Abstract
Interleukin (IL)-23, a new member of the IL-12 family, plays a central role in the Th17 immune response and in autoimmune diseases. It is clear that activated macrophages and dendritic cells produce IL-23, but the molecular mechanisms whereby inflammatory signals stimulate IL-23 expression are not fully understood. We demonstrate that induction of IL-23 p19 gene expression by LPS depends on the TLR4 and MyD88 pathways. All three MAPK pathways (ERK, JNK, and p38) that are activated by lipopolysaccharide (LPS) stimulation were shown to exert a positive effect on p19 expression. We cloned a 1.3-kb putative p19 promoter and defined its transcription initiation sites by the 5′-rapid amplification of cDNA ends method. By analyzing IL-23 p19 promoter mutants, we have identified a promoter region (−413 to +10) that contains several important elements, including NF-κB and AP-1. In addition to NF-κB, we have demonstrated that the proximal AP-1 site is important for p19 promoter activation. Mutation of the AP-1 site resulted in the loss of p19 promoter activation. Electrophoretic mobility shift assay (EMSA) analysis showed that c-Jun and c-Fos bind to the AP-1 site, which was confirmed by a chromatin immunoprecipitation assay. Furthermore, co-transfection of c-Jun and ATF2 synergistically induced p19 promoter activation, and c-Jun and ATF2 formed a protein complex, demonstrated by co-immunoprecipitation. Finally, LPS-stimulated peritoneal macrophages from IL-10-deficient mice expressed significantly higher IL-23 p19 than macrophages from wild type mice, and the addition of recombinant IL-10 strongly inhibited LPS-induced p19 expression. Thus, this study suggests that MyD88-dependent Toll-like receptor signaling induces IL-23 p19 gene expression through both MAPKs and NF-κB.
Interleukin (IL)2-23, a novel IL-12 family cytokine, is a heterodimeric molecule composed of IL-12/IL-23 p40, and p19, a peptide related to IL-12 p35 (1–3). Dendritic cells and macrophages responding to microbial infection rapidly secrete IL-23 (4, 5). IL-23 induces IFN-γ production by T cells, as does IL-12. However, IL-23 induces proliferation of memory T cells, whereas IL-12 preferentially stimulates proliferation of naive T cells (1, 6, 7). The IL-23 receptor is a heterodimer composed of IL-12Rβ1, a subunit of the IL-12 receptor, and a unique IL-23R protein (8). Despite some similarities between IL-12 and IL-23, experiments using p40-, p35-, and p19-deficient mice demonstrated that IL-12 and IL-23 have distinct functions in vivo. In particular, IL-23 is important for Th17 cell development (9). Mice lacking IL-23 p19 were resistant to collagen-induced arthritis, experimental autoimmune encephalomyelitis, and inflammatory bowel disease, because the generation of Th17 cells is impaired in the absence of IL-23 (6, 10, 11). However, IL-12-deficient mice were susceptible to collagen-induced arthritis, experimental autoimmune encephalomyelitis, and inflammatory bowel disease. In contrast, IL-12, rather than IL-23, is essential for Th1 cell differentiation (2, 12, 13). However, either IL-23 or IL-12 deficiency significantly compromises the host defense against various pathogens.
The synthesis of IL-12/IL-23 p40 is induced by different Toll-like receptor (TLR) ligands (14–17) and can be synergistically enhanced by pretreatment with IFN-γ (18). Expression of IL-12/IL-23 p40 is strictly controlled at the transcriptional level by multiple transcription factors, including NF-κB, IRF-1, IRF-8, PU.1, and AP-1 (14–16, 19, 20). Interestingly, studies using ERK MAPK inhibitors showed that the ERK MAPK pathway negatively regulates IL-12/IL-23 p40 gene expression (21). In contrast, the regulation of IL-23 p19 at the molecular level is not fully understood, although two recent studies suggest that the NF-κB subunit c-Rel is important for p19 gene expression in dendritic cells (22, 23).
The transcription factor AP-1 consists of a variety of dimers composed of members of the Jun, Fos, and ATF families of proteins (24). The Jun proteins can both homo- and heterodimerize with Fos members to form transcriptionally active complexes. The stimulation of macrophage TLR4 receptor rapidly activates not only the NF-κB pathway but also MAPK pathways, including JNK, ERK, and p38 (25). Many of the downstream targets of MAPK pathways are transcription factors that include c-Jun, ATF2, and Elk-1. It has been reported that AP-1 is important for IL-12/IL-23 p40 gene expression (14). However, it is still not clear how AP-1 controls IL-23 p19 gene expression.
We demonstrated that, in contrast to IL-12/IL-23 p40, which is negatively regulated by ERK, the TLR-activated ERK pathway is essential for IL-23 p19 gene expression. We identify an AP-1 element in the IL-23 p19 promoter region and demonstrate, by complementary approaches, that AP-1 is required for IL-23 p19 expression. In addition, we report that interleukin 10 inhibits IL-23 p19 gene expression. The clearer understanding of factors controlling IL-23 expression may assist efforts to control the induction and progression of autoimmune disease.
EXPERIMENTAL PROCEDURES
Mice and Cell Lines
C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). TLR4-deficent, MyD88-deficient, and IL-10 deficient mice are maintained in the barrier facility at Mount Sinai School of Medicine according to Institutional Animal Care and Use Committee guidelines. The mouse macrophage cell line RAW264.7 was obtained from the American Type Culture Collection and maintained in DMEM supplemented with 10% fetal bovine serum and 100 units/ml penicillin and streptomycin.
Preparation of Peritoneal Macrophages and Dendritic Cells
Wild type, TLR4-deficient, and MyD88-deficient mice were injected intraperitoneal with 2 ml of 5% thioglycollate medium for 3 days and sacrificed, and peritoneal macrophages were isolated by lavage. Cells were cultured in DMEM containing 10% fetal bovine serum and antibiotics. Mouse bone marrow-derived dendritic cells were generated from bone marrow stem cells obtained from the femurs of the mice, as described previously (26). After lysis of the red blood cells, 1 × 106 bone marrow cells were inoculated in each well of 24-well plates with complete DMEM in the presence of GM-CSF (10 ng/ml) for 7 days.
Plasmids
The mouse IL-23 p19 promoter from −1280 to +10 was amplified by PCR using primers with internal XhoI and HindIII restriction sites from C57BL/6 mouse genomic DNA. After sequence verification of the amplified DNA, the PCR product was digested with XhoI and HindIII and cloned into the pGL3 basic luciferase vector (Promega). Progressive deletion mutants were generated by PCR amplification in the same way. The NF-κB and AP-1 mutant p19 promoter constructs were generated by site-directed mutagenesis according to the manufacturer's (Stratagene) instructions. Dominant negative c-Jun, ATF2, and ERK2 plasmids were kindly provided by Dr. Constantin Bona (Mount Sinai School of Medicine). Dominant negative (DN) IκBα (IκBα, A32/36; this IκB mutant is not susceptible to phosphorylation at N-terminal serines 32 and 36, which have been replaced by alanine) and p65−/− cells were kindly provided by Dr. Adrian Ting (Mount Sinai School of Medicine). c-Jun, JunB, JunD, and c-Fos overexpression plasmids were kindly provided by Dr. Yusaku Nakabeppu.
Reagents
Antibodies for p65, p50, ATF-2, and c-Jun were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies for ERK and JNK were obtained from Cell Signaling Technology. Antibody for p19 was purchased from eBioscience.
5′-RNA Ligase-mediated Rapid Amplification of cDNA Ends (RLM-RACE)
RLM-RACE was employed to determine the transcriptional initiation sites of the mouse p19 gene using an assay kit (Invitrogen). Briefly, 5 μg of total RNA treated with LPS at different time points were dephosphorylated with calf intestinal phosphatase. The full-length capped mRNA was then treated with tobacco acid pyrophosphatase to remove the 5′ 7-methyl guanine cap of intact, mature mRNA molecules. RNA molecules that had 5′-phosphate groups, including degraded or unprocessed mRNAs lacking a 5′ cap, structural RNAs, and traces of contaminating genomic DNA, were dephosphorylated by calf intestinal phosphatase treatment and were therefore not ligated to the adapter primer sequence. The decapped mRNA was ligated with a 44-base GeneRacer RNA oligonucleotide (5′-cgacuggagcacgaggacacugacauggacugaaggaguagaaa-3′) using T4 RNA ligase. The ligated mRNA was reverse transcribed using SuperScript III RT and oligo(dT) primer to create RACE-ready first strand cDNA with known priming sites at the 5′-ends. The 5′-end of the p19 gene transcript were amplified using two nested sense primers corresponding to the RNA oligonucleotide sequence and two nested antisense primer specific to p19 mRNA (outer, 5′-tcccgctggtgcatgtgcgttcca-3′; inner, 5′-CTGGCTGGCTCTGTGATCTGCACCA-3′). PCR conditions were 94 °C for 2 min 1 cycle; 94 °C for 30 s, 66 °C for 30 s, 68 °C for 1 min for 20 cycles; 68 °C for 10 min. The PCR product was size-fractionated by 1.2% agarose gel electrophoresis and purified. The purified PCR products were cloned and sequenced. The transcription start sites of the p19 gene were defined by aligning the 5′-RLM-RACE sequences with the p19 gene sequence.
Transfection
RAW264.7 cells were transiently transfected using Superfect (Qiagen). For each transfection, 2.5 μg of plasmid was mixed with 100 μl of DMEM (without serum and antibiotics) and 10 μl of Superfect reagent. The mixture was incubated at room temperature for 10 min, 600 μl of DMEM complete medium was added and immediately placed onto the cells in 6-well plates, and luciferase activity was measured 30 h later. When indicated, different TLR ligands were added to the culture 12 h before harvest. The cells were harvested with reporter lysis buffer (Promega), and 20 μl of extract was assayed for luciferase activity, as described (27). Cells were co-transfected with a β-galactosidase reporter plasmid to normalize experiments for transfection efficiency.
Electrophoretic Mobility Shift Assay
RAW264.7 cell nuclear extracts were prepared as described previously (28). EMSA probes were prepared by annealing complementary single-stranded oligonucleotides with 5′-ACTG overhangs (MWG Biotechnologies, Inc.) and were labeled by filling in with [α-32P]dGTP and [α-32P]dCTP using Klenow enzyme. Labeled probes were purified with Nuctrap purification columns (Roche Applied Science). EMSAs were performed as described previously, using 105 cpm of labeled probe and 5 μg of nuclear extracts/reaction. DNA binding complexes were separated by electrophoresis on a 5% polyacrylamide-Tris/glycine-EDTA gel, which was dried and exposed to x-ray film.
Quantitative Reverse Transcription (qRT)-PCR Analysis
Total RNA was extracted using the RNeasy Plus kit (Qiagen, Valencia, CA), and the cDNA was generated with an oligo(dT) primer, followed by analysis using iCycler PCR with SYBR Green PCR Master Mix (Applied Biosystems). Results were normalized based on the expression of ubiquitin. The IL-23 p19, IL-12 p35, and ubiquitin primer sequences are listed in Table 1.
TABLE 1.
Oligonucleotides used
Cytokine Release
IL-23 and IL-12 production in macrophage cell culture was quantified by an enzyme-linked immunosorbent assay (R&D Systems).
Chromatin Immunoprecipitation Assay
The experiments were performed using a chromatin immunoprecipitation assay kit from Upstate Biotechnology according to the manufacturer's protocol. RAW264.7 cells were cultured in complete Dulbecco's modified Eagle's medium, activated with LPS (1 μg/ml) for 4 h, fixed with 1% formaldehyde for 30 min at room temperature, and immunoprecipitated with either anti-p65, p50, anti-c-Jun, or ATF antibody. The immunoprecipitated DNA was amplified by PCR with primers spanning the murine IL-23 p19 promoter region.
Western Blot
Cell lysates (30 μg of total protein) and prestained molecular weight markers were subjected to SDS-PAGE and transferred to nitrocellulose. The membranes were blocked with 5% nonfat milk in TBST (0.5% Triton X-100, Tris-buffered saline), incubated with various antibodies (1:1000) for 2 h, washed with TBST, and stained with peroxidase-conjugated IgG second antibody (1:5000). Immunoreactivity was visualized by enhanced chemiluminescence (ECL kit; Santa Cruz Biotechnology). All antibodies were purchased from Cell Signaling (Danvers, MA).
RESULTS
IL-23 p19 Expression Is Dependent on TLR4 and MyD88
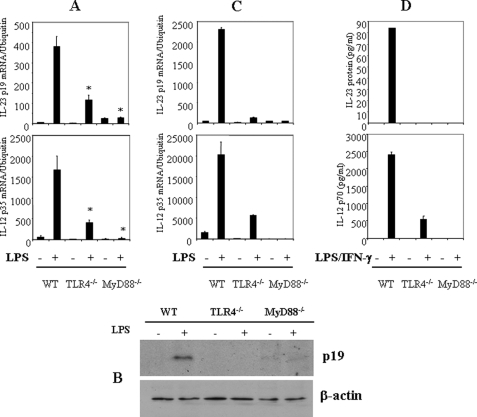
To evaluate the importance of TLR signaling pathways for IL-23 p19 expression, thioglycollate-elicited peritoneal macrophages from wild type (WT), TLR4−/−, and MyD88−/− mice were activated with LPS (1 μg/ml) for either 4 h (mRNA) or 24 h (protein). LPS strongly induced IL-23 p19 mRNA expression in macrophages from WT mice, but IL-23 p19 mRNA expression was significantly impaired in macrophages from TLR4−/− and MyD88−/− mice (Fig. 1A), as was LPS-stimulated expression of IL-12 p35 (Fig. 1A). In agreement with the inhibition of IL-23 p19 mRNA, macrophages from TLR4−/− and MyD88−/− mice did not synthesize IL-23 p19 protein (Fig. 1B). Bone marrow-derived dendritic cells behaved similarly (Fig. 1C). In addition, IL-23 and IL-12 protein production induced by incubation with LPS plus IFN-γ was significantly reduced in TLR4−/− and MyD88−/− macrophages (Fig. 1D), although LPS itself did not induce detectable amounts of IL-23. Thus, in both macrophages and dendritic cells, LPS-induced IL-23 p19 expression is dependent upon the TLR4-MyD88 pathway.
FIGURE 1.
IL-23 p19 expression is dependent upon TLR4 and MyD88. A, thioglycollate-elicited peritoneal macrophages from WT (C57BL/6), TLR4−/−, and MyD88−/− mice were activated with LPS (1 μg/ml) for 4 h. Total RNA was analyzed by qRT-PCR for p19 and p35. *, p < 0.01 versus WT LPS (+). B, the peritoneal macrophages described in A were activated with LPS for 16 h, and brefeldin A was added for 6 h. The whole cell lysates were collected for immunoblotting analysis using anti-p19 antibody. C, bone marrow-derived dendritic cells from WT, TLR4−/−, and MyD88−/− mice were activated with LPS (1 μg/ml) for 4 h. Total RNA was analyzed by qRT-PCR for p19 and p35. *, p < 0.01 versus WT LPS (+). D, the peritoneal macrophages described in A were activated with LPS plus IFN-γ for 24 h, and supernatants were assayed for IL-23 and IL-12 p70. The data are representative of three similar experiments.
Cloning of the 1.3-kb Predicted p19 Promoter and Mapping of the Transcription Initiation Sites
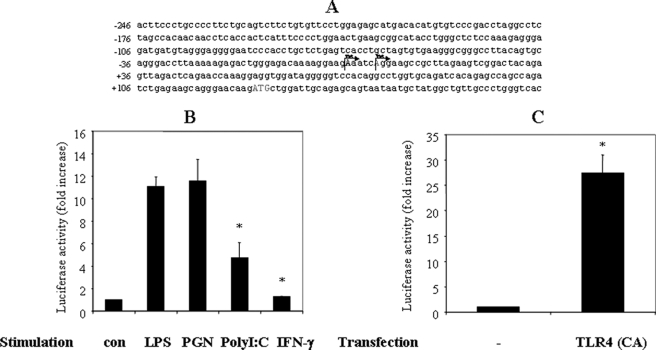
To study transcriptional regulation of the IL-23 p19 gene, we decided to define its promoter region (see “Experimental Procedures” for details) and to map its transcription initiation sites (TIS). Total RNA generated from control and LPS-treated mouse macrophages was used to map the TIS of the p19 gene by 5′-RLM-RACE. In the 25 clones we analyzed, two sequences were identified that differed in the sequence at the 5′-end, which define two TIS in the p19 gene (Fig. 2A).
FIGURE 2.
TLR signaling regulates p19 promoter activation. A, schematic of the p19 promoter with TIS that were determined by RLM-RACE. The promoter coordinates (from −246 down to +106) are relative to the most dominant TIS1, located at +1. B, RAW264.7 cells were transfected with the p19 luciferase reporter plasmid. The cells were incubated for 18–24 h, activated for 12 h with various TLR ligands (LPS (1 μg/ml), PGN (25 μg/ml), poly(I-C) (50 μg/ml), or IFN-γ (10 ng/ml)), after which luciferase activity from cell extracts was quantified. Results were normalized for β-galactosidase activity from a control (con) β-galactosidase expression plasmid. *, p < 0.01 versus LPS (+). C, RAW264.7 cells were co-transfected with the p19 luciferase reporter plasmid and a constitutive form of TLR4 for 30 h. *, p < 0.01 versus control. Luciferase activity was analyzed as described in A. The results are representative of three similar experiments.
To explore further the regulation of IL-23 p19 expression by the TLR signaling pathway, we cloned 1.3 kb of the IL-23 p19 promoter into the luciferase reporter plasmid pGL3. To quantify promoter activity of this region, we transfected the p19 promoter luciferase reporter construct into RAW264.7 cells for 18 h and activated the cells with LPS or PGN for 12 h. LPS and PGN induced an approximately 10-fold increase of luciferase activity (Fig. 2B), demonstrating that p19 promoter was within the cloned fragment. Extending the result with LPS stimulation, we found that co-transfection of a constitutively active TLR4 construct also strongly induced p19 promoter activation (Fig. 2C).
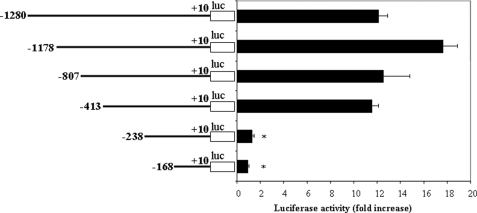
As a first step toward localizing important control elements within the p19 promoter, a set of 5′ deletion mutants coupled to the luciferase reporter was constructed and analyzed for its effect on luciferase activity. RAW264.7 cells were transfected with the series of deletion mutants, incubated for 18 h, and activated with LPS for 12 h, after which luciferase activity was quantified and normalized to the β-galactosidase activity from a co-transfected control plasmid. Gradual deletion of the sequence from nucleotide −1280 through −413 resulted in minimal changes in promoter activity (Fig. 3). However, complete loss of promoter activity was observed when elements between positions −413 and −168 were deleted (Fig. 3). Thus, the results of deletion analysis of the p19 promoter indicated that sequence from −413 to +10 of the IL-23 p19 promoter was sufficient to confer transcriptional activity.
FIGURE 3.
Deletion analysis of p19 promoter activity. The constructs with progressive deletions of 5′-flanking region of the murine p19 promoter were used to transfect RAW264.7 cells, and 18–24 h after transfection, the cells were activated with LPS (1 μg/ml) for 12 h. Luciferase activity was quantified and normalized for β-galactosidase. *, p < 0.01 versus −413. The data are representative of three similar experiments.
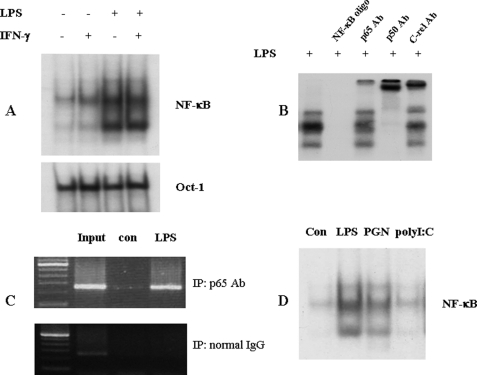
The promoter deletion studies clearly demonstrated the importance of the proximal IL-23 p19 promoter region in macrophages, which is consistent with p19 promoter analysis reported recently. The p19 promoter sequence between −413 and −238 has several putative binding sites for NF-κB, AP-1, and interferon regulatory factor transcription factors. First, we analyzed the contribution of the NF-κB site to the activation of the IL-23 p19 promoter. RAW264.7 cells were activated with LPS, IFN-γ, or IFN-γ and LPS for 4 h, and nuclear protein was extracted for EMSA with a probe spanning nucleotides −228 to −198 of the p19 promoter region. A protein-oligonucleotide complex induced by LPS and unaffected by IFN-γ was observed (Fig. 4A). Furthermore, NF-κB p65, p50, and c-Rel antibodies altered the electrophoretic mobility of the DNA-protein complex (Fig. 4B), demonstrating that NF-κB subunits bind to the site. To confirm these results, we performed chromatin co-immunoprecipitation experiments. PCR analysis showed that NF-κB p65 antibody precipitated the p19 promoter region (−413 to +10) from RAW264.7 cells activated with LPS for 4 h (Fig. 4C). As would be predicted from the results in Fig. 2, PGN also strongly induced NF-κB·DNA complex formation, whereas poly(I-C) had no clear effect (Fig. 4D).
FIGURE 4.
NF-κB p65 binds to the p19 promoter region. A, LPS induces NF-κB DNA complex formation. RAW264.7 cells were activated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or LPS plus IFN-γ for 4 h, and nuclear protein was extracted for EMSA. 5 μg of nuclear protein were added to the 32P-labeled probe (−228 to −198 of the p19 promoter). Extract and probe were incubated with 0.5 μg of poly(dI-dC) at room temperature for 30 min. B, p65 antibody attenuates DNA-protein complex formation. RAW264.7 cells were activated with LPS (1 μg/ml) for 4 h, and nuclear protein was extracted for EMSA. 5 μg of nuclear protein were incubated with 2 μg of p65, p50, or c-Rel antibody and the 32P-labeled probe used in A. C, chromatin co-immunoprecipitation assays. RAW264.7 cells were activated with LPS for 4 h. Formaldehyde (1% final concentration) was added for 30 min at room temperature. Cells were harvested, and the chromatin was sheared by sonication and purified by immunoprecipitation (IP) with an anti-p65 antibody. After reversal of cross-linking, DNA was amplified by PCR with primers spanning the p19 promoter. D, TLR ligands induce an NF-κB DNA complex. RAW264.7 cells were activated with LPS, PGN, and poly(I-C) for 4 h, and nuclear protein was extracted for EMSA. 5 μg of nuclear protein were added to the 32P-labeled probe used in A. con, control.
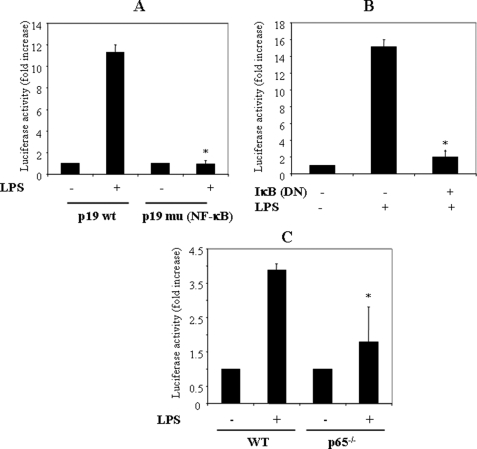
To analyze the functional importance of the NF-κB site, we transfected a p19 promoter luciferase plasmid (−413 to +10) with a mutated NF-κB site into RAW264.7 cells for 18 h, activated with LPS for 12 h, and quantified luciferase activity as before. The p19 promoter with the mutated NF-κB site was not activated under these conditions (Fig. 5A). Thus, in agreement with two recent studies on the p19 promoter NF-κB site, we find that the NF-κB site is essential for IL-23 p19 promoter activation (22, 23). To confirm this result, we co-transfected the p19 promoter and DN IκBα into RAW264.7 cells and activated with LPS. As predicted, we found that DN IκBα significantly inhibited p19 promoter activation (Fig. 5B). In addition, we found that LPS stimulation of the p19 promoter transfected into p65−/− mouse embryo fibroblasts resulted in little p19 promoter activation compared with WT cells (Fig. 5C). These results show that NF-κB is important for p19 promoter activation.
FIGURE 5.
NF-κB p65 is required for p19 promoter activation. A, mutation of the NF-κB site abolishes p19 promoter activity. RAW264.7 cells were transfected with either WT p19 promoter or a p19 promoter with a mutated NF-κB site for 18–24 h, the cells were activated with LPS (1 μg/ml) for 12 h, and luciferase activity was quantified and normalized for β-galactosidase. *, p < 0.01 versus p19 WT LPS (+). B, a dominant negative form of IκBα significantly reduced p19 promoter activation. RAW264.7 cells were co-transfected with the p19 promoter and DN-IκBα for 18–24 h, the cells were activated with LPS for 12 h, and extracts were quantified for luciferase activity and normalized for β-galactosidase. *, p < 0.01 versus LPS only. C, p19 promoter activity was significantly reduced in p65−/− mouse embryo fibroblasts. Wild type and p65−/− fibroblasts were transfected with the p19 promoter luciferase plasmid for 10–24 h and activated with LPS for 12 h, and extracts were quantified for luciferase and normalized for β-galactosidase. *, p < 0.01 versus p19 WT LPS (+).
Blocking MAPK ERK, JNK, and p38 Pathways Impairs IL-23 p19 Expression
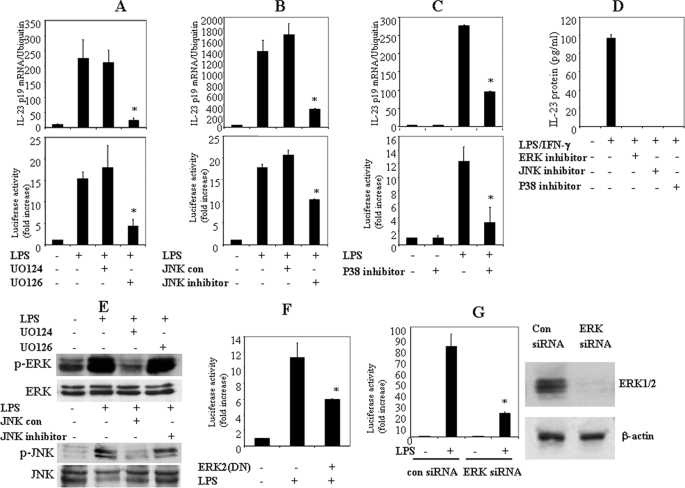
ERK MAPK negatively regulates IL-12/IL-23 p40 gene expression (21). To explore whether ERK regulates p19 similarly, we pretreated the macrophage cell line RAW264.7 with the ERK inhibitor UO126 (10 μm) and the inactive analogue UO124 (10 μm) for 30 min, after which the cells were activated with LPS (1 μg/ml) for 4 h, and total RNA was extracted for qRT-PCR. As reported previously, UO126 treatment significantly enhanced LPS-induced IL-12/IL-23 p40 expression (data not shown). In contrast, UO126 inhibition of ERK activation abolished LPS-induced IL-23 p19 expression (Fig. 6A). Similarly, JNK inhibitor also significantly suppressed p19 mRNA expression (Fig. 6B). In addition, the p38 inhibitor SB203580 significantly reduced p19 mRNA expression (Fig. 6C). Thus, these results show that MAPK activation is important for p19 gene expression.
FIGURE 6.
MAPKs are involved in the activation of the p19 promoter. A, an ERK inhibitor blocks LPS-induced p19 mRNA and p19 promoter activation. RAW264.7 cells were pretreated with either the ERK inhibitor UO126 or the inactive analogue UO124 (10 μm) for 1 h, the cells were activated with LPS for 4 h, and RNA was analyzed by qRT-PCR for p19 mRNA. For p19 promoter analysis, RAW264.7 cells were transfected with the p19 promoter-luciferase reporter construct for 18–24 h, treated with either UO124 or UO126 for 1 h, and activated with LPS for 12 h. Luciferase activity was quantified and normalized for β-galactosidase. The data are representative of two similar experiments. B, JNK inhibitor reduces LPS-induced p19 mRNA and p19 promoter activation. RAW264.7 cells were pretreated with either JNK inhibitor or control for 1 h, the cells were activated with LPS for 4 h, and p19 mRNA was quantified by qRT-PCR. For p19 promoter analysis, RAW264.7 cells were transfected with the p19 promoter for 18–24 h, incubated with either JNK control or JNK inhibitor for 1 h, and activated with LPS for 12 h, and luciferase activity in extracts was quantified and normalized for β-galactosidase. The data are representative of two similar experiments. C, p38 inhibitor inhibits LPS-induced p19 mRNA and p19 promoter activation. RAW264.7 cells were pretreated with SB203580 for 1 h, the cells were activated with LPS for 4 h, and RNA was quantified by qRT-PCR for p19 mRNA. For p19 promoter analysis, RAW264.7 cells were transfected with the p19 promoter for 18–24 h, incubated with either JNK control or JNK inhibitor for 1 h, and activated with LPS for 12 h, and luciferase activity in extracts was quantified and normalized for β-galactosidase. The data are representative of two similar experiments. D, thioglycollate-elicited peritoneal macrophages from C57BL/6 mice were pretreated with UO126, JNK inhibitor, or SB20350 for 1 h and activated with LPS plus IFN-γ for 24 h, and supernatants were assayed for IL-23. E, MAPK inhibitors block MAPK activation. RAW264.7 cells were pretreated with either ERK or JNK inhibitors for 1 h and activated with LPS for 10 min, and cell lysates were analyzed by immunoblotting (IB) for phosphorylated and total ERK and JNK. F, dominant negative ERK2 significantly reduces p19 promoter activation. RAW264.7 cells were co-transfected with the p19 promoter and DN ERK2 for 18–24 h and activated with LPS for 12 h, extracts were quantified for luciferase, and results were normalized for β-galactosidase. *, p < 0.01 versus LPS only. G, knockdown of ERK expression by siRNA significantly inhibits p19 promoter activation. Left, RAW264.7 cells were co-transfected with the p19 promoter and ERK siRNA or control siRNA for 18–24 h and activated with LPS for 12 h, extracts were quantified for luciferase, and results were normalized for β-galactosidase. *, p < 0.01 versus LPS only. The data are representative of two similar experiments. Right, 293T cells were transfected with control siRNA or ERK siRNA for 48 h. The whole cell lysates were immunoblotted with anti-ERK antibody.
We next explored if the inhibition of p19 mRNA by disruption of ERK, JNK, and p38 pathways was due to inhibition of promoter activity. After transfection of the p19 promoter luciferase reporter plasmid into RAW264.7 cells for 18 h, the cells were activated with LPS in the presence of either ERK inhibitor UO126 (10 μm), JNK inhibitor (10 μm), or p38 inhibitor SB203580 (10 μm) for 12 h. UO126 and JNK inhibitor significantly inhibited p19 promoter activation, whereas their inactive analogues had no effect (Fig. 6, A and B). In addition, the p38 inhibitor also significantly suppressed p19 promoter activation (Fig. 6C). To analyze the effect of MAPK inhibitors on IL-23 production, we pretreated thioglycollate-elicited peritoneal macrophages with UO126 (10 μm), JNK inhibitor (10 μm), and SB203580 (10 μm) for 30 min, after which the cells were activated with LPS (1 μg/ml) plus IFN-γ (10 ng/ml) for 24 h. Supernatants were collected, and IL-23 protein synthesis was determined by an enzyme-linked immunosorbent assay. The results showed that the MAPK inhibitors blocked IL-23 protein production (Fig. 6D). Western blotting experiments confirm the inhibition of ERK and JNK phosphorylation by their respective inhibitors (Fig. 6E). To confirm this result, RAW264.7 cells were co-transfected with a dominant negative form of Erk2 and a p19 luciferase reporter construct for 18 h, after which the cells were activated with LPS for 12 h prior to determination of luciferase activity. Co-transfection of DN ERK2 significantly inhibited p19 promoter activation (Fig. 6F). Furthermore, when we co-transfected RAW264.7 cells with a p19 luciferase reporter construct and ERK or control siRNA for 18 h, followed by activation with LPS, we found that the ERK siRNA significantly blocked p19 promoter activation induced by LPS (Fig. 6G). In summary, these results show that ERK, JNK, and p38 MAPKs regulate IL-23 p19 gene expression at the transcriptional level.
AP-1 Is Required for IL-23 p19 Promoter Activation
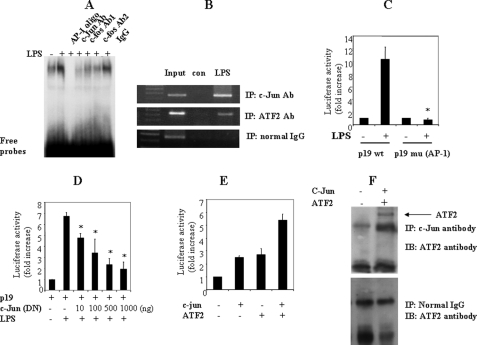
Since AP-1 family members are often found downstream of the MAPK pathway, we were interested in the function of a potential AP-1 site in the p19 promoter. To determine if this AP-1 site (−199 to −193) is important for p19 promoter activation, we mutated the putative AP-1 site in the p19 promoter region of the luciferase reporter construct. This construct was transfected into RAW264.7 cells for 18 h, and the cells were then activated with LPS for 12 h. We found that the AP-1 mutation abolished p19 promoter activation induced by LPS (Fig. 7C), suggesting that the AP-1 site is essential for LPS-induced p19 promoter activation. To show that AP-1 family proteins bind to this putative AP-1 site, RAW264.7 cells were activated with LPS for 4 h, and nuclear proteins were extracted for EMSA with a probe spanning −210 to −182 of the p19 promoter region, containing the AP-1 site. LPS clearly induced a protein-oligonucleotide complex (Fig. 7A), which was not observed with a probe (−210 to −182) containing a mutated AP-1 site (−199 to −193) (data not shown). Furthermore, antibodies against the AP-1 family members c-Jun and c-Fos blocked the formation of the DNA-protein complex (Fig. 7A). Chromatin immunoprecipitation experiments confirmed the binding of c-Jun to the p19 promoter. PCR analysis showed that c-Jun antibody precipitated the p19 promoter region (−413 to +10) from RAW264.7 cells activated with LPS for 4 h (Fig. 7B). In addition, co-transfection of DN c-Jun significantly reduced the p19 promoter activity (Fig. 7D). These results suggest that the AP-1 site (−199 to −193) of the p19 promoter is a binding site for c-Jun, which is essential for p19 promoter activation.
FIGURE 7.
AP-1 is important for p19 promoter activation. A, c-Jun binds to the AP-1 site in the p19 promoter. RAW264.7 cells were activated with LPS (1 μg/ml) for 6 h, and nuclear protein was extracted for EMSA. 5 μg of nuclear protein were incubated with 2 μg of anti-c-Jun and anti-c-Fos antibodies (Ab) and added to the 32P-labeled probe (−210 to −182 of the p19 promoter). B, chromatin co-immunoprecipitation (IP) assays were performed as in Fig. 4C, using c-Jun and ATF2 antibodies, and the immunoprecipitated DNA after cross-linker reversal was amplified by PCR with primers spanning the p19 promoter. C, mutation of the AP-1 site abolishes p19 promoter activation. RAW264.7 cells were transfected with either WT p19 promoter or the promoter with a mutated AP-1 site for 18–24 h and activated with LPS for 12 h, and luciferase activity in extracts was quantified and normalized for β-galactosidase. The data are representative of three similar experiments. D, dominant negative c-Jun significantly reduces the p19 promoter activity. RAW264.7 cells were co-transfected with the p19 promoter and DN c-Jun for 18–24 h and activated with LPS for 12 h, and extracts were assayed for luciferase activity, which was normalized for β-galactosidase. E, c-Jun and ATF2 synergistically induce p19 promoter activation. RAW264.7 cells were co-transfected with the p19 promoter, c-Jun, and ATF2 for 36 h. The extracts were assayed for luciferase activity, which was normalized for β-galactosidase. The data are representative of three similar experiments. F, c-Jun physically interacts with ATF2. 293T cells were transfected with c-Jun and ATF2 for 48 h. 500 μg of cell lysates were immunoprecipitated with c-Jun antibody or normal IgG and blotted (IB) with ATF2 antibody.
The AP-1 family is composed of different members of the c-Jun family (c-Jun, JunB, and JunD) and the c-Fos family (c-Fos, FosB, Fra1, and Fra2). To define which member(s) of the AP-1 family are involved in p19 promoter activation, we co-transfected into RAW264.7 cells different c-Jun/c-Fos family members individually or in combination with a p19 promoter plasmid. Interestingly, c-Jun moderately induced p19 promoter activation (Fig. 7E), but other members including JunB, JunD, and c-Fos had no significant effect (data not shown), even in the combination with c-Jun. This led us to search for potential molecules that are involved in this process. We have found that p38 MAPK was involved in p19 promoter activation, and it is well known that p38 MAPK can activate the transcription factor ATF2 and form a p38 MAPK·ATF2 signaling cascade complex. In addition, ATF2 has been reported to form a Jun·ATF-2 heterodimer that binds to the AP-1 site (29). Based on these results, we co-transfected ATF2 and c-Jun with the p19 promoter plasmid into RAW264.7 cells and quantified luciferase activity. p19 promoter activity was strongly enhanced by co-transfection of c-Jun and ATF2 (Fig. 7E). In addition, a co-immunoprecipitation assay using cell lysates from 293T cells transfected with c-Jun and ATF2 showed that c-Jun physically interacted with ATF2 (Fig. 7F). These results suggest that c-Jun and ATF2 form a protein complex that binds to the AP-1 site and activates the p19 promoter.
IL-10 Inhibits IL-23 p19 Gene Expression
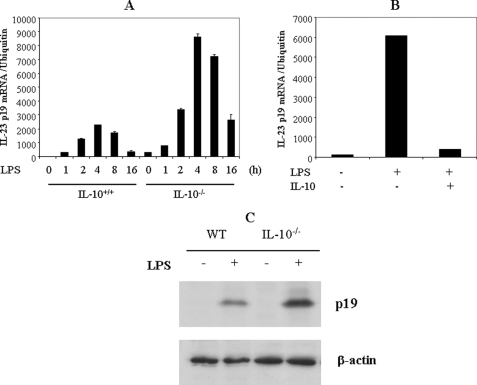
The IL-23/IL-17 axis plays an important role in murine models of several human autoimmune diseases, including experimental autoimmune encephalomyelitis, collagen-induced arthritis, and inflammatory bowel disease. Therefore, blocking IL-23 p19 gene expression may be helpful to treat autoimmune diseases. Since IL-10 has been reported to inhibit IL-12/IL-23 p40 expression, we wanted to determine if IL-10 affects IL-23 p19. To address this question, we activated thioglycollate-elicited peritoneal macrophages from both WT and IL-10−/− mice with LPS for various time intervals (1, 2, 4, 8, and 16 h) and quantified IL-23 p19 mRNA by qRT-PCR. We found that IL-23 p19 expression was significantly enhanced in IL-10−/− mice compared with WT mice (Fig. 8A). We then activated macrophages with LPS for either 4 h (RNA) or 24 h (protein) in the presence or absence of IL-10. Pretreatment with IL-10 inhibited IL-23 p19 expression in peritoneal macrophages (Fig. 8B). Similarly, IL-23 protein synthesis was also elevated in IL-10−/− mice, and the addition of IL-10 inhibited IL-23 p19 synthesis (Fig. 8C). These results indicate that IL-10 suppresses IL-23 p19 expression, suggesting a potential use of IL-10 to treat autoimmune diseases.
FIGURE 8.
IL-10 negatively regulates IL-23 p19 expression. A, enhanced IL-23 p19 expression in IL-10−/− macrophages. Thioglycollate-elicited peritoneal macrophages from WT and IL-10−/− mice were activated with LPS (1 μg/ml) for various time intervals, and total RNA was analyzed by qRT-PCR for IL-23 p19. B, IL-10 inhibits p19 mRNA expression. Thioglycollate-elicited peritoneal macrophages from IL-10−/− mice were activated with LPS (1 μg/ml) in the presence of IL-10 (10 ng/ml) for 4 h, and total RNA was extracted and analyzed by qRT-PCR for IL-23 p19. *, p < 0.01 versus LPS only. C, IL-10 inhibits IL-23 synthesis. Both WT and IL-10−/− peritoneal macrophages were activated with LPS for 16 h, and brefeldin A was added for 6 h. The whole cell lysates were collected for immunoblotting analysis using anti-p19 antibody.
DISCUSSION
IL-23 is a new member of the IL-12 family of heterodimeric cytokines that also includes IL-12 and IL-27. Interestingly, although members of the IL-12 family share ligands and receptor subunits, they play distinct roles in innate and adaptive immune responses. IL-12 is required for Th1 cell differentiation, whereas IL-23 is important for the survival and expansion of Th17 cells. IL-27 synergizes with IL-12 to induce IFN-γ production from naive T cells, but IL-27 suppresses Th17 differentiation. The detailed elucidation of the function of IL-12 family members will help us to understand better innate and adaptive immunity and to improve therapeutic approaches to autoimmune diseases. Since IL-23 is central for the Th17 immune response, which plays a pathological role in murine models of several autoimmune diseases, we believe that a detailed dissection of the molecular regulation of IL-23 is needed.
The transcriptional regulation of the IL-23 p19 gene has been studied recently. Two studies demonstrated that NF-κB c-Rel-deficient dendritic cells and macrophages had defects in LPS-induced IL-23 p19 gene expression, suggesting that c-Rel is essential for p19 induction (22, 23). Both studies demonstrated that a mutation of the proximal NF-κB binding site abolishes IL-23 p19 promoter activation. Interestingly, a binding site for NF-κB has also been reported in the promoter region of IL-12/IL-23 p40 (20), the other subunit of IL-23, suggesting that NF-κB regulates similarly both subunits of IL-23. The NF-κB family is composed of five members, RelA (p65), RelB, c-Rel, p52, and p50, which form hetero- and homodimeric complexes in the cytoplasm. TLR stimulation leads to phosphorylation, ubiquitination, degradation of IκB, and release of NF-κB. In agreement with the two reports, our study also suggests that the proximal NF-κB site in the p19 promoter region is important for IL-23 p19 gene expression. This concept is strongly supported by the fact that LPS-induced p19 gene expression is significantly impaired in macrophages from TLR4−/− and MyD88−/− mice.
In the present study, we have shown by EMSA and chromatin immunoprecipitation assays that both c-Jun and c-Fos can bind to the AP-1 site of the p19 promoter region. Furthermore, a mutation of the AP-1 site results in the loss of p19 promoter activation. These results suggest that AP-1 activated by the TLR4-MyD88-dependent pathway is essential for IL-23 p19 gene activation. Interestingly, AP-1 was also reported to be important for IL-12/IL-23 p40 gene activation (14). However, transfection assays showed that only c-Jun moderately induced p19 promoter activation and that c-Fos, JunB, and JunD had no effect, even with co-transfection of c-Jun. However, c-Jun and ATF-2 synergistically induced p19 promoter activation through physical interaction. ATF2 is a member of the ATF/CREB transcription factor family. MAPKs, JNK, ERK, and p38 phosphorylate and thus activate ATF2 (30). Previous study suggested that ATF2 can interact with c-Jun at the IFN-β promoter (29). Al-Salleeh et al. (31)reported that ATF2 is involved in IL-23 p19 gene expression induced by Theiler's virus (TMEV) infection. They have found that ATF2 binds to the endogenous p19 promoter and that ATF2 short hairpin RNA reduced p19 promoter activity in RAW264.7 cells following challenge with TMEV. Our evidence shows that the protein complex of c-Jun with ATF-2 is essential for IL-23 p19 gene expression.
Experiments using inhibitors of MAPKs ERK, JNK, and p38 revealed the functional importance of MAPK activation for p19 gene expression. It was reported previously that the ERK pathway negatively regulates IL-12/IL-23 p40 gene expression (21), whereas negative regulation of the p35 subunit is calcium-dependent, not ERK-dependent. In contrast to IL-12/IL-23 p40, we found that the ERK inhibitor UO126 blocked both IL-23 p19 promoter activation and mRNA expression, whereas the inactive analogue UO124 had no effect. Knockdown of ERK with siRNA further confirmed the importance of ERK in the activation of p19. These results suggest that ERK has a distinct role in the regulation of two subunits of IL-23, although the biological significance for this differential regulation for IL-23 is still not clear.
Previously, we have reported that IFN-γ, a Th1 cytokine, interacts synergistically with various TLR ligands to induce several proinflammatory cytokines, including IL-12/IL-23 p40, the other subunit of IL-23 (18). We have shown previously that IFN-γ synergizes with LPS and PGN to induce IL-12/IL-23 p40 promoter activation (18). However, IFN-γ significantly inhibits Th17 cell differentiation. Interestingly, we found that IFN-γ has no effect on IL-23 p19 promoter activation. Although there is a typical putative ISRE site in the p19 promoter, IFN-γ did not induce protein-DNA complex formation by EMSA using a probe containing the putative ISRE site. The results suggest that IFN-γ has differential role in the regulation of the two subunits of IL-23, although both p19 and p40 are activated by the TLR signaling pathway.
IL-23 is important for the in vivo generation of Th17 cells, which play a central role in the pathogenesis of autoimmune and inflammatory diseases (32–34), including experimental autoimmune encephalitis, collagen-induced arthritis, and inflammatory bowel disease (9), and which secrete IL-17 and IL-22. Some previous observations regarding IL-12 may relate to IL-23, since both cytokines share the IL-12 p40 subunit (6, 9, 10). Supporting this view is recent evidence that mice deficient in both IL-10 and IL-23 p19 do not develop colitis (11). Expression of IL-23 is elevated in patients with Crohn disease, and IL-17 expression is elevated in the serum and intestinal mucosa of patients with Crohn disease (35, 36). In addition, IL-23 and IL-17 are elevated in IL-10-deficient and RAG1-deficient models of murine colitis (11, 37). Indeed, lamina propria mononuclear cells from patients with Crohn disease treated with anti-IL-12 p40 antibody showed greatly reduced synthesis of IL-23 and IL-17 (38). The evidence clearly demonstrates that IL-23 and IL-17 are proinflammatory and contribute to development of several autoimmune diseases (6, 32).
IL-10, a 37-kDa homodimer first described as a factor inhibiting cytokine synthesis by a Th2 clone, inhibits IFN-γ expression in Th1 cells (39). Various cells produce IL-10, including Th2 and Tr1 T cell subsets, macrophages, and dendritic cells (40). IL-10 signals through IL-10 receptor, which is expressed on a variety of cells. IL-10 inhibits Th1 cell differentiation and macrophage production of IL-12 (40). Recently, we have reported that IL-10 inhibits Th17 cell differentiation, although the molecular mechanism is still not clear (41). In the present study, we found that IL-10-deficient macrophages expressed significantly higher levels of IL-23 p19 and that recombinant IL-10 suppressed LPS-induced IL-23 p19 mRNA expression. These results suggest that IL-10 may play a negative role in the control of Th17 immune response by regulating IL-23. A recent study suggests that Bcl3 may be involved in IL-10 regulation of IL-23 p19 (42). Mühlbauer et al. (42) found that LPS-induced Bcl3 expression was strongly impaired in IL-10-deficient dendritic cells. In addition, Bcl3 overexpression decreased LPS-induced IL-23 p19 expression in IL-10-deficient dendritic cells. However, the exact molecular mechanism for the regulation of IL-23 p19 gene expression by IL-10 still needs to be explored.
In summary, the present study suggests that IL-23 p19 expression in macrophages and dendritic cells is controlled by TLR signaling via transcription factors AP-1 and NF-κB. MAPKs ERK, JNK, and p38 are also important for p19 promoter activation. In addition, IL-10 strongly suppresses IL-23 p19 expression. The results should stimulate further exploration of IL-23 regulation in the development of autoimmune diseases.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 DK072201 (to H. X.).
- IL
- interleukin
- TLR
- Toll-like receptor
- IFN
- interferon
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- DMEM
- Dulbecco's modified Eagle's medium
- DN
- dominant negative
- RLM
- RNA ligase-mediated
- RACE
- rapid amplification of cDNA ends
- PGN
- peptidoglycan
- EMSA
- electrophoretic mobility shift assay
- qRT
- quantitative reverse transcription
- WT
- wild type
- LPS
- lipopolysaccharide
- TIS
- transcription initiation site(s)
- siRNA
- small interfering RNA
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Oppmann B., Lesley R., Blom B., Timans J. C., Xu Y., Hunte B., Vega F., Yu N., Wang J., Singh K., Zonin F., Vaisberg E., Churakova T., Liu M., Gorman D., Wagner J., Zurawski S., Liu Y., Abrams J. S., Moore K. W., Rennick D., de Waal-Malefyt R., Hannum C., Bazan J. F., Kastelein R. A. (2000) Immunity 13, 715–725 [DOI] [PubMed] [Google Scholar]
- 2.Hunter C. A. (2005) Nat. Rev. Immunol. 5, 521–531 [DOI] [PubMed] [Google Scholar]
- 3.Kastelein R. A., Hunter C. A., Cua D. J. (2007) Annu. Rev. Immunol. 25, 221–242 [DOI] [PubMed] [Google Scholar]
- 4.McKenzie B. S., Kastelein R. A., Cua D. J. (2006) Trends Immunol. 27, 17–23 [DOI] [PubMed] [Google Scholar]
- 5.Langrish C. L., McKenzie B. S., Wilson N. J., de Waal Malefyt R., Kastelein R. A., Cua D. J. (2004) Immunol. Rev. 202, 96–105 [DOI] [PubMed] [Google Scholar]
- 6.Cua D. J., Sherlock J., Chen Y., Murphy C. A., Joyce B., Seymour B., Lucian L., To W., Kwan S., Churakova T., Zurawski S., Wiekowski M., Lira S. A., Gorman D., Kastelein R. A., Sedgwick J. D. (2003) Nature 421, 744–748 [DOI] [PubMed] [Google Scholar]
- 7.Belladonna M. L., Renauld J. C., Bianchi R., Vacca C., Fallarino F., Orabona C., Fioretti M. C., Grohmann U., Puccetti P. (2002) J. Immunol. 168, 5448–5454 [DOI] [PubMed] [Google Scholar]
- 8.Parham C., Chirica M., Timans J., Vaisberg E., Travis M., Cheung J., Pflanz S., Zhang R., Singh K. P., Vega F., To W., Wagner J., O'Farrell A. M., McClanahan T., Zurawski S., Hannum C., Gorman D., Rennick D. M., Kastelein R. A., de Waal Malefyt R., Moore K. W. (2002) J. Immunol. 168, 5699–5708 [DOI] [PubMed] [Google Scholar]
- 9.Langrish C. L., Chen Y., Blumenschein W. M., Mattson J., Basham B., Sedgwick J. D., McClanahan T., Kastelein R. A., Cua D. J. (2005) J. Exp. Med. 201, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy C. A., Langrish C. L., Chen Y., Blumenschein W., McClanahan T., Kastelein R. A., Sedgwick J. D., Cua D. J. (2003) J. Exp. Med. 198, 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen D., Cheung J., Scheerens H., Poulet F., McClanahan T., McKenzie B., Kleinschek M. A., Owyang A., Mattson J., Blumenschein W., Murphy E., Sathe M., Cua D. J., Kastelein R. A., Rennick D. (2006) J. Clin. Invest. 116, 1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G., Pflanz S., Kastelein R. A. (2003) Immunity 19, 641–644 [DOI] [PubMed] [Google Scholar]
- 13.Trinchieri G. (2003) Nat. Rev. Immunol. 3, 133–146 [DOI] [PubMed] [Google Scholar]
- 14.Zhu C., Rao K., Xiong H., Gagnidze K., Li F., Horvath C., Plevy S. (2003) J. Biol. Chem. 278, 39372–39382 [DOI] [PubMed] [Google Scholar]
- 15.Wang I. M., Contursi C., Masumi A., Ma X., Trinchieri G., Ozato K. (2000) J. Immunol. 165, 271–279 [DOI] [PubMed] [Google Scholar]
- 16.Plevy S. E., Gemberling J. H., Hsu S., Dorner A. J., Smale S. T. (1997) Mol. Cell Biol. 17, 4572–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X., Neurath M., Gri G., Trinchieri G. (1997) J. Biol. Chem. 272, 10389–10395 [DOI] [PubMed] [Google Scholar]
- 18.Zhao J., Kong H. J., Li H., Huang B., Yang M., Zhu C., Bogunovic M., Zheng F., Mayer L., Ozato K., Unkeless J., Xiong H. (2006) J. Biol. Chem. 281, 10073–10080 [DOI] [PubMed] [Google Scholar]
- 19.Masumi A., Tamaoki S., Wang I. M., Ozato K., Komuro K. (2002) FEBS Lett. 531, 348–353 [DOI] [PubMed] [Google Scholar]
- 20.Murphy T. L., Cleveland M. G., Kulesza P., Magram J., Murphy K. M. (1995) Mol. Cell Biol. 15, 5258–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng G. J., Goodridge H. S., Harnett M. M., Wei X. Q., Nikolaev A. V., Higson A. P., Liew F. Y. (1999) J. Immunol. 163, 6403–6412 [PubMed] [Google Scholar]
- 22.Carmody R. J., Ruan Q., Liou H. C., Chen Y. H. (2007) J. Immunol. 178, 186–191 [DOI] [PubMed] [Google Scholar]
- 23.Mise-Omata S., Kuroda E., Niikura J., Yamashita U., Obata Y., Doi T. S. (2007) J. Immunol. 179, 6596–6603 [DOI] [PubMed] [Google Scholar]
- 24.Jochum W., Passegué E., Wagner E. F. (2001) Oncogene 20, 2401–2412 [DOI] [PubMed] [Google Scholar]
- 25.Guha M., Mackman N. (2001) Cell. Signal. 13, 85–94 [DOI] [PubMed] [Google Scholar]
- 26.Inaba K., Inaba M., Romani N., Aya H., Deguchi M., Ikehara S., Muramatsu S., Steinman R. M. (1992) J. Exp. Med. 176, 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong H., Li H., Kong H. J., Chen Y., Zhao J., Xiong S., Huang B., Gu H., Mayer L., Ozato K., Unkeless J. C. (2005) J. Biol. Chem. 280, 23531–23539 [DOI] [PubMed] [Google Scholar]
- 28.Xiong H., Zhu C., Li H., Chen F., Mayer L., Ozato K., Unkeless J. C., Plevy S. E. (2003) J. Biol. Chem. 278, 2271–2277 [DOI] [PubMed] [Google Scholar]
- 29.Falvo J. V., Parekh B. S., Lin C. H., Fraenkel E., Maniatis T. (2000) Mol. Cell Biol. 20, 4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzio M., Polentarutti N., Bosisio D., Prahladan M. K., Mantovani A. (2000) J. Leukocyte Biol. 67, 450–456 [DOI] [PubMed] [Google Scholar]
- 31.Al-Salleeh F., Petro T. M. (2008) J. Immunol. 181, 4523–4533 [DOI] [PubMed] [Google Scholar]
- 32.Weaver C. T., Harrington L. E., Mangan P. R., Gavrieli M., Murphy K. M. (2006) Immunity 24, 677–688 [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y., Danilenko D. M., Valdez P., Kasman I., Eastham-Anderson J., Wu J., Ouyang W. (2007) Nature 445, 648–651 [DOI] [PubMed] [Google Scholar]
- 34.Liang S. C., Tan X. Y., Luxenberg D. P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L. A. (2006) J. Exp. Med. 203, 2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt C., Giese T., Ludwig B., Mueller-Molaian I., Marth T., Zeuzem S., Meuer S. C., Stallmach A. (2005) Inflamm. Bowel Dis. 11, 16–23 [DOI] [PubMed] [Google Scholar]
- 36.Fujino S., Andoh A., Bamba S., Ogawa A., Hata K., Araki Y., Bamba T., Fujiyama Y. (2003) Gut 52, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kullberg M. C., Jankovic D., Feng C. G., Hue S., Gorelick P. L., McKenzie B. S., Cua D. J., Powrie F., Cheever A. W., Maloy K. J., Sher A. (2006) J. Exp. Med. 203, 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuss I. J., Becker C., Yang Z., Groden C., Hornung R. L., Heller F., Neurath M. F., Strober W., Mannon P. J. (2006) Inflamm. Bowel Dis. 12, 9–15 [DOI] [PubMed] [Google Scholar]
- 39.Fiorentino D. F., Bond M. W., Mosmann T. R. (1989) J. Exp. Med. 170, 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore K. W., de Waal Malefyt R., Coffman R. L., O'Garra A. (2001) Annu. Rev. Immunol. 19, 683–765 [DOI] [PubMed] [Google Scholar]
- 41.Gu Y., Yang J., Ouyang X., Liu W., Li H., Yang J., Bromberg J., Chen S. H., Mayer L., Unkeless J. C., Xiong H. (2008) Eur. J. Immunol. 38, 1807–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mühlbauer M., Chilton P. M., Mitchell T. C., Jobin C. (2008) J. Biol. Chem. 283, 14182–14189 [DOI] [PMC free article] [PubMed] [Google Scholar]