and a LEU2 marker into CW1, and the ability of the transformants to grow in the presence of 5-FOA was determined. Starting from a cell density of 4.0 A600, cell cultures were 5-fold serially diluted, and 10-μl aliquots of each dilution were spotted onto the designated plates containing 5-FOA. Plates were incubated at 30 °C for 3–5 days. The transformants evicted the maintenance plasmid with a URA3 marker in the presence of 5-FOA and thus could not grow on the selection medium unless a functional cytoplasmic ValRS was encoded by the test plasmid.
Complementation Assays for Mitochondrial Function
CW1 was cotransformed with a test plasmid (carrying a LEU2 marker) and a second maintenance plasmid (carrying a HIS3 marker) that expresses only the cytoplasmic form of ValRS (due to a mutation in the ATG1 initiator codon). In the presence of 5-FOA, the first maintenance plasmid (carrying a URA3 marker) was evicted from the cotransformants, whereas the second maintenance plasmid was retained. Thus, all cotransformants survived 5-FOA selections due to the presence of the cytoplasmic ValRS derived from the second maintenance plasmid. The mitochondrial phenotypes of the cotransformants were further tested on YPG plates at 30 °C, with results documented on day 3 after plating. Because a yeast cell cannot survive on glycerol without functional mitochondria, the cotransformants did not grow on the YPG plates unless a functional mitochondrial ValRS was generated from the test plasmid.
Western Blot Analysis
The protein expression patterns of the constructs used in the complementation assays were determined by a chemiluminescence-based Western blot analysis. INVSc1 (Invitrogen) was first transformed with the constructs of interest, and total protein extracts were prepared from each transformant. Aliquots of the protein extracts (40 μg) were loaded onto a mini gel (8 × 10 cm) containing 10% polyacrylamide and electrophoresed at 100 V for 1–2 h. After electrophoresis, the resolved proteins were transferred using a semidry transfer device to a polyvinylidene fluoride membrane in a buffer containing 30 mm glycine, 48 mm Tris base (pH 8.3), 0.037% SDS, and 20% methanol. The membrane was probed with a horseradish peroxidase-conjugated anti-FLAG tag antibody (Sigma) and then exposed to x-ray film after the addition of the appropriate substrates.
Aminoacylation Assay
Aminoacylation reactions were carried out at 25 °C in a buffer containing 10 mm Tris-HCl (pH 7.9), 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol, 4 mm ATP, 0.1 mg/ml bovine serum albumin, 0.1 mm E. coli or brewers' yeast tRNA (Hoffmann-La Roche, Basel, Switzerland), and 30 μm valine (1.7 μm [3H]valine; Moravek Biochemicals, Brea, CA). The specific activity of [3H]valine used was 35.0 Ci/mmol. Purification of the His6-tagged proteins was as described previously (32). Determination of active protein concentrations by active-site titration was as described (33). The final concentration of ValRS used in the reaction was 5, 10, or 50 nm. Reactions were quenched by spotting 10-μl aliquots of the reaction mixture onto Whatman filters soaked in 5% trichloroacetic acid and 1 mm valine. The filters were washed three times, for 15 min each, in ice-cold 5% trichloroacetic acid before liquid scintillation counting. Data were obtained from at least three independent experiments and averaged. Determination of the kinetic parameters, Km and kcat, of the purified enzymes for yeast tRNAVal was as described (15).
RESULTS
Converting E. coli ValRS into a Functional Yeast Cytoplasmic Enzyme
Previous studies showed that a single VAS1 gene specifies both the mitochondrial and cytoplasmic forms of ValRS through alternative use of two in-frame AUG initiator codons, AUG1 and AUG47 (10, 31). Hence, the mitochondrial precursor form essentially possesses the same polypeptide sequence as its cytoplasmic counterpart, except for an N-terminal MTS that is subsequently cleaved away by a matrix-processing peptidase upon being imported into mitochondria. Comparison of E. coli and yeast (the cytoplasmic form or the mature mitochondrial form) ValRSs showed that the catalytic cores of these two enzymes are significantly homologous to each another (∼42% identity), but the yeast enzyme has an N-terminal polypeptide extension of ∼97 residues that is absent from its E. coli counterpart (Fig. 1). This Ad has recently been shown to act in cis as a nonspecific TRBD to facilitate tRNA binding and aminoacylation by the enzyme (15).
FIGURE 1.
Comparison of E. coli and yeast ValRS. Although the catalytic cores of E. coli and yeast ValRSs are significantly homologous to each other, the yeast cytoplasmic (cyt) protein contains an N-terminal domain of ∼97 residues that is absent from its E. coli counterpart. The polypeptide sequence of the mitochondrial (mit) precursor form of yeast ValRS is essentially identical to that of its cytoplasmic isoform except for a mitochondrial targeting signal attached at its N terminus.
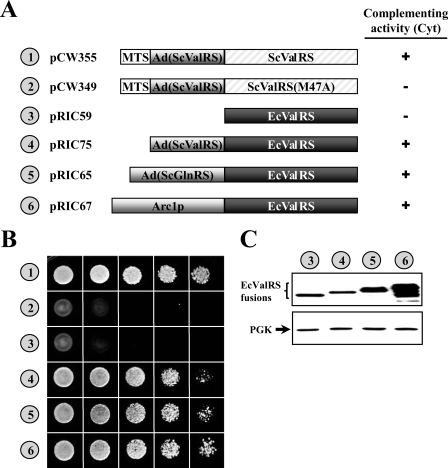
To test whether the E. coli enzyme can functionally substitute for its yeast homolog in vivo, the wild-type E. coli valS gene (encoding ValRS) was cloned into a yeast shuttle vector, and the ability of the resultant construct to rescue the growth defect of a yeast vas1− strain on 5-FOA was tested. As shown in Fig. 2, the construct containing E. coli valS failed to restore the growth phenotype of the knock-out strain CW1 on 5-FOA (Fig. 2B, number 3), suggesting that the cytoplasmic activity of yeast VAS1 cannot be substituted with the bacterial enzyme in vivo. However, fusion of a nonspecific TRBD, such as Arc1p or the Ads of yeast GlnRS and ValRS, to the E. coli enzyme enabled the otherwise nonfunctional bacterial enzyme to act as a yeast enzyme; each of these fusions effectively rescued the growth defect of the knock-out strain on 5-FOA (Fig. 2B, numbers 4–6). Constructs pCW355 and pCW349 served as positive and negative controls, respectively, in the assay (Fig. 2B, numbers 1 and 2). Construct pCW355 carries a wild-type VAS1 gene that expresses both cytoplasmic and mitochondrial forms of yeast ValRS, whereas construct pCW349 carries an initiator mutant of VAS1 (ATG47 to GCG) that expresses only the mitochondrial form of yeast ValRS.
FIGURE 2.
Converting E. coli ValRS into a functional yeast cytoplasmic enzyme. CW1, a yeast VAS1 knock-out strain, was transformed with constructs encoding various ValRSs, and the ability of these constructs to rescue the cytoplasmic defect of the knock-out strain was tested. A, schematic summary of constructs and their complementation activities. The symbols + and − indicate positive and negative complementation, respectively. Cyt, cytoplasmic. B, complementation assays for cytoplasmic ValRS activity on a 5-FOA plate. C, assay of protein expression by Western blotting. Upper panel, ValRS; lower panel, phosphoglycerate kinase (PGK) (as a loading control). Numbers 1–6 (circled) in B and C represent the constructs shown in A.
To examine whether E. coli ValRS and its fusions are properly expressed in yeast, we next examined the relative expression levels of these constructs by Western blotting using an anti-FLAG tag antibody. As shown in Fig. 2C, E. coli ValRS and its fusions were all properly expressed in yeast. Fusion of Arc1p or the Ad of yeast GlnRS to E. coli ValRS significantly enhanced its expression level (2- and 7-fold, respectively), whereas fusion of the Ad of yeast ValRS to the E. coli enzyme slightly reduced its expression level (1.5-fold). Evidently, not all TRBDs enhanced the protein expression level of E. coli ValRS in yeast. This observation also reinforces the idea that the negative phenotype of E. coli ValRS in the complementation assay (Fig. 2, number 3) was not caused by a lower level of protein expression.
Converting E. coli ValRS into a Functional Yeast Mitochondrial Enzyme
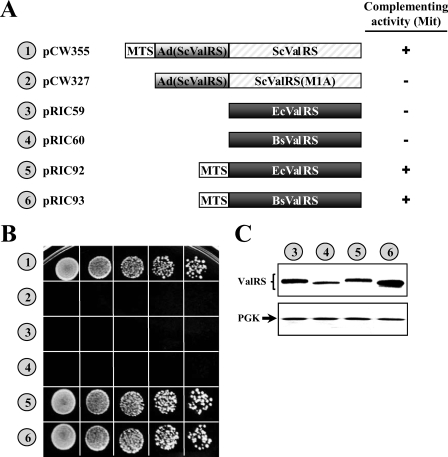
In contrast to the cytoplasmic aaRSs, the mitochondrial enzymes are generally believed to be more bacterium-like in terms of their genetic origin and sequence homology. To investigate whether the mitochondrial activity of yeast VAS1 can be functionally substituted with E. coli valS, a DNA segment coding for the MTS of the mitochondrial precursor form of yeast ValRS (base pairs +1 to +138 relative to the ATG1 initiator codon) was inserted in-frame at the 5′ end of E. coli valS to facilitate the mitochondrial import of the encoded bacterial protein, and the mitochondrial complementation activity of the resulting construct was tested. As shown in Fig. 3, although the E. coli enzyme per se (without fusion of an MTS) failed to restore the growth phenotype of CW1 on YPG, fusion of an MTS to the bacterial enzyme enabled it to rescue the mitochondrial defect of the knock-out strain (Fig. 3B, numbers 3 and 5). Interestingly, ValRS of B. subtilis, a Gram-positive bacterium that is genetically highly divergent from E. coli, could also be made a functional yeast mitochondrial enzyme by fusion of the MTS (Fig. 3B, numbers 4 and 6).
FIGURE 3.
Conversion of E. coli ValRS into a functional yeast mitochondrial enzyme. CW1, a yeast VAS1 knock-out strain, was transformed with constructs encoding various ValRSs, and the ability of these constructs to rescue the mitochondrial defect of the knock-out strain was tested. A, schematic summary of constructs and their complementation activities. Mit, mitochondrial. ScValRS(M1A) specifies only the cytoplasmic form of yeast ValRS due to a mutation at the AUG1 initiator codon. B, complementation assays for mitochondrial ValRS activity on a YPG plate. C, assay of protein expression by Western blotting. Upper panel, ValRS; lower panel, phosphoglycerate kinase (PGK) (as a loading control). Numbers 1–6 (circled) in B and C represent the constructs shown in A.
Because the MTS of the fusion enzymes was cleaved away upon being imported into mitochondria (Fig. 3C, numbers 3–6), it is therefore likely that the MTS did not participate in the aminoacylation reactions catalyzed by the bacterial enzymes. Constructs pCW355 and pCW327 served as positive and negative controls, respectively, in the assay (Fig. 3B, numbers 1 and 2). Construct pCW355 carries a wild-type VAS1 gene that expresses both cytoplasmic and mitochondrial forms of yeast ValRS, whereas construct pCW327 carries an initiator mutant of VAS1 (ATG1 to GCG) that expresses only the cytoplasmic form of yeast ValRS.
Enhancing the Aminoacylation Activity of E. coli ValRS with a TRBD
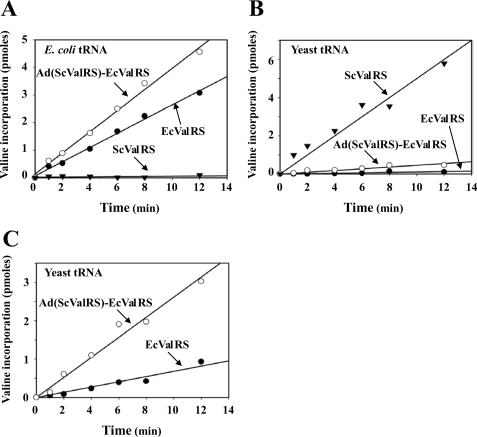
To investigate whether the TRBDs shown in Fig. 2A enhance the aminoacylation activity of E. coli ValRS in vitro, His6-tagged E. coli ValRS, Ad(ScValRS)-EcValRS, and yeast ValRS were purified to homogeneity using nickel-nitrilotriacetic acid column chromatography and then assayed using unfractionated E. coli and yeast tRNAs as the substrates. Aminoacylation was carried out at 25 °C at a final concentration of 5, 10, or 50 nm ValRS. As shown in Fig. 4A, yeast ValRS could barely charge E. coli tRNA in vitro compared with the E. coli enzyme under the conditions used. Likewise, E. coli ValRS had a relatively poor charging activity against yeast tRNA in vitro compared with the yeast enzyme (Fig. 4B). This finding is not surprising, considering the fact that the E. coli and yeast enzymes have coevolved with their respective cognate tRNAs and therefore developed certain levels of species specificity or barriers in tRNA recognition.
FIGURE 4.
Aminoacylation assays. A, aminoacylation assay using crude E. coli tRNA as the substrate. The ability of ScValRS, EcValRS, and Ad(ScValRS)-EcValRS to incorporate [3H]valine into E. coli tRNAVal was assayed at a final concentration of 5 nm. The relative amounts of [3H]valine that were incorporated into tRNA were subsequently determined by a liquid scintillation counter. B, aminoacylation assay using crude yeast tRNA as the substrate. The aminoacylation activities of the ValRSs were assayed at a final concentration of 10 nm. C, aminoacylation assay using crude yeast tRNA as the substrate. The aminoacylation activities of the ValRSs were assayed at a final concentration of 50 nm.
As for the aminoacylation activity of the fusion enzyme (Ad(ScValRS)-EcValRS), our results showed that fusion of the Ad of yeast ValRS to the E. coli enzyme enhanced its aminoacylation activity against yeast tRNA by ∼3.8-fold (Fig. 4B) but had only a minor effect on its aminoacylation activity against E. coli tRNA (∼1.4-fold increase) (Fig. 4A). To illustrate better the difference in aminoacylation activities against yeast tRNA between E. coli ValRS and the fusion enzyme, the protein concentration used for the assay was increased by 5-fold (from 10 to 50 nm). As shown in Fig. 4C, the fusion enzyme showed significantly higher aminoacylation activity against yeast tRNA than did E. coli ValRS, which might explain why the fusion enzyme was able to rescue the growth defect of CW1 on 5-FOA, whereas E. coli ValRS was not (Fig. 2). It should be noted that although the fusion enzyme had a significantly higher aminoacylation activity against yeast tRNA than did the E. coli enzyme, this level of activity was still far lower than that of the yeast enzyme (Fig. 4B).
To gain further insights into the molecular mechanism of enzyme catalysis, the Km and kcat values of E. coli ValRS and Ad(ScValRS)-EcValRS for yeast tRNAVal were determined. As it turned out, fusion of the Ad of yeast ValRS to the E. coli enzyme significantly enhanced its tRNA-binding affinity (∼5.3-fold increase) and had little effect on its turnover number. Ad(ScValRS)-EcValRS has a kcat/Km value 5-fold higher than that of E. coli ValRS (see Table 1). In sum, these results suggest that TRBDs such as the Ad of yeast ValRS can significantly improve the aminoacylation activity of E. coli ValRS against yeast tRNA.
TABLE 1.
Kinetic parameters for aminoacylation of tRNAVal by EcValRS and Ad(ScValRS)-EcValRS
Each value is determined from a hyperbolic fit of two independent data sets.
| ValRS variant | Km | kcat | kcat/Km | Relative efficiency |
|---|---|---|---|---|
| μm | s−1 | m−1s−1 | ||
| EcValRS | 2.08 ± 0.08 | (2.9 × 10−2) ± 0.007 | 1.39 × 104 | 1.0 |
| Ad(ScValRS)-EcValRS | 0.39 ± 0.05 | (2.7 × 10−2) ± 0.002 | 6.92 × 104 | 5.0 |
Merging Cytoplasmic and Mitochondrial ValRS Activities into a Single Gene
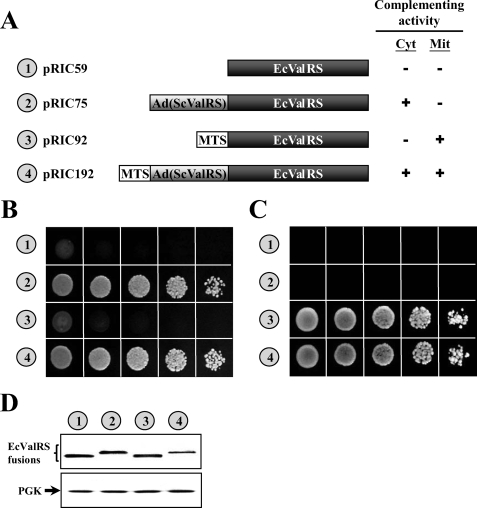
Because E. coli ValRS can be converted to a functional yeast cytoplasmic or mitochondrial enzyme, we wondered whether the E. coli ValRS gene can be converted into a dual functional yeast gene specifying both cytoplasmic and mitochondrial activities. To investigate this possibility, a DNA segment (base pairs −300 to +432 relative to the ATG1 initiator codon of yeast VAS1) that contains the VAS1 promoter and the sequence coding for the MTS (residues 1–46) and Ad (residues 47–144) of yeast ValRS was fused in-frame to the 5′ end of the open reading frame of E. coli valS, and the complementation activities of the resultant construct were tested. As shown in Fig. 5, the fusion construct successfully rescued the growth defects of the VAS1 knock-out strain on both 5-FOA and YPG, suggesting that, as with VAS1, the fusion construct specifies both activities, with the mitochondrial activity being provided by the longer form (MTS-Ad(ScValRS)-EcValRS) and the cytoplasmic activity by the shorter form (Ad(ScValRS)-EcValRS). Because the MTS of the mitochondrial precursor form (the longer form) was cleaved away upon being imported into mitochondria, making the processed mitochondrial form indistinguishable in size from its cytoplasmic counterpart, only one protein band with a molecular mass close to Ad(ScValRS)-EcValRS (∼119 kDa) was identified by Western blotting (Fig. 5D).
FIGURE 5.
Merging cytoplasmic and mitochondrial ValRS activities into a single gene. CW1 was transformed with constructs encoding E. coli ValRS and its fusions, and the ability of these constructs to rescue the cytoplasmic and mitochondrial defects of the knock-out strain was tested. A, schematic summary of constructs and their complementation activities. Cyt, cytoplasmic; Mit, mitochondrial. B, complementation assays for cytoplasmic ValRS activity on a 5-FOA plate. C, complementation assays for mitochondrial ValRS activity on a YPG plate. D, assay of protein expression by Western blotting. Upper panel, ValRS; lower panel, phosphoglycerate kinase (PGK) (as a loading control). Numbers 1–4 (circled) in B–D represent the constructs shown in A.
DISCUSSION
For a given amino acid specificity, there are generally two distinct nuclear aaRS genes in yeast, one encoding the cytoplasmic form and the other its mitochondrial counterpart. Occasionally, one of the two homologous genes has been lost during evolution, whereas the other has picked up the lost function and become functional in both compartments. Some examples of this type include ALA1 (6, 7), GRS1 (8), HTS1 (9), and VAS1 (10). Our results presented herein argue that a bacterial aaRS gene can be converted into a dual functional yeast gene by fusion of a DNA sequence coding for both an MTS and a TRBD (Fig. 5). In addition, our results underline the necessity of obtaining a nonspecific TRBD for efficient binding and aminoacylation of yeast cytoplasmic tRNA by the bacterial enzyme (Fig. 4 and Table 1).
Although the fusion enzyme Ad(ScValRS)-EcValRS has relatively poor aminoacylation activity against yeast tRNA compared with the wild-type yeast enzyme (Fig. 4), the fusion enzyme can still effectively rescue the growth defect of CW1 on 5-FOA (Fig. 2). A similar scenario was observed in E. coli GlnRS, where fusion of Arc1p to the E. coli enzyme only slightly improved its aminoacylation activity against yeast tRNA yet enabled this bacterial enzyme to rescue the growth defect of a yeast GLN4 (encoding GlnRS) knock-out strain (13). Therefore, it appears that a mutant or chimeric enzyme with aminoacylation activity as low as 1–2% relative to the native yeast enzyme is sufficient to retain a near wild-type growth phenotype for the knock-out strain. This finding is particularly interesting in light of the fact that although these TRBDs possess only nonspecific tRNA-binding activity, they can promote the formation of an active complex with yeast cognate tRNA by the prokaryotic enzyme and not an abortive complex that is often seen with noncognate or mutant tRNA. This observation also reinforces the notion that kcat discrimination (specificity of tRNA aminoacylation) plays a key role in the formation of Val-tRNAVal.
The binding affinity of a tRNA synthetase for its cognate tRNAs is generally characterized by dissociation constants on the order of 0.1–1 μm under physiological conditions (34). This relatively poor affinity ensures that the synthetases (or tRNAs) turn over rapidly during aminoacylation. In this sense, it is interesting to point out that the binding affinities of the Ads of yeast GlnRS and ValRS for tRNA also fall into this range, making them useful as a cis-acting tRNA-binding cofactor during aminoacylation (15). Although the Ads of GluRS and MetRS are also rich in positively charged residues and important for aminoacylation, they do not function as a TRBD. Instead, these Ads specifically interact with a tRNA-binding cofactor, Arc1p, which, in turn, recruits tRNA to the vicinity of the enzymes for aminoacylation (35). A functionally similar tRNA-recruiting domain was identified in an auxiliary protein associated with the mammalian multisynthetase complex (36).
ValRS from mammalian cells is exclusively isolated as a high molecular mass complex with the elongation factor EF-1H (26–28). Like yeast ValRS, the mammalian enzyme also contains a strong affinity for the polyanionic carrier, heparin-Ultrogel. However, the mammalian enzyme exhibits additional hydrophobic properties (37). Sequence analysis revealed that mammalian ValRS has conserved the positively charged N-terminal extension (residues 200–298) that distinguishes yeast ValRS from its bacterial counterparts while acquiring an additional hydrophobic domain (residues 1–199) that is responsible for interacting with the four subunits of elongation factor EF-1H (α, β, γ, and δ subunits) (38). A very recent report showed that, like the Ad of yeast ValRS, the positively charged N-terminal extension (residues 200–298) of the mammalian enzyme is also a nonspecific TRBD (15).
However, regardless of the detailed interpretation, the most interesting findings reported here are the capability of a nonspecific TRBD to enhance the formation of an active synthetase-tRNA complex and the conversion of an E. coli aaRS gene to a dual functional yeast gene encoding both cytoplasmic and mitochondrial activities. To our knowledge, this appears to be the first example wherein an engineered bacterial aaRS gene can provide both cytoplasmic and mitochondrial aminoacylation activities in yeast. In addition, our findings also suggest that a nonspecific TRBD is necessary for proper functioning of eukaryotic cytoplasmic aaRSs but not for functioning of eukaryotic mitochondrial or bacterial enzymes. Perhaps this is one of the major reasons why so many yeast cytoplasmic aaRSs possess an N- or C-terminal appendage, whereas most yeast mitochondrial aaRSs do not.
This work was supported by Grant NSC97-2311-B-008-002-MY3 from the National Science Council, Taipei, Taiwan (to C.-C. W.).
- aaRS
- aminoacyl-tRNA synthetase
- Ad
- appended domain
- Ec
- E. coli
- EF
- elongation factor
- 5-FOA
- 5-fluoroorotic acid
- GlnRS
- glutaminyl-tRNA synthetase
- MTS
- mitochondrial targeting signal
- Sc
- Saccharomyces cerevisiae
- TRBD
- tRNA-binding domain
- ValRS
- valyl-tRNA synthetase
- YPG
- yeast extract peptone glycerol.
REFERENCES
- 1.Carter C. W., Jr. (1993) Annu. Rev. Biochem. 62, 715–748 [DOI] [PubMed] [Google Scholar]
- 2.Martinis S. A., Schimmel P. (1996) in Escherichia coli and Salmonella Cellular and Molecular Biology (Neidhardt F. C. ed) 2nd Ed., pp. 887–901, American Society for Microbiology, Washington, D. C [Google Scholar]
- 3.Giegé R., Sissler M., Florentz C. (1998) Nucleic Acids Res. 26, 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelchat M., Lapointe J. (1999) Biochem. Cell Biol. 77, 343–347 [PubMed] [Google Scholar]
- 5.Dietrich A., Weil J. H., Maréchal-Drouard L. (1992) Annu. Rev. Cell Biol. 8, 115–131 [DOI] [PubMed] [Google Scholar]
- 6.Tang H. L., Yeh L. S., Chen N. K., Ripmaster T., Schimmel P., Wang C. C. (2004) J. Biol. Chem. 279, 49656–49663 [DOI] [PubMed] [Google Scholar]
- 7.Chang K. J., Lin G., Men L. C., Wang C. C. (2006) J. Biol. Chem. 281, 7775–7783 [DOI] [PubMed] [Google Scholar]
- 8.Chang K. J., Wang C. C. (2004) J. Biol. Chem. 279, 13778–13785 [DOI] [PubMed] [Google Scholar]
- 9.Natsoulis G., Hilger F., Fink G. R. (1986) Cell 46, 235–243 [DOI] [PubMed] [Google Scholar]
- 10.Chatton B., Walter P., Ebel J. P., Lacroute F., Fasiolo F. (1988) J. Biol. Chem. 263, 52–57 [PubMed] [Google Scholar]
- 11.Souciet G., Menand B., Ovesna J., Cosset A., Dietrich A., Wintz H. (1999) Eur. J. Biochem. 266, 848–854 [DOI] [PubMed] [Google Scholar]
- 12.Mirande M. (1991) Prog. Nucleic Acids Res. Mol. Biol. 40, 95–142 [DOI] [PubMed] [Google Scholar]
- 13.Wang C. C., Schimmel P. (1999) J. Biol. Chem. 274, 16508–16512 [DOI] [PubMed] [Google Scholar]
- 14.Wang C. C., Morales A. J., Schimmel P. (2000) J. Biol. Chem. 275, 17180–17186 [DOI] [PubMed] [Google Scholar]
- 15.Chang C. P., Lin G., Chen S. J., Chiu W. C., Chen W. H., Wang C. C. (2008) J. Biol. Chem. 283, 30699–30706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaminska M., Deniziak M., Kerjan P., Barciszewski J., Mirande M. (2000) EMBO J. 19, 6908–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminska M., Shalak V., Mirande M. (2001) Biochemistry 40, 14309–14316 [DOI] [PubMed] [Google Scholar]
- 18.Francin M., Kaminska M., Kerjan P., Mirande M. (2002) J. Biol. Chem. 277, 1762–1769 [DOI] [PubMed] [Google Scholar]
- 19.Francin M., Mirande M. (2006) Biochemistry 45, 10153–10160 [DOI] [PubMed] [Google Scholar]
- 20.Simos G., Segref A., Fasiolo F., Hellmuth K., Shevchenko A., Mann M., Hurt E. C. (1996) EMBO J. 15, 5437–5448 [PMC free article] [PubMed] [Google Scholar]
- 21.Godinic V., Mocibob M., Rocak S., Ibba M., Weygand-Durasevic I. (2007) FEBS J. 274, 2788–2799 [DOI] [PubMed] [Google Scholar]
- 22.Schimmel P., Wang C. C. (1999) Trends Biochem. Sci. 24, 127–128 [DOI] [PubMed] [Google Scholar]
- 23.Lund E., Dahlberg J. E. (1998) Science 282, 2082–2085 [DOI] [PubMed] [Google Scholar]
- 24.Sarkar S., Azad A. K., Hopper A. K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirande M., Lazard M., Martinez R., Latreille M. T. (1992) Eur. J. Biochem. 203, 459–466 [DOI] [PubMed] [Google Scholar]
- 26.Bec G., Kerjan P., Zha X. D., Waller J. P. (1989) J. Biol. Chem. 264, 21131–21137 [PubMed] [Google Scholar]
- 27.Venema R. C., Peters H. I., Traugh J. A. (1991) J. Biol. Chem. 266, 12574–12580 [PubMed] [Google Scholar]
- 28.Motorin Y. A., Wolfson A. D., Löhr D., Orlovsky A. F., Gladilin K. L. (1991) Eur. J. Biochem. 201, 325–331 [DOI] [PubMed] [Google Scholar]
- 29.Martinis S. A., Plateau P., Cavarelli J., Florentz C. (1999) EMBO J. 18, 4591–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francklyn C., Perona J. J., Puetz J., Hou Y. M. (2002) RNA 8, 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C. C., Chang K. J., Tang H. L., Hsieh C. J., Schimmel P. (2003) Biochemistry 42, 1646–1651 [DOI] [PubMed] [Google Scholar]
- 32.Chen S. J., Ko C. Y., Yen C. W., Wang C. C. (2009) J. Biol. Chem. 284, 818–827 [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. R., Ashford J. S., Bruton C. J., Jakes R., Koch G. L., Hartley B. S. (1975) Biochemistry 14, 1–4 [DOI] [PubMed] [Google Scholar]
- 34.Schimmel P. R., Söll D. (1979) Annu. Rev. Biochem. 48, 601–648 [DOI] [PubMed] [Google Scholar]
- 35.Simos G., Sauer A., Fasiolo F., Hurt E. C. (1998) Mol. Cell 1, 235–242 [DOI] [PubMed] [Google Scholar]
- 36.Shalak V., Kaminska M., Mitnacht-Kraus R., Vandenabeele P., Clauss M., Mirande M. (2001) J. Biol. Chem. 276, 23769–23776 [DOI] [PubMed] [Google Scholar]
- 37.Bec G., Waller J. P. (1989) J. Biol. Chem. 264, 21138–21143 [PubMed] [Google Scholar]
- 38.Bec G., Kerjan P., Waller J. P. (1994) J. Biol. Chem. 269, 2086–2092 [PubMed] [Google Scholar]