Abstract
The cis-trans peptidylprolyl isomerase Pin1 plays a critical role in regulating a subset of phosphoproteins by catalyzing conformational changes on the phosphorylated Ser/Thr-Pro motifs. The phosphorylation-directed ubiquitination is one of the major mechanisms to regulate the abundance of p27Kip1. In this study, we demonstrate that Pin1 catalyzes the cis-trans conformational changes of p27Kip1 and further mediates its stability through the polyubiquitination mechanism. Our results show that the phosphorylated Thr-187-Pro motif in p27Kip1 is a key Pin1-binding site. In addition, NMR analyses show that this phosphorylated Thr-187-Pro site undergoes conformational change catalyzed by Pin1. Moreover, in Pin1 knock-out mouse embryonic fibroblasts, p27Kip1 has a shorter lifetime and displays a higher degree of polyubiquitination than in Pin1 wild-type mouse embryonic fibroblasts, suggesting that Pin1 plays a critical role in regulating p27Kip1 degradation. Additionally, Pin1 dramatically reduces the interaction between p27Kip1 and Cks1, possibly via isomerizing the cis-trans conformation of p27Kip1. Our study thus reveals a novel regulatory mechanism for p27Kip1 stability and sheds new light on the biological function of Pin1 as a general regulator of protein stability.
Cellular differentiation and cell cycle inhibition are tightly controlled via sensitive molecular mechanisms. p27Kip1, a member of the Cip/Kip family, is an essential cell cycle inhibitor that functions largely during the G0/G1 phase where it promotes the assembly of the cyclin D1-CDK4 complex and inhibits the kinase activity of the cyclin E-CDK2 complex in the G1-S phase (1–4). Several review articles have elegantly summarized and discussed the detailed cellular functions of p27Kip1 (1–6). p27Kip1 is also a phosphoprotein with multiple Ser/Thr phosphorylation sites, including Ser-10, Ser-178, and Thr-187, followed by a proline residue. Hence, these motifs are potential substrate sites for proline-directed kinases (5, 6). Compared with Ser-178, which has not yet been well studied, the phosphorylation of Ser-10 and Thr-187 has been well characterized to be important for the regulation of p27Kip1 function. For instance, Ser-10 has been found to be the major phosphorylation site of p27Kip1 (7) and to play an important role in regulating cell migration (8–10), although the regulation of Ser-10 phosphorylation is still not completely defined (11, 12).
In contrast to Ser-10 and Thr-178, Thr-187 is the best characterized phosphorylation site on p27Kip1 and is known to regulate the complex formation of p27Kip1-cyclin E-CDK2 (1–2). In addition, it is also widely accepted that Thr-187 plays a crucial role in determining the abundance of mature p27Kip1 proteins. The phosphorylation of Thr-187 directs p27Kip1 to an SCFSkp2 ubiquitin ligase complex (consisting of Skp2-Skp1-Cks1-Cul1-Roc1), which in turn promotes the polyubiquitination and degradation of p27Kip1 (13, 14). The crystal structure of the Skp1-Skp2-Cks1-p27Kip1 phosphopeptide complex shows that p27Kip1 binds both Cks1 and Skp2 and that the C terminus of Skp2 and Cks1 forms the substrate recognition core of the SCF complex (15). Furthermore, the structure of this complex has revealed that the phosphorylation of Thr-187 in p27Kip1 is recognized by the phosphate-binding site of Cks1, indicating that Cks1 is not only a facilitator but also an indispensable component in p27Kip1 degradation machinery (15).
Pin1 is a unique peptidyl-prolyl isomerase (PPIase)2 that recognizes only the phosphorylated Ser/Thr motif preceding a proline residue (16). In addition, Pin1 is very prominent in isomerizing the cis-trans conformation of prolyl-peptidyl bonds in its substrates, resulting in either the modification of their function (e.g. c-Jun (17), β-catenin (18), Bax (19), and Notch1 (20)) or modulation of their stability (e.g. cyclin D1 (21), p53 (22, 23), and NF-κB (24)). Loss of Pin1 in mice results in several phenotypes similar to those of cyclin D1-null mice (21) and neuronal degenerative phenotypes (25–28), suggesting the conformational changes mediated by Pin1 may be crucial for the normal functioning of cells. Additionally, Pin1 also plays important roles in cancer and other cellular events, which have been extensively discussed in several recent review articles (29–33).
In this study, we show that Pin1 binds to p27Kip1, mainly through the phosphorylated Thr-187-Pro motif, and causes subsequent prolyl isomerization of this cell cycle protein. Moreover, we also find that Pin1 can protect p27Kip1 from degradation. Importantly, we demonstrate that by catalyzing conformational changes in p27Kip1, Pin1 hinders its association with Cks1, resulting in a reduction of polyubiquitination of p27Kip1 and protecting its degradation by SCFSkp2 complexes. Our results suggest that the cis-trans isomerization catalyzed by Pin1 represents a novel regulatory mechanism during post-phosphorylation of proteins and polyubiquitination-directed degradation pathways.
EXPERIMENTAL PROCEDURES
Constructs, Reagents, and Antibodies
Full-length cDNAs for p27Kip1, Cks1, and Skp2 were cloned from a HeLa cDNA library and inserted into the pXJ-40-FLAG vector and/or pXJ-40-GFP vector (a gift from Dr. B. C. Low, Department of Biological Sciences, National University of Singapore) or into p3x-FLAG-CMV vector (Sigma). Recombinant proteins of the full-length human Pin1, and its WW and PPIase domains, were expressed from the pET42b(+) vector (Novagen). All mutant constructs were generated using the QuikChange® site-directed mutagenesis kit (Stratagene). Cycloheximide, FLAG-M2 beads, and MG132 were purchased from Sigma. Okadaic acid was purchased from Santa Cruz Biotechnology. Antibodies used for Western blotting and pulldown assays were as follows: polyclonal antibody against p27Kip1 (C-19), monoclonal antibody against p27Kip1 (F-8), polyclonal antibodies against Ser(P)-10-p27Kip1, Skp2 (H-435), Cks1 (FL-79), and ubiquitin (P4D1) (Santa Cruz Biotechnology); polyclonal antibody against Thr(P)-187-p27Kip1 (Zymed Laboratories Inc.); monoclonal antibodies against α-tubulin and FLAG (Sigma).
Cell Culture and Transfection
Human embryonic kidney (HEK) epithelial 293T cells and Pin1-WT (wild-type) and Pin1-KO (knock-out) mouse embryonic fibroblasts (MEFs; a gift from Dr. K. P. Lu, Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin unless otherwise noted. For overexpression analysis, all constructs into MEFs were transfected using Lipofectamine (Invitrogen) according to the manufacturer's protocol. HEK 293T cells were transfected using the calcium phosphate method. Following transfection, cells were harvested in mammalian lysis buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 10% glycerol, 1% Triton X-100, 1 mm EDTA, supplemented with protease inhibitors and phosphatase inhibitors, including 1 μm pepstatin, 1 μm leupeptin, 50 μm β-glycerophosphatase, 1 mm okadaic acid, 1 mm Na3VO4).
GST Pulldown and Co-immunoprecipitation Assays
For GST pulldown assay, recombinant GST-full-length Pin1, -WW, or -PPIase domain proteins were conjugated to glutathione-Sepharose 4B beads. Beads were rocked with lysates from cells overexpressing WT or mutant p27Kip1 constructs for 3 h at 4 °C, followed by five washes with mammalian lysis buffer. The bound proteins were eluted with SDS-loading dye and resolved by 12–15% SDS-PAGE, followed by Western blotting. Calf intestinal phosphatase (CIP) treatments were performed as described previously (23). CIP, 1 unit/ml (Promega), was added to the cell lysates for 30 min at 30 °C, which were then subjected to GST pulldown. For co-immunoprecipitation, FLAG-p27Kip1 and mutant constructs were overexpressed in HEK 293T cells. Cell lysates were then cleared by centrifugation at 13,000 rpm for 10 min, and the resulting supernatants were incubated with FLAG-M2 beads for 3–5 h at 4 °C. The bound proteins were then analyzed by Western blotting.
Protein Stability Assay
Pin1-WT and -KO MEF cells were transfected with FLAG-p27Kip1 WT or mutant constructs. Within 24 h of transfection, cycloheximide (100 μg/ml) was added to the cells to block protein synthesis. Cells were then harvested at 4-h intervals, followed by Western blotting.
In Vivo Ubiquitination Assay
FLAG-p27Kip1 and Myc-ubiquitin constructs were co-transfected into Pin1-WT and -KO MEF cells, respectively. At 24 h following transfection, cells were treated with 10 μm MG132 to inhibit protein degradation. After 3 h of drug treatment, cells were harvested. The supernatants of cell lysates were then incubated with FLAG-M2 beads at 4 °C for 3 h, followed by five washes with mammalian lysis buffer and Western blotting using anti-ubiquitin antibody. The membranes were subsequently stripped and re-blotted with anti-p27Kip1 antibody to evaluate the amount of p27Kip1 that had bound to the beads. Reciprocal experiments were carried out by co-overexpressing FLAG-ubiquitin and GFP-p27Kip1 in Pin1-WT and KO MEF cells. After MG312 treatment, cell lysates were subjected to pulldown assays using FLAG-M2 beads. Consequently, the beads were cooked at 95 °C for 5 min. Western blotting analyses were done using either anti-p27Kip1 or anti-ubiquitin antibody.
Conformational Change by NMR Spectroscopy
All NMR experiments were performed using a Bruker 800-MHz NMR spectrometer at 25 °C. All spectra were recorded for a 2.0 mm peptide concentration dissolved in a 20 mm phosphate buffer (90% H2O and 10% D2O, pH 6.5) in the presence or absence of 0.03 mm Pin1. For all experiments, 256 × 512 complex points were acquired with spectral widths of 7200 × 9600 Hz in t1 × t2 dimensions and a relaxation delay of 1 s. ROESY spectra (34) were acquired at a mixing time of 110 ms, a spin-lock field strength of 4 kHz, and 16 scans. The mixing times were 30, 50, 70, 90, and 110 ms. TOSCY experiments (35) were carried out at a mixing time of 75 ms and 8 scans.
RESULTS
Interaction of Pin1 and Phosphorylated p27Kip1
Observations from our several preliminary experiments led us to speculate that Pin1 may associate with p27Kip1 (supplemental Fig. S1). Sequence analysis also showed that p27Kip1 contains three potential Pin1-binding Ser/Thr-Pro motifs as follows: S10P, S178P, and T187P (Fig. 1A). To confirm that Pin1 could interact with p27Kip1, we performed glutathione S-transferase (GST) pulldown assays using recombinant GST-Pin1, GST-Pin1-W34A (impaired WW domain-binding site), and GST-Pin1-R68A/R69A mutants (impaired PPIase domain-binding sites) or GST proteins to pull down FLAG-p27Kip1 protein overexpressed in HEK 293T cells. As shown in Fig. 1B, our results indicate that GST-Pin1 interacts with FLAG-p27Kip1, but not GST control, indicating that Pin1 can bind p27Kip1 in vitro (Fig. 1B, 1st and 2nd lanes). It is also known that both WW and PPIase domains of Pin1 can bind phosphorylated Ser/Thr-Pro motifs (36). Therefore, we further performed GST pulldowns using Pin1-W34A and Pin1-R68A/R69A mutants to identify the region of Pin1 that is responsible for p27Kip1 binding. Notably, neither GST-Pin1-W34A nor GST-Pin1-R68A/R69A mutant associates strongly with p27Kip1 protein compared with GST-Pin1-WT, suggesting that both the WW and PPIase domains are required for an efficient Pin1-p27Kip1 interaction (Fig. 1B, 3rd and 4th lanes). To further test for a direct interaction of p27Kip1 with Pin1, which does not rely on any bridging proteins, cell lysates with overexpressed FLAG-p27Kip1 were subjected to heating at 95 °C for 10 min before GST pulldown because p27Kip1 is an intrinsically heat-stable protein (13). As shown in Fig. 1B (5th and 6th lanes), the Pin1-p27Kip1 interaction is intact after the heat treatment, indicating that p27Kip1 directly interacts with Pin1 in vitro. In addition, using an antibody specifically against phosphorylated p27Kip1, we also found that Pin1 interacts with phosphorylated p27Kip1 (Fig. 1B).
FIGURE 1.
Pin1 binds the Ser/Thr-Pro motifs of p27Kip1 through its WW and PPIase domain. A, schematic illustration of the potential Pin1-binding sites in p27Kip1, which includes the S10P (Ser-10-Pro), S178P (Ser-178-Pro), and T187P (Thr-187-Pro) motifs. NLS, nuclear localization signal. B, immunoblotting analysis following the use of GST-Pin1, GST-Pin1-W34A (W34A mutant), and GST-Pin1-R2A (R68A/R69A mutants) to pull down FLAG-p27Kip1. GST beads were used as a control. GST and GST-Pin1 proteins were stained as loading controls. C, immunoblotting (IB) analyses following the use of GST-Pin1 or GST beads to pull down either FLAG-p27Kip1 or -p27Kip1 mutants, in the absence (left panel) or presence (right panel) of CIP. OA, okadaic acid; CIP, calf intestinal phosphatase. D, co-immunoprecipitation (IP) assay using FLAG-M2 beads. GFP-Pin1-S16A was co-immunoprecipitated with FLAG-p27Kip1, but not a FLAG-p27Kip1-3A mutant. E, GFP-Pin1-S16A was co-immunoprecipitated with either the FLAG-p27Kip1-WT or the FLAG-p27Kip1-S10A mutant but not with the FLAG-p27Kip1-T187A mutant.
To identify the preferred Pin1-binding motif in p27Kip1, we constructed a series of p27Kip1 mutants by substituting Ser/Thr for Ala at its three putative Pin1-binding sites. We then performed GST pulldown and co-immunoprecipitation assays. Fig. 1C shows that Pin1 binds WT, all single mutants, and one double mutant of p27Kip1. However, the triple mutation (3A) of p27Kip1 totally abolishes this association (Fig. 1C, left panel), indicating that Ser-10, Ser-178, and Thr-187 of p27Kip1 may all be involved in Pin1-p27Kip1 binding in vitro. Given that Pin1 only binds phosphoproteins, and to confirm that the binding of Pin1 to p27Kip1 is phosphorylation-dependent, we incubated cell lysates with CIP before performing GST pulldown assays. As shown in Fig. 1C (right panel), the interactions between Pin1 and p27Kip1 WT/mutants are completely disrupted by CIP treatment, suggesting that the binding of Pin1 to p27Kip1 is phosphorylation-dependent. By next co-overexpressing FLAG-p27Kip1 and GFP-Pin1-S16A, a stronger binding derivative of Pin1 (37), the co-immunoprecipitation assays further demonstrated that Pin1 binds p27Kip1 WT but not the triple mutant (3A) in vivo (Fig. 1D), consistent with the GST pulldown results (Fig. 1C). Interestingly, we observed that the p27Kip1 S10A mutant co-immunoprecipitated significantly with Pin1 (Fig. 1E) but that there was almost no interaction between Pin1 and the p27Kip1 T187A mutant. Taken together, these results suggest that Thr-187 could be the major Pin1-binding site in p27Kip1 in vivo (Fig. 1E).
A single Ala residue substitution at the Ser/Thr-Pro motifs of p27Kip1 did not totally eliminate the interaction between Pin1 and p27Kip1 in GST pulldown assays (Fig. 1C). Hence, to further elucidate the binding property of the Pin1-p27Kip1 interaction, we developed phosphopeptide chips for binding analysis. To this end, three N-terminal biotinylated phosphopeptides derived from Ser/Thr-Pro motifs in p27Kip1 were synthesized (Fig. 2B). Subsequently, we immobilized these three biotinylated-p27Kip1 peptides on glass slides coated with avidin to generate peptide chips, as described previously (38, 39). To determine the affinity of Pin1 to these peptides, different concentrations of Cy3-labeled Pin1 were titrated on chips. The fluorescence reading for each spot was then extracted from the unbound background intensities and fitted to binding curves for individual Pin1-peptide interaction (Fig. 2A). A smaller Kd value indicates a stronger binding. As shown in Fig. 2B, Pin1 binds most strongly to the phosphorylated Thr-187 peptide, with a dissociation constant (Kd) of ∼7.2 μm. In contrast, Pin1 has a weaker affinity for phosphorylated Ser-10 or Ser-178 peptides with Kd values of ∼16 μm. Therefore, taken together both in vitro and in vivo binding assays confirm that the phosphorylated Thr-187 residue on p27Kip1 is the major Pin1-binding site.
FIGURE 2.
Phosphorylated Thr-187 is the major Pin1-binding site on p27Kip1. A, fitted binding curves for Pin1 recombinant protein bound to three phosphorylated peptides derived from p27Kip1. B, phosphorylated peptide sequences and Kd and S.D. values calculated from the binding curve shown in A, indicating the affinity of Pin1 for phosphorylated peptides derived from p27Kip1.
Pin1 Accelerates Conformational Changes in p27Kip1
Pin1 is a peptidylprolyl isomerase and accelerates conformational changes between the cis and trans forms of phosphorylated polypeptides. In addition to its binding of cognate substrates, the best way to demonstrate the function of Pin1 is to measure these catalytically driven conformational changes. To this end, we analyzed the isomerase activity of Pin1 against nonphosphorylated (Fig. 3A) and phosphorylated Thr-187-Pro peptides (GSVEGpTPKKPGA, where boldface indicates positions 6 and 8, respectively), in the absence of (Fig. 3B) or presence of Pin1 (Fig. 3C). Because of the slow exchange between cis and trans conformations of proline, several residues in both peptides displayed two distinct sets of 1H signals in the ROESY and total correlation spectroscopy spectra (Fig. 3, B and C). The cis and trans populations of both peptides were ∼10 and 90%, respectively, as estimated from the one-dimensional 1H spectrum. In a normal situation, the exchange between the cis and trans conformations was so slow on an NMR time scale; therefore, no cross-peaks between the two conformations were observed by NMR (<0.1 s−1, see Fig. 3B). In contrast, in the presence of Pin1, the proline isomerization rate of the phosphorylated peptide is greatly enhanced by Pin1. Cross-peaks from conformational exchange were also identified (Fig. 3B). Both cross-peaks and diagonal peaks of Thr-6 and Lys-8 amide protons from the phosphorylated Thr-187 peptides were also found (Fig. 3B). On the other hand, the nonphosphorylated peptide displayed no exchange peaks, even in the presence of Pin1 (Fig. 3A). Hence, Pin1 only accelerates the isomerization of the phosphorylated Thr-187 peptide in p27Kip1 (Fig. 3C) but not the nonphosphorylated control (Fig. 3A).
FIGURE 3.
Pin1 catalyzes conformational changes in phosphorylated p27Kip1. A, selected region of a two-dimensional ROESY spectrum of a nonphosphorylated Thr-187 peptide (AGSVEGTPKKPGLR) at a concentration of 2.4 mm and in the presence of 0.03 mm Pin1 (mixing time 110 ms) is shown. There is no cross-peak found (as indicated by arrows). B and C, selected regions of a two-dimensional ROESY spectrum of a phosphorylated Thr-187 peptide (GSVEGpTPKKPGA, where boldface indicates positions 6 and 8, respectively) at a concentration of 2.0 mm are shown in the absence (B) or in the presence (C) of 0.03 mm Pin1 (mixing time 110 ms). Negative and positive peaks are indicated by arrows. Diagonal peaks from cis and trans conformers are indicated by cc and tt, respectively. Exchange peaks resulting from Pin1-catalyzed isomerization are labeled ct and tc. Note that rotating frame nuclear Overhauser enhancement and exchange cross-peaks are indicated by arrows. Diagonal peaks of Thr (T6) and Lys (K8) amide protons from the phosphorylated Thr-187 peptides are identified by arrows. D and E, ratios of cross-peak and diagonal peak intensities for the cis and trans conformations of Thr-6 on rotating frame nuclear Overhauser enhancement mixing times and its isomerization rates are indicated; D, from the cis to trans (kctcat) conformation; E, from the trans to cis (ktccat) conformation. F, illustration of the conformational change of phosphorylated Thr-187 peptides by Pin1. The peptide models were generated using PyMOL. In the absence of Pin1, the isomerization rates between the cis and trans conformations are very slow; however, in the presence of Pin1, the isomerization rates are greatly enhanced by Pin1. The rates of Thr-6 are presented.
Taking the intensities of cross-peaks and diagonal peaks of Thr-6 and Lys-8 amide protons of phosphopeptide, we further calculated the isomerization rates from the cis to trans (kctcat) and from the trans to cis (ktccat) conformations of Thr-6 (Fig. 3, D and E). The values of kctcat- and ktccat-obtained intensities for Thr-6 were 1.08 and 0.12 s−1, respectively. Similarly, the values of kctcat- and ktccat-obtained intensities for Thr-8 were 1.05 and 0.15 s−1, respectively (data not shown). The enhanced cis-trans conformational exchange rate by more than 10-fold suggests that the isomerization rates catalyzed by Pin1 are significantly faster than those without Pin1 (Fig. 3F).
Pin1 Protects p27Kip1 from Degradation
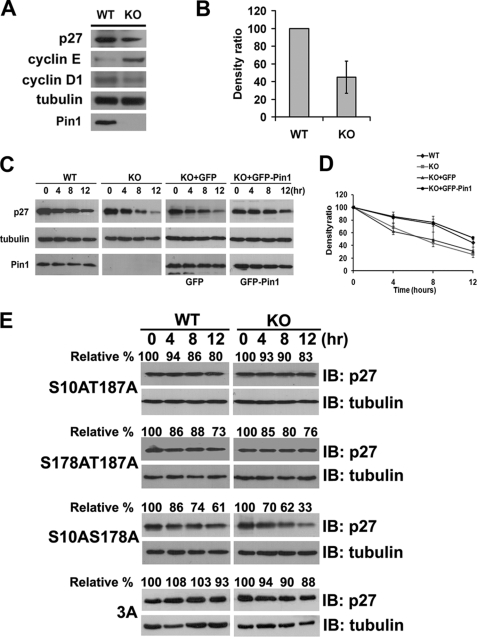
Phosphorylation on Thr-187 was known to be very important for p27Kip1 degradation (6, 40). Our current results show that Pin1 catalyzes p27Kip1 through its phosphorylated Thr-187-Pro motif (Fig. 3). We next speculated whether Pin1 is involved in regulating p27Kip1 stability. To test this possibility, we measured endogenous p27Kip1 levels and found them to be dramatically lower, by about 50%, in Pin1-KO MEFs compared with Pin1-WT MEFs (Fig. 4, A and B); as previously reported, in our Pin1-KO MEFs, the cyclin D1 levels are markedly decreased (21), and cyclin E is significantly increased (41) (Fig. 4A). Consistently, we also find in our present experiments that a knockdown of Pin1 using small interfering RNA causes a decrease of the p27Kip1 levels in HEK 293T cells (supplemental Fig. S2). This observation indicates that Pin1 is able to stabilize p27Kip1 protein. Subsequently, we compared the half-life of endogenous p27Kip1 in both Pin1-WT and -KO MEFs exposed to cycloheximide. As shown in Fig. 4C, the endogenous p27Kip1 levels in Pin1-KO MEFs are significantly less stable than those in Pin1-WT MEFs. After 12 h of cycloheximide inhibition, endogenous p27Kip1 is markedly reduced by up to ∼76% in the absence of Pin1. In contrast, only a 50% reduction of p27Kip1 is evident in Pin1-WT MEFs (Fig. 4, C and D). To demonstrate that the destabilization of p27Kip1 is a direct effect of the absence of Pin1, we investigated the turnover rate of p27Kip1 by re-introducing GFP-Pin1 into Pin1-KO MEFs. As shown in Fig. 4, C and D, the stability of p27Kip1 is markedly enhanced by re-overexpression of GFP-Pin1, but not GFP alone, in Pin1-KO MEFs, suggesting that Pin1 plays a critical role in modulating p27Kip1 stability.
FIGURE 4.
Pin1 regulates p27Kip1 stability. A, immunoblotting analysis of the endogenous p27Kip1, cyclin E, and cyclin D1 levels in Pin1-WT and -KO MEFs. B, quantification of endogenous p27Kip1 levels shown in A, normalized to tubulin levels. C, protein stability assay of endogenous p27Kip1 in both Pin1-WT and -KO MEFs. Cells were starved for 36 h before their arrest at G0 phase. The cells were then treated with cycloheximide, harvested at 4-h intervals (left panel), and analyzed by immunoblotting. To confirm the function of Pin1 in regulating p27Kip1 stability, GFP-Pin1 and GFP vectors were re-introduced into Pin1-KO MEF cells, respectively, followed by a protein stability assay. D, densitometric analysis of the degradation assays from C, normalized to tubulin levels. E, protein stability assay of exogenous FLAG-p27Kip1 or its mutants in either Pin1-WT or -KO MEFs. Cells were treated with cycloheximide after 24 h of transfection and then harvested at 4-h intervals, followed by immunoblotting (IB) analyses. Tubulin was used as a loading control. KO, knock-out.
To clearly determine whether Pin1 specifically regulates p27Kip1 stability through the phosphorylated Thr-187 site, we investigated this stability in the presence or absence of Pin1 using the p27Kip1 double mutants, S10A/S178A (potential Thr(P)-187), S10A/T187A (potential Ser(P)-178), and S178A/T187A (potential Ser(P)-10), instead of single mutants of p27Kip1. We found that the stability of the two mutants containing T187A and the triple mutant (3A) showed no significant difference in the presence or absence of Pin1 (Fig. 4E). Furthermore, even though Pin1 may potentially interact with Ser(P)-10 (Fig. 1), the S178A/T187A mutant (potential Ser(P)-10) has no significant difference in turnover rate between Pin1-WT (73% remaining) and -KO MEFs (76% remaining), suggesting that Pin1 has little or no effect on p27Kip1 degradation through the Ser-10 site. Most importantly, the double mutant S10A/S178A (potential Thr(P)-187) displays a shorter half-life in the absence of Pin1, degrading 66% of its total level (Fig. 4E), underscoring the significance of Thr-187 of p27Kip1 for Pin1 function.
Pin1 Plays a Role in the Ubiquitination of p27Kip1
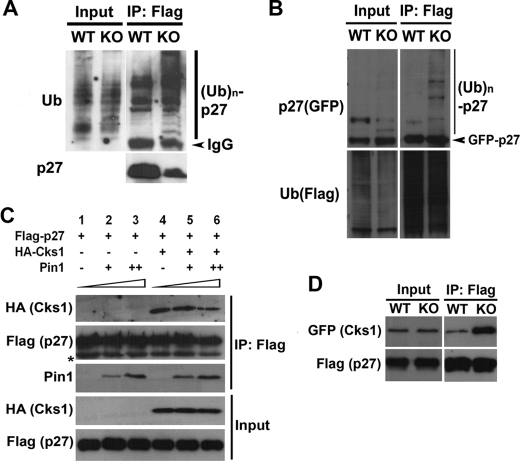
To further explore the molecular mechanisms underlying Pin1 regulation of p27Kip1 stability, we evaluated whether Pin1 is in fact involved in the p27Kip1 ubiquitination machinery. To this end, we performed in vivo ubiquitination assays in which following the treatment of MG132, FLAG-p27Kip1 and Myc-ubiquitin proteins were co-immunoprecipitated from Pin1-WT and -KO MEF cell lysates, respectively. Western blotting analyses revealed that p27Kip1 is strongly polyubiquitinated in Pin1-KO MEFs but only modestly so in Pin1-WT MEFs (Fig. 5A). To rule out the possibility that the polyubiquitination bands detected were not because of a contamination by the polyubiquitinated p27Kip1-associated proteins, we performed a reciprocal pulldown experiment by co-expressing the FLAG-ubiquitin and GFP-p27Kip1 in Pin1-WT and KO MEF cells. Consistent with previous results shown in Fig. 5A, loss of Pin1 significantly enhanced the polyubiquitination of p27Kip1 (Fig. 5B). These results are in a good agreement with our previous findings that in the presence of Pin1, p27Kip1 protein accumulates at a higher level (Fig. 4A and supplemental Fig. S2), whereas in the absence of Pin1, the half-life of p27Kip1 is much shorter because of a more rapid degradation rate (Fig. 4C).
FIGURE 5.
Pin1 is involved in p27Kip1 polyubiquitination. A, in vivo ubiquitination (Ub) assay of FLAG-p27Kip1 in Pin1-WT and -KO MEFs. At 24 h following transfection, cells were treated with 10 μm MG132 to inhibit protein degradation. After 3 h of MG132 treatment, cells were harvested, and lysates were then incubated with FLAG-M2. Western blots were carried out using anti-FLAG or anti-ubiquitin antibody. IP, immunoprecipitation; KO, knock-out. B, reciprocal experiments were carried out as in A, after FLAG beads pulldown, and the beads were cooked at 95 °C for 5 min, and Western blots were done by blotting with either anti-p27Kip1 or anti-ubiquitin antibody. C, effects of Pin1 on co-immunoprecipitation of HA-Cks1 and FLAG-p27Kip1. Immunoblotting analysis of HA-Cks1 bound to FLAG-p27Kip1 in the presence of different concentrations of recombinant Pin1 protein are shown. D, co-immunoprecipitation of HA-Cks1 and FLAG-p27Kip1 in the presence or absence of Pin1. Immunoblotting analysis of HA-Cks1 bound to FLAG-p27Kip1 in Pin1-WT and -KO MEFs, respectively, is shown.
The crystallographic structure of Skp2-Cks1-p27Kip1 complex shows that Cks1 directly binds to the phosphorylated Thr-187 site in p27Kip1 and promotes its degradation (15). On the other hand, we find in our present study that Pin1 enhances p27Kip1 stability also through the phosphorylated Thr-187. Given that these two proteins bind to the same site on p27Kip1 but exert an opposite effect, we hypothesized that Pin1 might compete with Cks1 for binding to p27Kip1. To test this hypothesis, a gradient concentration of recombinant human Pin1 proteins was added to co-immunoprecipitation lysates in which FLAG-p27Kip1-T187D (a mutant that mimics the phosphorylated-Thr-187 of p27Kip1) and HA-Cks1 were co-overexpressed. As shown in Fig. 5C, a lower level of Cks1 remains bound to p27Kip1 in the presence of a higher concentration of Pin1, suggesting that Pin1 competes with Cks1 for binding to p27Kip1. Moreover, we further confirmed this observation in vivo by co-overexpressing FLAG-p27Kip1-T187D and HA-Cks1 in Pin1-WT and -KO MEFs. Our co-immunoprecipitation results show that a markedly higher level of Cks1 is precipitated by p27Kip1 in the Pin1-null background compared with the WT background (Fig. 5D), suggesting that the interaction of p27Kip1 with Cks1 is significantly impaired in the presence of Pin1. Collectively, however, our results illustrate that Pin1 stabilizes p27Kip1, possibly due to its inhibition of the association of Cks1 with p27Kip1.
DISCUSSION
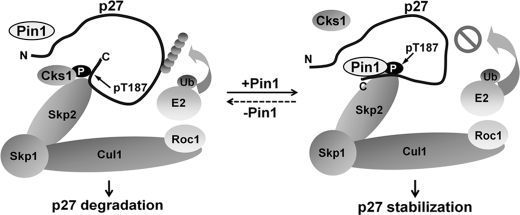
We report herein that Pin1 plays a protective role during the turnover of p27Kip1 in a phosphorylation-dependent manner. We describe a novel molecular mechanism by which Pin1 alters the conformation of phosphorylated p27Kip1, thereby suppressing the association of Cks1 and p27Kip1 and preventing p27Kip1 degradation via the SCF complex (Fig. 6). Our study thus sheds new light on the biological function of Pin1 as an important regulator of specific protein abundance and also uncovers evidence of a cross-talk between post-phosphorylation regulation and the ubiquitination-mediated degradation of proteins.
FIGURE 6.
A putative mechanism for Pin1 in the regulation of p27Kip1. Cks1 binds to phosphorylated p27Kip1 and promotes its degradation through ubiquitination (Ub). However, to regulate p27Kip1 stability at specific time points, Pin1 binds phosphorylated p27Kip1 and catalyzes its conformational change, which hinders the association of p27Kip1 with Cks1. This process in turn inhibits the polyubiquitination and degradation of p27Kip1.
Pin1 provides a novel mechanism for p27Kip1 ubiquitin-proteasome degradation. It has been extensively shown that the degradation of p27Kip1 is regulated by its subcellular compartmentalization (11, 42), phosphorylation status (43–44), and the availability of components in the degradation complex (13–15). In this study, we show that the ubiquitination status of p27Kip1 is noticeably increased in the absence of Pin1, indicating that the role of Pin1 is significant in regulating this process. Our NMR studies clearly show that the phosphorylated p27Kip1 undergoes a conformational change mediated by Pin1, suggesting that the post-phosphorylation modification may be crucial for preventing the recognition of p27Kip1 by its degradation machinery. Cks1 is an important factor in p27Kip1 degradation (13–15), and we further find that Pin1 disrupts the binding of Cks1 to phosphorylated p27Kip1. Therefore, one of the possible mechanisms could be that the conformational changes of p27Kip1 catalyzed by Pin1 is unfavorable for the binding of Cks1, resulting in a reduced interaction of p27Kip1 with the SCF complex and an enhancement of p27Kip1 stability (Figs. 5 and 6). From the structure of Skp1-Skp2-Cks1-p27Kip1 complex, we further found that Cks1 indeed interacts with a trans conformation of the Thr-187-Pro motif in p27Kip1 (15), suggesting that Cks1 may have a structural preference to its target. In the presence of Pin1, the cis-trans isomerization rate of Thr-187-Pro motif is increased by more than 10-fold. This result suggests that the conformational change catalyzed by Pin1 is a critical regulatory mechanism in mediating the interaction of Cks1 and p27Kip1. Another possible mechanism could be that the Pin1 enhances the dephosphorylation of p27Kip1 by its downstream phosphatase, resulting in a reduction of the binding to Cks1 and rescuing p27Kip1 stability. It has been reported that Pin1 can regulate the conformation and dephosphorylation of its substrates (25, 29). Pin1 facilitates the dephosphorylation of Cdc25 and Tau by PP2A, a conformation-specific proline-directed phosphatase that effectively dephosphorylates only the trans-Ser/Thr-Pro motifs (29). It would be interesting to further investigate whether Pin1 can regulate the dephosphorylation of p27Kip1 in the future. The third possible mechanism is that Pin1 competes with Cks1 or other components of the SCF complex for binding to phosphorylated p27Kip1 (Figs. 5 and 6). Indeed, we also find that GST-Pin1 can pulldown Skp2 in vitro (data not shown), indicating that Pin1 may become incorporated into the p27Kip1 degradation complex and is likely to have a regulatory role. Our current study provides the first experimental evidence of a novel “post-phosphorylation” regulation mechanism for the polyubiquitination of p27Kip1. Therefore, overall the post-phosphorylation mechanisms mediated by prolyl isomerases may be a crucial determinant of the abundance of some proteins such as β-catenin (18), cyclin D1 (21), and NF-κB (24) in vivo.
There are three Ser/Thr-Pro motifs on p27Kip1, Ser-10, Ser-178, and Thr-187, which are potential Pin1-binding sites. Among these, Thr-187 is the most well characterized site for p27Kip1 degradation, as revealed in a number of previous studies (13, 40, 45). Using peptide chips and co-immunoprecipitation assays, our current results further indicate that phosphorylated Thr-187 of p27Kip1 is the most favored Pin1-binding site. At the same time, we also find that this interaction is fully abolished only when all three of these Ser/Thr-Pro sites are mutated, indicating that all Ser/Thr-Pro motifs, at least partially, might be involved in the Pin1-p27Kip1 interaction. Although Ser-10 is reported to be the major phosphorylated site in p27Kip1 (6, 7), our stability assay shows that the stability of S178A/T187A mutant has no significant difference in Pin1-WT and -KO MEF cells (Fig. 4E), suggesting that the interaction of Pin1 with phosphorylated Ser-10 may not have a significant effect on p27Kip1 stability. On the other hand, because Pin1 can associate both sites, it would also be interesting to test whether Pin1 can induce conformational changes on Ser(P)-10 or Ser(P)-178 peptides in the future. In addition, FOXO4 is known to regulate the p27Kip1 level through transcriptional regulation. Recently, it was reported that the overexpression of Pin1 inhibits FOXO4 transcriptional activity resulting in an impairment of p27Kip1 expression (46). To confirm this, we also tested the turnover rate of p27Kip1 in HEK 293T cells with or without ectopically overexpressed Pin1 (supplemental Fig. S2), but we observed no ectopic effects of Pin1 on p27Kip1 stability. However, because of a high endogenous level of Pin1 in HEK 293T cells, additional ectopic expression may not have significant effects, similar to the results shown previously (46). On the other hand, we show that the loss of Pin1 in cells significantly enhances the polyubiquitination levels of p27Kip1 resulting in reduced stability (Fig. 4, A and C). Furthermore, we also demonstrated that there is little or no detectable FOXO4 in our MEFs (supplemental Fig. S3), suggesting the regulatory function of Pin1 on p27Kip1 degradation is not likely to be mediated through FOXO4 in our study.
Isomerization of the phosphorylated Ser/Thr-Pro motifs by Pin1 is a key mechanism underlying a post-phosphorylation regulation in many proteins. In general, Pin1 catalyzes the conformational changes of its substrates and thereby alters their properties, e.g. transcriptional activity, protein-protein interaction, and subcellular localization. The cis-trans isomerization of proline is thus likely to be a key regulatory switch in signal transduction. Given the cumulative evidence to date, we also propose that by switching the cis-trans conformations of the proline residue, Pin1 plays a pivotal role in the protein degradation machinery. For instance, Pin1 stabilizes cyclin D1 (21), NF-κB (24), β-catenin (18), p53 (22, 23), p73 (47), and Tax (48). On the other hand, Pin1 negatively regulates protein stability, including that of c-Myc (49), SRC-3 (50), IRF3 (51), cyclin E (41), Daxx (52), and SMRT (53). Additionally, many of these substrates undergo ubiquitination-mediated proteosomal degradation. The turnover of Pin1 substrates is generally through the cis-trans conformational changes. In this study, we report that Pin1 stabilizes p27Kip1 by a direct involvement in its degradation machinery, which adds weight to a general role for Pin1 in regulating protein stability. Hence, the post-phosphorylation isomerization by Pin1 may act as a molecular switch that determines the fate of its substrates. However, the reason why Pin1 can act as both a stabilizer and a de-stabilizer is unclear at present. Future studies focusing on the general function of Pin1 in ubiquitination-mediated protein degradation may help to elucidate this issue.
The molecular mechanisms underlying the regulation of p27Kip1 by Pin1 are likely to be highly complex given the roles that both proteins play in different phases of the cell cycle. As an example of this, the overexpression of Pin1 in mammalian cells leads to a G2 arrest, whereas its inhibition causes mitotic arrest (16). Moreover, Pin1 regulates the turnover of c-Myc and cyclin E (49, 41), both of which play critical roles in the G1/S phase transition, and the cyclin E protein has been shown to be further destabilized by Pin1 in MEFs (41). On the other hand, we have also reported that Pin1 can regulate cyclin D1 through both transcriptional and translational mechanisms (17–18, 21). Pin1 directly stabilizes cyclin D1 and regulates its localization; in the absence of Pin1, the cyclin D1 protein levels are markedly reduced (21). Furthermore, the cell cycle reentry of Pin1-KO MEFs is retarded in response to serum starvation (55). Taken together, these results suggest that Pin1 plays a pivotal role in the G0/G1-S transition. You et al. (56) have reported in their previous study that in response to IGF-1 treatment, Pin1-KO MEFs display a delayed entry into S phase. Conversely, IGF-1 was found to stimulate Pin1 expression, resulting in an increased expression of cyclin D1 and the phosphorylation of pRb, thus further promoting the G0/G1-S transition (56). The transcription factor E2F is known to be regulated by pRb, the hyperphosphorylation of which releases E2F1 thereby activating the downstream essential genes for the G1/S phases of the cell cycle. In addition, E2F1 cannot only bind to the p27Kip1 promoter (57), but also the Pin1 promoter (58) and activates the expression of these two proteins. On the other hand, the Cdk inhibitor p27Kip1 can prevent pRb phosphorylation by inhibiting the activities of cyclin D1/Cdk4 and cyclin E/Cdk2 (59, 60). Interestingly, the phosphorylation of Thr-187 on p27Kip1 by cyclin E/Cdk2 and the subsequent recognition by the ubiquitin ligase Skp2 SCF proteasome complex are the predominant mechanisms that regulate the protein abundance of p27Kip1. In this study, we show that the phosphorylation-dependent ubiquitination of p27Kip1 is highly controlled by Pin1, further highlighting the complexity of the cell cycle regulatory processes in which Pin1 and p27Kip1 function.
Interestingly and unexpectedly, Pin1 knock-out mice are viable and undergo relatively normal development despite several age-dependent and cell-proliferative abnormalities (21). Accordingly, Pin1-KO MEFs are slightly slower growing than their wild-type counterparts but otherwise show no significant differences. Conversely, overexpression of Pin1 not only confers transforming properties on epithelial cells but also enhances the transformed phenotypes of Neu/Ras activated mammary epithelial cells, indicating an important role of Pin1 in tumor formation (58). On the other hand, it is also surprising that the inactivation of the Thr-187-dependent p27Kip1 turnover pathway has no severe impact on cell cycle regulation, as revealed by studies of p27Kip1T187A knock-in mice. In addition, these mice are viable and display only modest cell-proliferative alterations (61). More interestingly, the p27Kip1Thr-187A can also be down-regulated in activated K-ras-induced lung tumors, and the p27Kip1Thr-187A mice have a same tumor-dependent death rate as the p27Kip1 wild-type mice, implying that an alternative degradation pathway other than Skp2-dependent mechanism plays a significant role in regulating p27Kip1 stability (54). Taken together, these observations suggest, at least in part, that there are negative feedback mechanisms in different phases of the cell cycle that control p27Kip1 degradation and Pin1 isomerase activity. Given our current data showing that Pin1 also interacts with and stabilizes p27Kip1, further studies of the temporal and spatial regulation of phosphorylated p27Kip1 mediated by Pin1 may provide a better understanding of cell cycle control. Also, it would be interesting to cross Pin1 knock-out mice with cyclin D1, cyclin E, or p27Kip1 transgenic/knock-out mice to dissect each of their roles in this complicated cell cycle progression.
In conclusion, our current study elucidates a novel molecular mechanism by which phosphorylated p27Kip1 is further regulated by the peptidylprolyl isomerase Pin1. This may underscore the significance of prolyl isomerization in the post-phosphorylation regulation and polyubiquitination-directed degradation of proteins in the cell.
Supplementary Material
Acknowledgments
We thank K. P. Lu for providing invaluable Pin1 antibody, plasmids, and MEFs. We also thank H. Y. Sun and C. Lu for peptide synthesis and technical assistance with the peptide arrays. We also thank the anonymous reviewers for their invaluable comments. We are grateful to the members of Y. C. Liou laboratory for useful discussions and to K. Perrem, B. L. Tan, and B. C. Low for excellent advice during the preparation of the manuscript.
This work was supported by Grant 06/1/21/19/473 from the Biomedical Research Council, the Agency for Science, Research, and Technology, and in part by Facility Research Council Grant R-154-000-403-112 from the National University of Singapore (to Y. C. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Results, and Figs. S1–S3.
- PPIase
- peptidylprolyl isomerase
- MEF
- mice embryonic fibroblast
- CIP
- calf intestinal phosphatase
- WT
- wild type
- GST
- glutathione S-transferase
- HEK
- human embryonic kidney
- ROESY
- rotating frame Overhauser effect spectroscopy
- SCF
- SKP1-CUL1-F-box
- pRb
- retinoblastoma protein.
REFERENCES
- 1.Borriello A., Cucciolla V., Oliva A., Zappia V., Della Ragione F. (2007) Cell Cycle 6, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 2.Reed S. I. (2002) Cell Cycle 1, 389–390 [DOI] [PubMed] [Google Scholar]
- 3.Viglietto G., Motti M. L., Fusco A. (2002) Cell Cycle 1, 394–400 [DOI] [PubMed] [Google Scholar]
- 4.Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 5.Kaldis P. (2007) Cell 26, 241–244 [DOI] [PubMed] [Google Scholar]
- 6.Vervoorts J., Lüscher B. (2008) Cell. Mol. Life Sci. 65, 3255–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishida N., Kitagawa M., Hatakeyama S., Nakayama K. (2000) J. Biol. Chem. 275, 25146–25154 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen L., Besson A., Heng J. I., Schuurmans C., Teboul L., Parras C., Philpott A., Roberts J. M., Guillemot F. (2006) Genes Dev. 20, 1511–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldassarre G., Belletti B., Nicoloso M. S., Schiappacassi M., Vecchione A., Spessotto P., Morrione A., Canzonieri V., Colombatti A. (2005) Cancer Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 10.McAllister S. S., Becker-Hapak M., Pintucci G., Pagano M., Dowdy S. F. (2003) Mol. Cell. Biol. 23, 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodier G., Montagnoli A., Di Marcotullio L., Coulombe P., Draetta G. F., Pagano M., Meloche S. (2001) EMBO J. 20, 6672–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotake Y., Nakayama K., Ishida N., Nakayama K. I. (2005) J. Biol. Chem. 280, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 13.Ganoth D., Bornstein G., Ko T. K., Larsen B., Tyers M., Pagano M., Hershko A. (2001) Nat. Cell Biol. 3, 321–324 [DOI] [PubMed] [Google Scholar]
- 14.Seeliger M. A., Breward S. E., Friedler A., Schon O., Itzhaki L. S. (2003) Nat. Struct. Biol. 10, 718–724 [DOI] [PubMed] [Google Scholar]
- 15.Hao B., Zheng N., Schulman B. A., Wu G., Miller J. J., Pagano M., Pavletich N. P. (2005) Mol. Cell 20, 9–19 [DOI] [PubMed] [Google Scholar]
- 16.Lu K. P., Hanes S. D., Hunter T. (1996) Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 17.Wulf G. M., Ryo A., Wulf G. G., Lee S. W., Niu T., Petkova V., Lu K. P. (2001) EMBO J. 20, 3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryo A., Nakamura M., Wulf G., Liou Y. C., Lu K. P. (2001) Nat. Cell Biol. 3, 793–801 [DOI] [PubMed] [Google Scholar]
- 19.Shen Z. J., Esnault S., Schinzel A., Borner C., Malter J. S. (2009) Nat. Immunol. 10, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rustighi A., Tiberi L., Soldano A., Napoli M., Nuciforo P., Rosato A., Kaplan F., Capobianco A., Pece S., Di Fiore P. P., Del Sal G. (2009) Nat. Cell Biol. 11, 133–142 [DOI] [PubMed] [Google Scholar]
- 21.Liou Y. C., Ryo A., Huang H. K., Lu P. J., Bronson R., Fujimori F., Uchida T., Hunter T., Lu K. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zacchi P., Gostissa M., Uchida T., Salvagno C., Avolio F., Volinia S., Ronai Z., Blandino G., Schneider C., Del Sal G. (2002) Nature 419, 853–857 [DOI] [PubMed] [Google Scholar]
- 23.Zheng H., You H., Zhou X. Z., Murray S. A., Uchida T., Wulf G., Gu L., Tang X., Lu K. P., Xiao Z. X. (2002) Nature 419, 849–853 [DOI] [PubMed] [Google Scholar]
- 24.Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 25.Liou Y. C., Sun A., Ryo A., Zhou X. Z., Yu Z. X., Huang H. K., Uchida T., Bronson R., Bing G., Li X., Hunter T., Lu K. P. (2003) Nature 424, 556–561 [DOI] [PubMed] [Google Scholar]
- 26.Pastorino L., Sun A., Lu P. J., Zhou X. Z., Balastik M., Finn G., Wulf G., Lim J., Li S. H., Li X., Xia W., Nicholson L. K., Lu K. P. (2006) Nature 440, 528–534 [DOI] [PubMed] [Google Scholar]
- 27.Ryo A., Togo T., Nakai T., Hirai A., Nishi M., Yamaguchi A., Suzuki K., Hirayasu Y., Kobayashi H., Perrem K., Liou Y. C., Aoki I. (2006) J. Biol. Chem. 281, 4117–4125 [DOI] [PubMed] [Google Scholar]
- 28.Lim J., Balastik M., Lee T. H., Nakamura K., Liou Y. C., Sun A., Finn G., Pastorino L., Lee V. M., Lu K. P. (2008) J. Clin. Invest. 118, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- 30.Takahashi K., Uchida C., Shin R. W., Shimazaki K., Uchida T. (2008) Cell. Mol. Life Sci. 65, 359–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh E. S., Means A. R. (2007) Nat. Rev. Cancer 7, 381–388 [DOI] [PubMed] [Google Scholar]
- 32.Shaw P. E. (2007) EMBO Rep. 8, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryo A., Wulf G., Lee T. H., Lu K. P. (2009) Trends Biochem. Sci. 34, 162–165 [DOI] [PubMed] [Google Scholar]
- 34.Bax A., Davis D. G. (1985) J. Magn. Reson. 63, 207–213 [Google Scholar]
- 35.Bax A., Davis D. G. (1985) J. Magn. Reson. 65, 355–360 [Google Scholar]
- 36.Yaffe M. B., Schutkowski M., Shen M., Zhou X. Z., Stukenberg P. T., Rahfeld J. U., Xu J., Kuang J., Kirschner M. W., Fischer G., Cantley L. C., Lu K. P. (1997) Science 278, 1957–1960 [DOI] [PubMed] [Google Scholar]
- 37.Lu P. J., Zhou X. Z., Liou Y. C., Noel J. P., Lu K. P. (2002) J. Biol. Chem. 277, 2381–2384 [DOI] [PubMed] [Google Scholar]
- 38.Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H., Lu C. H., Uttamchandani M., Xia Y., Liou Y. C., Yao S. Q. (2008) Angew. Chem. Int. Ed. Engl. 47, 1698–1702 [DOI] [PubMed] [Google Scholar]
- 40.Sheaff R. J., Groudine M., Gordon M., Roberts J. M., Clurman B. E. (1997) Genes Dev. 11, 1464–1478 [DOI] [PubMed] [Google Scholar]
- 41.Yeh E. S., Lew B. O., Means A. R. (2006) J. Biol. Chem. 281, 241–251 [DOI] [PubMed] [Google Scholar]
- 42.Connor M. K., Kotchetkov R., Cariou S., Resch A., Lupetti R., Beniston R. G., Melchior F., Hengst L., Slingerland J. M. (2003) Mol. Biol. Cell 14, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J., Zubovitz J., Petrocelli T., Kotchetkov R., Connor M. K., Han K., Lee J. H., Ciarallo S., Catzavelos C., Beniston R., Franssen E., Slingerland J. M. (2002) Nat. Med. 8, 1153–1160 [DOI] [PubMed] [Google Scholar]
- 44.Shin I., Yakes F. M., Rojo F., Shin N. Y., Bakin A. V., Baselga J., Arteaga C. L. (2002) Nat. Med. 8, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 45.Kossatz U., Dietrich N., Zender L., Buer J., Manns M. P., Malek N. P. (2004) Genes Dev. 18, 2602–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenkman A. B., de Keizer P. L., van den Broek N. J., van der Groep P., van Diest P. J., van der Horst A., Smits A. M., Burgering B. M. (2008) Cancer Res. 68, 7597–7605 [DOI] [PubMed] [Google Scholar]
- 47.Mantovani F., Piazza S., Gostissa M., Strano S., Zacchi P., Mantovani R., Blandino G., Del Sal G. (2004) Mol. Cell 14, 625–636 [DOI] [PubMed] [Google Scholar]
- 48.Jeong S. J., Ryo A., Yamamoto N. (2009) Biochem. Biophys. Res. Commun. 381, 294–299 [DOI] [PubMed] [Google Scholar]
- 49.Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T., Counter C. M., Nevins J. R., Means A. R., Sears R. (2004) Nat. Cell Biol. 6, 308–318 [DOI] [PubMed] [Google Scholar]
- 50.Yi P., Wu R. C., Sandquist J., Wong J., Tsai S. Y., Tsai M. J., Means A. R., O'Malley B. W. (2005) Mol. Cell. Biol. 25, 9687–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. (2006) Nat. Immunol. 7, 598–605 [DOI] [PubMed] [Google Scholar]
- 52.Ryo A., Hirai A., Nishi M., Liou Y. C., Perrem K., Lin S. C., Hirano H., Lee S. W., Aoki I. (2007) J. Biol. Chem. 282, 36671–36681 [DOI] [PubMed] [Google Scholar]
- 53.Stanya K. J., Liu Y., Means A. R., Kao H. Y. (2008) J. Cell Biol. 183, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmerbeul I., Garrett-Engele C. M., Kossatz U., Chen X., Firpo E., Grünwald V., Kamino K., Wilkens L., Lehmann U., Buer J., Geffers R., Kubicka S., Manns M. P., Porter P. L., Roberts J. M., Malek N. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14009–14014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujimori F., Takahashi K., Uchida C., Uchida T. (1999) Biochem. Biophys. Res. Commun. 265, 658–663 [DOI] [PubMed] [Google Scholar]
- 56.You H., Zheng H., Murray S. A., Yu Q., Uchida T., Fan D., Xiao Z. X. (2002) J. Cell. Biochem. 84, 211–216 [DOI] [PubMed] [Google Scholar]
- 57.Wang C., Hou X., Mohapatra S., Ma Y., Cress W. D., Pledger W. J., Chen J. (2005) J. Biol. Chem. 280, 12339–12343 [DOI] [PubMed] [Google Scholar]
- 58.Ryo A., Liou Y. C., Wulf G., Nakamura M., Lee S. W., Lu K. P. (2002) Mol. Cell. Biol. 22, 5281–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng M., Olivier P., Diehl J. A., Fero M., Roussel M. F., Roberts J. M., Sherr C. J. (1999) EMBO J. 18, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherr C. J., Roberts J. M. (2004) Genes Dev. 18, 2699–2711 [DOI] [PubMed] [Google Scholar]
- 61.Malek N. P., Sundberg H., McGrew S., Nakayama K., Kyriakides T. R., Roberts J. M., Kyriakidis T. R. (2001) Nature 413, 323–327 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.