Abstract
ABC50 is an ATP-binding cassette (ABC) protein, which, unlike most ABC proteins, does not possess membrane-spanning domains. ABC50 interacts with eukaryotic initiation factor 2 (eIF2), which plays a key role in translation initiation and its control. ABC50 binds to ribosomes, and this interaction requires both the N-terminal domain and at least one ABC domain. Knockdown of ABC50 by RNA interference impaired translation of both cap-dependent and -independent reporters, consistent with a positive role for ABC50 in the function of eIF2, which is required for both types of translation initiation. Mutation of the Walker box A or B motifs in both ABC regions of ABC50 yielded a mutant protein that exerted a dominant-interfering phenotype with respect to protein synthesis and translation initiation. Importantly, although dominant-interfering mutants of ABC50 impaired cap-dependent translation, translation driven by certain internal ribosome entry segments was not inhibited. ABC50 is located in the cytoplasm and nucleoplasm but not in the nucleolus. Thus, ABC50 is not likely to be directly involved in early ribosomal biogenesis, unlike some other ABC proteins. Taken together, the present data show that ABC50 plays a key role in translation initiation and has functions that are distinct from those of other non-membrane ABC proteins.
ABC50 was first reported as a protein whose expression is increased following treatment of synoviocytes with tumor necrosis factor α (1). ABC50 was subsequently identified independently as a protein that co-purified extensively with eukaryotic initiation factor 2 (eIF2)2 (2). In common with other members of the ATP-binding cassette (ABC) family of proteins, ABC50 contains two ATP-binding cassettes (also termed nucleotide-binding domains (NBDs)) (1). Unlike most other members of the group, however, it lacks recognizable trans-membrane domains.
Sequence analysis revealed that ABC50 is a close relative of the yeast protein Gcn20p, which is required for the control by amino acids of the yeast eIF2 kinase, Gcn2p, which is activated by binding to uncharged tRNA molecules (3). Gcn20p is thought to cooperate with Gcn1p to bring uncharged tRNAs to Gcn2p during the elongation process; this couples the availability of amino acids for tRNA charging to the control of Gcn2p (4). However, Gcn20p and ABC50 differ in important respects. For example, whereas Gcn20p associates with ribosomes that are engaged in elongation, ABC50 apparently binds ribosomes involved in initiation as well as elongation (2). Its association with ribosomes is stimulated by ATP. In addition, although Gcn20p and ABC50 are similar in their ABC domains, they differ markedly in their N termini. Since it is only the N terminus of Gcn20p that is required to support the function of Gcn2p in yeast (4), it seems likely that ABC50 and Gcn20p play distinct roles.
Tyzack et al. (2) have provided initial data indicating that ABC50 stimulates the formation of complexes between eIF2, GTP, and the initiator methionyl-tRNA in vitro. It did so without affecting the binding of guanine nucleotides to eIF2, indicating that the effect is likely to be on the association of initiator methionyl-tRNA with eIF2. The available data thus suggested that ABC50 might play a positive role in the initiation of protein synthesis. However, no data for this have previously been presented. Similarly, the manner in which ABC50 binds to ribosomes, the significance of its ABC domains, and other features remained unclear.
The two NBDs of ABC proteins are involved in nucleotide binding/hydrolysis and contain a number of conserved features, including the Walker box A and B motifs and the “ABC signature motif” (usually LSGGQ) (5, 6). The NBDs of eukaryotic ABC proteins “dimerize” such that the two ATP-binding/hydrolytic sites involve Walker box A of one NBD and the ABC signature motif of the other.
Certain other non-membrane ABC proteins are known to be involved in translation or its control (7). Indeed, three of the eukaryotic ABCF classes contain proteins involved in the control of protein translation. Class I proteins are exemplified by ABC50 (also termed ABCF1). Class III proteins (exemplified by yeast Gcn20p) can interact with the ribosome in an ATP-dependent manner (4). The proteins of Class IVA (elongation factor 3) mediate translation elongation in certain fungi. eEF3 stimulates binding of the eEF1·GTP·aminoacyl-tRNA ternary complex to the ribosomal A site by facilitating the release of the deacylated tRNA from the E site, thus stimulating protein synthesis (8, 9). On the other hand, Class IVB contains proteins thought to be important for the export of mRNAs from the nucleus in yeast (10).
The ABCE1 gene product was originally identified for its inhibition of ribonuclease L (11) and is hence also termed RLI1. Yeast Rli1p associates with 40 S ribosomal subunits in vivo and can interact with eIF3 and eIF5 independently of ribosomes (12). The available data indicate that ABCE1 is involved in both ribosome biogenesis and mRNA translation and shuttles between cytoplasm and nucleus, possibly as a nucleocytoplasmic transporter (13–17).
Here, we report the first detailed investigation into the function and interactions of ABC50. The data described here identify features of ABC50 that are required for its interaction with ribosomes. Most importantly, we provide the first evidence that ABC50 is required for efficient translation initiation in living cells and show that the requirement for ABC50 differs between cap-dependent and internal ribosome entry segment (IRES)-dependent translation. These and other data indicate that the function of ABC50 is distinct from those of other ABC proteins.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human embryonic kidney 293 cells were grown in 6- or 10-cm plates in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% (w/v) fetal bovine serum (Invitrogen). Transient transfections were carried out by calcium phosphate precipitation of 10 μg of DNA in BES-buffered saline pH 6.96 with cells at a density of ∼6 × 105 to 8 × 105 cells/6-cm plate or 1.5 × 106 to 2 × 106 cells/10-cm plate (18).
For siRNA transfections, we used a strain of easily transfectable HeLa cells, generously provided by Prof. J. Pouysségur (CNRS, Nice, France). Lipid-mediated transfection was used to introduce siRNAs into these cells. Cells (in 6-well plates) were transfected when they reached 30–50% confluence in 1-ml antibiotic-free DMEM. A preincubated mixture (containing 160 pmol of siRNA in 177 μl of DMEM plus 3 μl of Oligofectamine® (Invitrogen) in 12 μl of DMEM) was added to the cells. After 5 h, cells were washed twice with phosphate-buffered saline (PBS), and 2 ml of fresh antibiotic-free DMEM with serum was added. Cells were then grown for 96 h and then harvested. Forty-eight hours after transfection and when required, HeLa cells were transfected with 3 μg of a bicistronic construct containing either the green fluorescent protein (GFP) and chloramphenicol acetyltransferase (CAT) reporter genes or the dsRED and GFP reporter genes. To do this, cells were transfected with GenePORTER 2 (GTS) transfection reagent according to the manufacturer's instructions. For imaging of RPL27, HeLa YFP-RPL27 cells were used (19).
Cell Harvesting
After treatment, cells were washed once with PBS and harvested in 400 μl of harvesting buffer (20 mm HEPES·KOH (pH 7.4), 50 mm KCl, 50 mm β-glycerophosphate, 0.2 mm EDTA, 10% glycerol, 1% (v/v) Triton X-100, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, and 1 μg/ml each of leupeptin, antipain, and pepstatin). Cell debris and nuclei were spun down for 1 min at 12,000 × g, and the supernatant was transferred to a new tube.
Cell Fractionation
HeLa cells were separated into cytoplasmic and nuclear fractions, and the latter were further fractionated into nucleoplasmic and nucleolar fractions as previously described (20). Proteins from these fractions, normalized to the same cell number, were applied to a 12% bisacrylamide gel and Western blotting was performed following the standard methods.
Immunoprecipitation
For anti-GFP and anti-CAT immunoprecipitations, 2 μl of antibody (Roche Applied Science and 5′Prime-3′Prime, respectively) was added to cell extracts and incubated for 1 h at 4 °C. Subsequently, protein G-Sepharose beads (20 μl of packed beads per immunoprecipitation) in wash buffer (harvesting buffer without Triton X-100) were added and allowed to bind the antibody for 2 h at 4 °C. The beads were then washed three times with 1 ml of this buffer and finally resuspended in SDS-PAGE loading buffer.
Plasmids and Primers
Plasmids pET28c-ABC50 and pCMV5-HA-ABC50 were created as shown in Ref. 21. The mutations in pCMV5-HA-ABC50 were created using the QuikChange® kit from Stratagene.
The bicistronic constructs containing the cricket paralysis virus (CrPV) IRES, hepatitis C virus (HCV) IRES, and encephalomyocarditis virus (EMCV) IRES were constructed within the pcDNA3 vector backbone. First, reporter pcDNA3 vectors that contain the CrPV IRES-GFP, HCV IRES-GFP, EMCV IRES-GFP, or GFP alone within the EcoRI and XbaI sites were generated. Second, the dsRED reporter gene was PCR-amplified using primers that contain 5′ KpnI and 3′ BamHI sites. The PCR dsRED fragment was subsequently cloned within the KpnI and BamHI sites of the pcDNA3 vector.
DNA sequencing was performed by the Sequencing Service (School of Life Sciences, University of Dundee, Scotland) using DYEnamic ET terminator chemistry (Amersham Biosciences) on Applied Biosystems automated DNA sequencers.
Gel Electrophoresis and Immunoblotting
SDS-polyacrylamide gels containing 12.5% (w/v) acrylamide were used. The proteins were transferred onto Immobilon-P polyvinylidene difluoride membranes (Millipore) or nitrocellulose (for samples from the cellular fractionation) and detected by Western blot analysis. Blots were visualized by ECL (Amersham Biosciences).
Antibodies
Antibody for eIF2α was obtained from New England Biolabs. HA and GFP antibodies were purchased from Roche Applied Science, actin 20-33 antibody from Sigma, dsRED antibody from Clontech, CAT antibody from 5′Prime-3′Prime, and antibodies for S6 and rpL28 from Cell Signaling and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. Rabbit anti-ABC50 antibodies were obtained as detailed earlier (21). Mouse anti-fibrillarin antibody 72B9 (22) was a kind gift from Prof. E. M. Tan (Scripps Research Institute). Mouse anti-nucleoporin p62 antibody 414 was purchased from Covance. Mouse anti-α-tubulin DM1A was from Sigma. Rabbit anti-fibrillarin for Western blotting was a kind gift from Dr. F. Fuller-Pace (Department of Molecular and Cellular Pathology, University of Dundee). Sheep anti-calregulin antibody was from Santa Cruz Biotechnology. The anti-ABC50 antibody has been described previously (21).
Sucrose Cushion or Density Gradient Centrifugation
These procedures were performed as described in Ref. 2.
Production of Recombinant Proteins in Escherichia coli
Plasmid pET28c-ABC50 was introduced into E. coli strain BL21 (DE3) by transformation. One liter of culture was grown at 37 °C to an optical density at 600 nm (A600 nm) of 0.6–0.8, and at this point, isopropyl 1-thio-β-d-galactopyranoside was added to 1 mm. Incubation was carried out for 4 h at room temperature. Cells were pelleted by centrifugation and resuspended in 20 ml of buffer A (20 mm HEPES (pH 7.9), 10% (v/v) glycerol, 100 mm KCl, 5 mm MgCl2, 20 mm imidazole. The cells were lysed by sonication, and the lysed extract was centrifuged at 4300 rpm for 30 min at 4 °C. The His-tagged protein was purified by immobilized metal affinity chromatography by using Ni2+-NTA technology (23) and eluted in buffer A, 500 mm imidazole.
UV Cross-linking of [γ-32P]ATP to ABC50
UV cross-linking of [γ-32P]ATP to recombinant His-tagged ABC50 expressed and purified was performed as follows. Reactions (20 μl) in SDG100 buffer lacking the proteinase/phosphatase inhibitors (50 mm HEPES/KOH, pH 7.6, 2 mm MgCl2, 100 mm KCl), containing ∼4 μg of His-ABC50, [γ-32P]ATP (5 μCi/185 Bq), and 100 μm ATP (for the control), were incubated on ice for 5 min. The reactions were then exposed for 30 min at 4 °C to UV radiation (1.2 J/cm2) in a Spectronics Corp. Spectrolinker XL-1500 UV cross-linker. 5× SDS-PAGE sample buffer was added to each reaction, and the samples were boiled for 5 min and resolved by SDS-PAGE. The gels were stained, dried, and subjected to autoradiography. As a “negative” control for potential phosphorylation by contaminating protein kinases and/or autophosphorylation, additional reactions were performed as above without exposure to UV irradiation.
Cellular Imaging Analysis
HeLa cells (ATCC) were cultured in DMEM supplemented with 10% fetal calf serum. For immunofluorescence, cells were cultured on borosilicate glass coverslips (thickness number 1; VWR) to 80% confluence. The coverslips were rinsed with PBS (three times), fixed in freshly prepared 4% (v/v) paraformaldehyde (Sigma; dissolved in PBS) for 10 min at room temperature, and then permeabilized in 0.5% Triton X-100 (Sigma) in PBS for 5 min at room temperature. The cells were rinsed three times in PBS and immunolabeled as previously described (24). In some experiments, immunofluorescence analysis of ABC50 was performed using HeLa cells stably expressing ribosomal protein L27 (19). Images presented here are maximal intensity projections of Z-stacks. Cells were imaged on a Zeiss Axiovert S100 Delta-Vision Restoration microscope (Applied Precision) using a Zeiss Planachromat (60 × 1.40 nuclear aperture objective) and a CCD-1300-Y/HS camera (Roper Scientific). Fifty optical sections separated by 0.2 μm were recorded for each field, and the images were processed by constrained iterative deconvolution using SoftWorx (Applied Precision). All of the images shown in this paper are single optical sections.
siRNA Experiments
The siRNAs used here correspond to sequences that target the 3′-untranslated region (UTR) of ABC50 mRNA and were purchased from Dharmacon. The siRNAs used to knock down ABC50 were as follows: sense, 5′-GGAAUGCUGCUGAACUUGAUU-3′; antisense, 3′-UUCCUUACGACGACUUGAACU-5′. The nonspecific siRNA used as a control was purchased from Dharmacon (Control IX; 47% GC content).
Protein Synthesis Assays
Cells were treated as usual. When indicated, cycloheximide (Sigma) was added to a final concentration of 10 μg/ml or DMSO to untreated cells for 40 min. Two hours before harvesting, 5 μCi of [35S]methionine/ml of medium was added. Cell extracts were prepared as described above, and 20 μl of the sample was spotted onto squares of 3MM paper, two for each treatment. Papers were boiled three times for 1 min in 5% trichloroacetic acid. They were then rinsed in 100% ethanol, dried, and put in scintillation vials with 3 ml of scintillation fluid. Incorporation of label was measured in a scintillation counter, and the results were normalized to the protein content of each sample measured with Bradford reagent (Bio-Rad).
Northern Blot Analysis
For Northern blot analyses, one dish of cells was collected for RNA isolation with RNeasy MiniKit (Qiagen). 7.5 μg of the RNA isolated was loaded per lane in a 3.6% formaldehyde-agarose gel. Northern blot hybridization with probes for ABC50 and GFP was performed as described in Ref. 25. The GFP probe was amplified by PCR using the following primers: forward, 5′-GAGGGCGAGGGCGATGCCA-3′; reverse, 5′-AGTTCACCTTGATGCCGTTC-3′. The ABC50 probe was amplified by PCR using the following primers: forward, 5′-TGAGCGCCAAGTGGCTTCAT-3′; reverse, 5′-AGCCCAGGCCAGCCAGGATC-3′.
Cap- and IRES-dependent Translation Assays
HEK293 or HeLa cells, as indicated, were transfected with a bicistronic construct containing the GFP reporter gene downstream from the β-globin gene 5′-UTR and CAT reporter gene downstream from a picornaviral (encephalomyocarditis virus) IRES (provided by Dr. A. A. M. Thomas, University of Utrecht). Cells were treated as usual, and 2 h before harvesting, 5 μCi of [35S]methionine was added per ml of medium. Cell extracts were prepared as described above. GFP and CAT proteins were immunoprecipitated from the cell extracts (600 μg of protein) by using anti-GFP (from Sigma) and anti-CAT (from 5′Prime-3′Prime) as described above. The samples were resolved on an SDS-polyacrylamide gel containing 12.5% acrylamide, which was fixed, washed with Amplify (GE Healthcare), dried, and then subjected to fluorography to detect the incorporation of [35S]methionine into GFP and CAT (26). To monitor CrPV and HCV IRES-mediated translation, HEK293 cells were transfected with bicistronic constructs containing the cDNAs encoding dsRED reporter, followed by the GFP reporter. Where indicated, the CrPV, EMCV, or HCV IRESs was cloned within the intercistronic region of the bicistronic construct. After transfection, cell lysates were prepared as described above, and the expression of dsRED and GFP reporter genes was analyzed by Western blot analysis.
RESULTS
Functional NBDs Are Not Required for the Association of ABC50 with Ribosomes
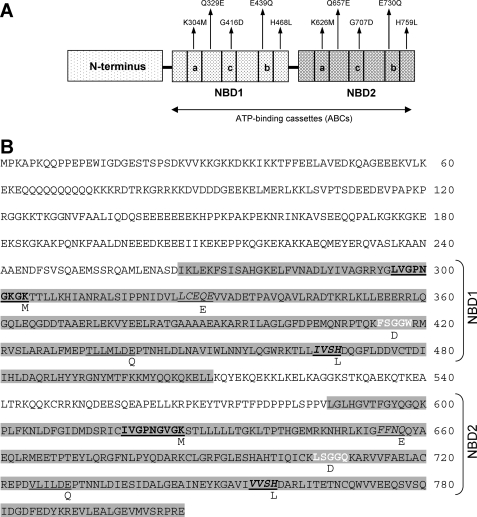
ABC50 has previously been shown to interact with ribosomes (2), and its NBDs have been shown to be required for this (21). To determine the role of canonical features of the NBDs in this interaction, we created a series of point mutants of ABC50, based on mutations known to interfere with functions of other ABC proteins. We generated the single and double mutants for the “ABC signature motif” in which a glycine was mutated to an aspartate in one or both nucleotide binding domains (G416D and G707D in the motifs FSGGW and LSGGQ, which conform to the standard ABC signature sequence; Fig. 1B). It has previously been shown, for example, that mutating the corresponding residue in P-glycoprotein to an aspartate still allows ATP binding but causes a loss of measurable ATPase activity (27). A second approach was to introduce the mutations H468L and H759L in the His-loop at each NBD. Mutations in this residue in the HlyB transporter abolish its ATPase activity yet preserve the ATP-binding capability of the protein (28). The data in Ref. 29 showed that mutating glutamine in the Gln-loop to either alanine or glutamate reduced the ATPase activity of P-glycoprotein by 10-fold. For this reason, we also generated mutations at these positions in ABC50 (Q329E and Q657E), in each NBD alone or in both (see also Fig. 1B). Finally, mutations in the Walker B motif of mouse P-glycoprotein decrease its ATPase activity and thus impair ATP hydrolysis (30). Neutralizing the charge on the conserved glutamates seems to inhibit ATP hydrolysis but has little effect on ATP binding, leading to the formation of a stable NBD dimer with ATP bound between the Walker A and signature motifs (31, 32). For this reason, glutamate-to-glutamine mutations in Walker box B at this site were created (E439Q and E730Q).
FIGURE 1.
Features of the ABC50 polypeptide. A, schematic representation of ABC50 protein. ABC50 contains an N-terminal region and two NBDs. The Walker A motif (a), signature motif (c), and Walker B motif (b) are shown. B, amino acid sequence of the ABC50 protein. Nucleotide binding domains 1 and 2 (NBD1 and NBD2) are highlighted in gray. The five types of conserved motifs that have been mutated in this work are indicated as follows: Walker A motif (boldface type, underlined), Walker B motif (underlined), signature motif (boldface type, white lettering), Gln-loop (italic type, underlined), and His-loop (boldface type, underlined, italic type). Mutations performed in this work (single or double targeting in the last case the same motif on both NBDs) are indicated above the schematic representation (A) and the wild type sequence (B).
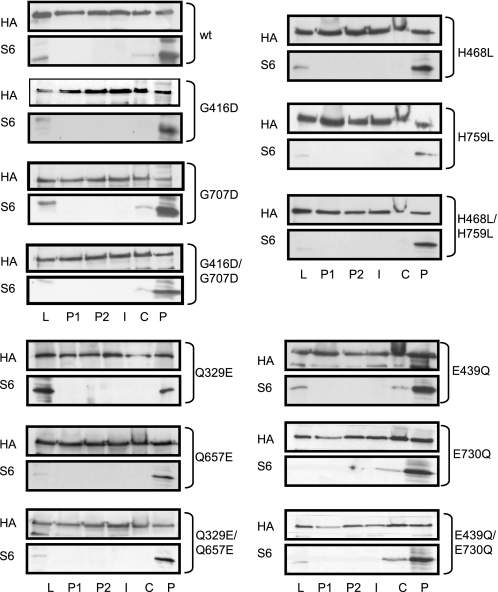
Each of these mutated proteins was able to bind to ribosomes, as indicated by its presence in the ribosomal pellet, indicated with a P in Fig. 2. Each appeared in this fraction to an extent similar to that of the wild type protein, indicating that the mutated features (and thus that ATP hydrolysis) are not essential for binding of ABC50 to ribosomes. The distribution of ectopically expressed HA-ABC50 is consistent with that we have previously observed for the endogenous protein (2).
FIGURE 2.
Ability of point mutants of ABC50 to bind to ribosomes. HEK293 cells were transfected with pCMV5-HA-based vectors encoding full-length ABC50, the single or double mutants G416D and G707D in the signature or C-motif, H468L and H759L in the His-loop, Q329E and Q657E in the Gln-loop, and E439Q and E730Q in Walker B motif. Cells were harvested in normal lysis buffer, and the supernatants were layered onto a 0.8 m sucrose cushion and centrifuged as described under “Experimental Procedures.” Immunoblotting for HA-ABC50 and S6 was performed on the following samples: initial cell lysate (L), upper layer (P1), second layer (P2), interface (I), cushion layer (C), pellet (P).
Binding of ABC50 to ATP
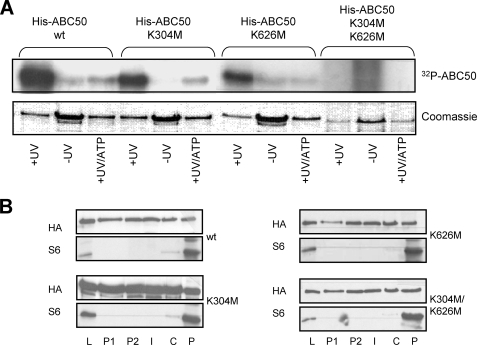
Since each ABC domain of ABC50 seemed to function independently in ribosome binding (21), we asked whether each could also bind ATP. To this end, we mutated Lys-304 and Lys-626 to methionine, individually or in combination. These residues were selected because the corresponding residues in the Walker A motif in eEF3 are essential for binding to ATP and the ensuing generation of ribosome-activated ATPase activity. Mutation of these residues in eEF3 abolishes its ribosome-dependent ATPase activity and ability to support poly(U)-directed polyphenylalanine synthesis and also impair cell growth (33). We expressed and purified recombinant wild type His6-ABC50 or the single and double mutated proteins in E. coli (pET28c-ABC50 WT, K304M, K626M, and K304M/K626M). These proteins were then incubated with [γ-32P]ATP and subjected to UV irradiation to cross-link bound nucleotide to the protein (Fig. 3A). As negative controls, no irradiation was performed, or unlabeled ATP was added as a competitor with the radiolabeled compound. When subjected to UV irradiation, WT ABC50 and the single point mutants became labeled with 32P, and this was competed away by the unlabeled ATP. For each of the single mutants, there was a marked reduction in labeling compared with wild type ABC50. There was no cross-linking of ATP to the mutant protein in which the lysines in both ATP-binding cassettes were mutated. This illustrates that each ABC domain binds ATP independently of the other, consonant with the properties of other ABC proteins.
FIGURE 3.
The Walker A motifs in ABC50 are required for ATP binding to ABC50 but not for ribosome binding. A, [γ-32P]ATP was cross-linked by UV irradiation (+UV) to recombinant wild type His-ABC50 and the K304M, K626M, or K304M/K626M variants (in the Walker A motifs of the two ATP-binding cassettes), which had been expressed in E. coli and purified as described under “Experimental Procedures.” As negative controls, samples were either not treated with UV (−UV) or irradiated in the presence of an excess of non-radiolabeled ATP in addition to the [γ-32P]ATP (+UV/ATP). [γ-32P]ATP labeling was visualized by autoradiography of the dried gels. The lower panel shows the Coomassie Blue-stained SDS-polyacrylamide gel from which the autoradiograph (upper panel) was obtained. B, HEK293 cells were transfected with the plasmid pCMV5-HA containing full-length ABC50 or the single or double mutants K304M and K626M. Cells were harvested, and the supernatant was layered onto a 0.8 m sucrose cushion and centrifuged as described under “Experimental Procedures.” Immunoblotting for ABC50 (HA) and the 40 S ribosomal protein S6 was performed on the following: L, initial cell lysate; P1, upper layer; P2, second layer; I, interface; C, cushion layer; P, pellet.
These mutant proteins were also expressed in HEK293 cells, and their abilities to bind ribosomes were analyzed. Each mutant, including the double mutant K304M/K626M, could still bind ribosomes (Fig. 3B), although their ability to bind ATP was compromised to a greater or lesser extent (Fig. 3A).
ABC50 Plays a Key Role in Translation Initiation
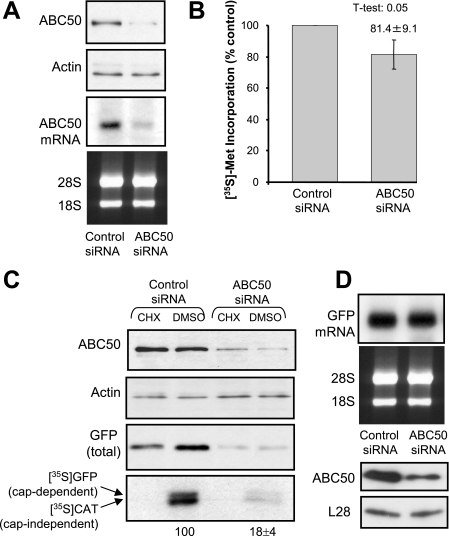
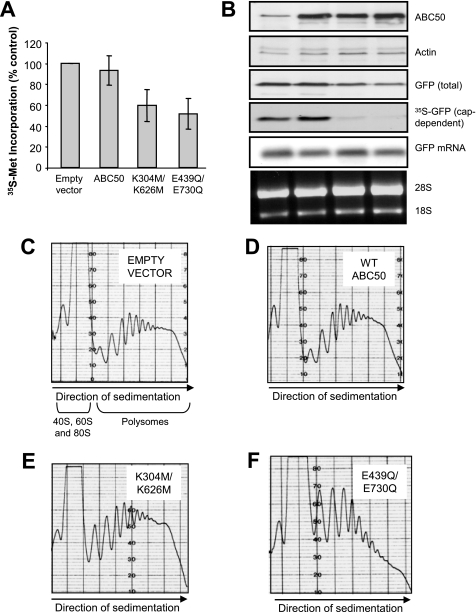
The ability of ABC50 to associate with eIF2, a core translation factor, and to stimulate its binding to Met-tRNA (2), suggested that ABC50 might play an important role in the process of translation initiation. To study this, we first used siRNA-mediated knockdown to reduce the cellular levels of ABC50. Extensive pilot experiments revealed that siRNA was an inefficient way of knocking down ABC50 levels in HEK293 cells, even when performed over extended times (up to 96 h) and when multiple treatments were carried out (data not shown). This may reflect either high levels of ABC50 in HEK293 cells or a long half-life for ABC50 in them. We therefore turned to an alternative cell type, HeLa cells, in which siRNA-mediated knockdown of other proteins works well (data not shown). As shown in Fig. 4A, siRNA against the ABC50 mRNA effectively decreased the amounts of ABC50 protein and mRNA to almost undetectable levels.
FIGURE 4.
siRNA-mediated knockdown of ABC50 impairs both cap-dependent and IRES-driven translation. A, HeLa cells were transfected with the indicated siRNAs as described under “Experimental Procedures.” At 96 h after transfection, cells were harvested and lysed. Levels of ABC50 and actin (loading control) were assessed by Western blot. In addition, after the same time period, total RNA was isolated from siRNA-transfected cells, and a Northern blot was performed to determine the levels of the ABC50 mRNA. 28 and 18 S rRNAs are shown as a loading control for the Northern blot. B, to assess the rate of protein synthesis, [35S]methionine was added to HeLa cells 96 h after transfection with the indicated siRNAs, and incubation was continued for a further 2 h. Cells were then lysed, and samples were processed to measure incorporation of label into 5% trichloroacetic acid-precipitable material. Incorporation was normalized to the total protein content of each sample. The mean ± S.D. of six independent experiments (each assayed in duplicate) is shown. C, a bicistronic vector containing both a GFP reporter gene downstream of the β-globin 5′-UTR and a CAT reporter gene downstream of the EMCV IRES was transfected into HeLa cells at 48 h after siRNA transfection. After a further 48 h, where indicated, cells were treated with cycloheximide (or DMSO as vehicle) for 40 min, and then [35S]methionine was added to the cells, which were incubated for a further 2 h. Cells were harvested, and the crude lysate was subjected to immunoprecipitation with GFP and CAT antibodies. The samples were resolved by SDS-PAGE, and [35S]methionine-labeled GFP and CAT were visualized by autoradiography. The radiolabeling of GFP and CAT was quantitated, and the results are shown below the relevant lane (n = 3; ±S.E.). The amounts of endogenous ABC50, actin, and total GFP were detected by Western blotting using the appropriate antibodies. D, RNA was isolated from cells transfected with siRNA/bicistronic vector, and a Northern blot was performed to assess the levels of GFP mRNA. 28 and 18 S rRNAs are shown as loading controls for the Northern blot. Levels of endogenous ABC50 and L28 were detected by Western blot (bottom).
Measurements of overall protein synthesis (by [35S]methionine incorporation) showed that this resulted in a modest, but still significant, inhibition of general protein synthesis (Fig. 4B). However, the effect on total protein synthesis was relatively small. Since the transfection efficiency of siRNAs is difficult to evaluate, the small magnitude of the effect could mean we only achieve efficient knockdown of ABC50 protein in those cells that are transfected optimally. We therefore tested the effect of knocking down ABC50 on the expression of reporter proteins encoded by a vector that we co-transfected with the siRNAs, anticipating that this might more reliably report the importance of ABC50 for protein synthesis. Furthermore, since a role of ABC50 in the function of eIF2 should affect both cap-dependent and -independent translation, we used a bicistronic vector which contains both a GFP reporter downstream of the β-globin 5′-UTR, to monitor cap-dependent mRNA translation, and a CAT reporter downstream of a picornaviral (EMCV) IRES. The latter monitors cap-independent mRNA translation. If ABC50 were only required for cap-dependent translation, for example, no effect on this “downstream” (IRES-driven) reporter would be observed.
The knockdown of ABC50 in HeLa cells markedly inhibited the synthesis of both the reporters (GFP and CAT; assessed by labeling with [35S]methionine), consistent with a role for ABC50 in general translation, rather than specifically in cap-dependent or IRES-mediated translation (Fig. 4C). There was also a drastic decrease in total GFP levels (Fig. 4C), again consistent with its impaired synthesis. These effects were not due to any difference in the levels of the reporter GFP-mRNA, which was the same in ABC50 siRNA- and control siRNA-treated cells (Fig. 4D). As expected, treatment of cells with the protein synthesis inhibitor cycloheximide completely blocked incorporation into both reporter proteins (Fig. 4C, bottom). Knocking down ABC50 levels therefore impairs mRNA translation, revealing a key role for ABC50 in protein synthesis. Since no difference in the levels of ribosomal RNA or ribosomal proteins (L28, Fig. 4D; S6, not shown) was detected in siRNA-ABC50 cells, loss of ABC50 does not appear to impair ribosome biogenesis.
Functional ATP-binding Domains Are Critical for the Role of ABC50 in mRNA Translation
By analogy with other ABC proteins, it was likely that the NBDs of ABC50 were required for its function in translation (7). To test this, we studied whether expressing several mutants of ABC50 in which both ABC domains are disrupted affected protein synthesis in HEK293 cells. Cells were transfected with pCMV5-HA vector encoding WT ABC50, the E439Q/E730Q, or the K304M/K626M mutants, and 24 h later, rates of protein synthesis were measured by the incorporation of [35S]methionine into total protein. Expression of WT ABC50 had little effect on general protein synthesis (Fig. 5A), whereas expressing mutants in which both Walker box A motifs or both Walker box B motifs are disrupted caused modest inhibition, to similar extents (Fig. 5A: K304M/K626M and E439Q/E730Q mutants). Similar effects were observed when cap-dependent protein synthesis was monitored by following incorporation of [35S]methionine into GFP (Fig. 5B). Expression of either the Walker box A or B mutant markedly inhibited GFP expression without affecting the levels of the reporter mRNA or the levels of 28 or 18 S ribosomal RNA (Fig. 5B). These data are entirely consistent with ABC50 playing a positive role in translation that is dependent upon the integrity of its NBDs.
FIGURE 5.
Effect of ABC50 and selected mutants on protein synthesis and polyribosome profiles. HEK293 cells were transfected with the plasmids pCMV5-HA (empty vector) or pCMV5-HA-ABC50 WT, K304M/K626M, or E439Q/E730Q without (A and C–F) or with a vector containing the GFP reporter cDNA downstream of the β-globin 5′-UTR (B). A, samples processed to measure incorporation of [35S]methionine into precipitated material after 5% trichloroacetic acid treatment as under “Experimental Procedures.” Incorporation was normalized to the total protein content of each sample. The mean ± S.D. of four independent experiments (assayed in duplicate) is shown. B, the crude lysate was subjected to immunoprecipitation with anti-GFP. Samples were resolved on a 12.5% SDS-polyacrylamide gel and visualized by autoradiography. The amounts of endogenous and ectopically expressed ABC50, actin, and total GFP were detected by Western blotting using appropriate antibodies. RNA was isolated from siRNA/bicistronic transfected cells, and a Northern blot was performed to assess the levels of GFP mRNA. 28 and 18 S are shown as a loading control for the Northern blot. C–F, sucrose density gradient analysis was performed as described under “Experimental Procedures.” Polyribosome profile analyses are shown for cells transfected with the empty vector pCMV5-HA (C), wild type HA-ABC50 (D), K304M/K626M (E), or E439Q/E730Q (F). Note that the top of the 80 S ribosome peak was “off scale” at the sensitivity used (which allows a clearer view of the polyribosome peaks). The sedimentation positions of 40, 60, and 80 S particles and of the polysomes are shown, as is the direction of sedimentation.
In order to assess in more detail the role of ABC50 in translation, we performed polyribosome profile analyses using sucrose density gradient centrifugation. In such analyses, the proportion of ribosomes that are engaged in active protein synthesis is indicated by the quantity of polyribosomes relative to non-polysomal ribosomes. As shown in Fig. 5C, a substantial proportion of the total ribosomes is found in polyribosomes in growing HEK293 cells. Expression of wild type ABC50 slightly increased the size of the peaks corresponding to polyribosomes of differing sizes (Fig. 5D). This is consistent with a positive role of ABC50 in translation initiation (the process by which ribosomes are recruited into polyribosomes).
Expressing the different Walker box mutants had differing effects on the ribosome distribution, although they have similar effects on overall protein synthesis (Fig. 5, A and B). Expressing the ABC50 mutant with K304M and K626M mutations (in both Walker box A motifs) seemed to modestly impair translation initiation, as indicated by the larger proportion of small polyribosomes in Fig. 5E as compared with the profile in Fig. 5, C and D. Mutation of both Walker box B motifs gave rise to a variant protein that exerted a more strongly interfering phenotype, as shown by the very substantial loss of large polyribosomes, and the marked increase in small polyribosomes (Fig. 5F) and in 80 S “monosomes” (which are “off scale” in the image shown). This finding suggests (i) that this mutant also interferes with translation initiation, (ii) that the Walker box B motifs are essential for the efficient function of ABC50, and (iii) that ABC50 normally plays an important positive role in the initiation of mRNA translation.
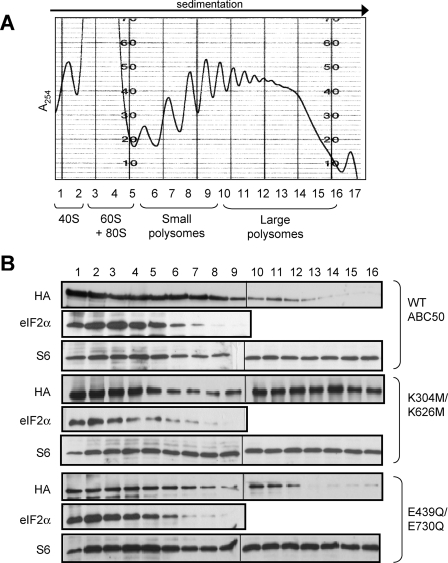
Given these findings, it was important to determine whether (i) the ABC50 mutants associated with polyribosomes and (ii) whether they affected the ability of eIF2 to do so. Fig. 6A shows a representative profile (ABC50 WT) for the UV absorbance and the small (S6) ribosomal subunit. Immunoblots were also developed with anti-HA (to detect the ectopically expressed ABC50 protein) and anti-eIF2α (for eIF2).
FIGURE 6.
Effects of expression of dominant-interfering mutants of ABC50 on the distribution of eIF2 across polyribosome profiles. A, sucrose density gradient analysis was performed as described under “Experimental Procedures.” The UV absorbance (A254) trace obtained for the gradient from cells expressing wild type HA-ABC50 is shown. The locations of the 40 and 60 S ribosomal subunits, the 80 S ribosomes, and the polyribosomes are indicated at the bottom. Fractions were collected (and the numbering is indicated). B, SDS-PAGE (15%)/immunoblot analysis of the sucrose density gradients performed from HEK293 cells ectopically expressing HA-ABC50 WT, K304M/K626M, or E439Q/E730Q, as indicated. The top panel shows the distribution of ectopically expressed ABC50 (anti-HA), and the bottom panel shows the distribution of endogenous eIF2 (anti-eIF2α; no signal was seen beyond fraction 9, so this section is not shown). The blots for the HA signal had to be performed using separate gels (indicated by the vertical line) run and analyzed in parallel. In each case, the distribution of S6 (S6 antibody; corresponding to the distribution of 40 and 80 S particles) is also shown.
The data (Fig. 6B) reveal that WT ABC50 is found in the region of the gradient containing 40, 60, and 80 S subunits/ribosomes and is also recovered, albeit to a lesser extent, in the polysomal part of the gradient. Strikingly, the Walker box A double mutant (K304M/K626M) showed a greatly enhanced association with polyribosomes relative to WT ABC50 (the quantity found in the upper part of the gradient was fairly similar for both proteins). One possible explanation for this is that intact Walker box As (i.e. ATP binding) are required for the release of ABC50 prior to entry of ribosomes into elongation. Expression of this mutant also appeared to decrease the amount of eIF2 associated with 60/80 S ribosomes but to leave the amount associated with 40 S subunits unaltered. The analysis for S6 revealed more of this protein in the small polysome region, consistent with the polysome profile data (Fig. 5E) which showed an increased proportion of small polysomes.
There was clearly less of the Walker box B double mutant in the polysomal region of the gradient. In part, this may reflect the decreased number of polyribosomes documented in Fig. 5E. However, the loss of ABC50 appears to be greater than that of ribosomes (as indicated by S6) as compared with cells expressing WT ABC50. This suggests that Walker box B (and, by implication, ATP hydrolysis) plays a positive role in the association of ABC50 with polysomal ribosomes. The data also indicate that its function in ABC50 is distinct from that of Walker box A, which is consistent with the fact that, although both motifs contribute to nucleotide binding, Walker box B also contributes the catalytic base for ATP hydrolysis (5). The analysis for S6 shows an increase in the signal for this protein in the fractions from the upper part of the gradient (Fig. 6B), consistent with an increase in 40 and 80 S particles, due to the decreased number of large polysomes (cf. Fig. 5F).
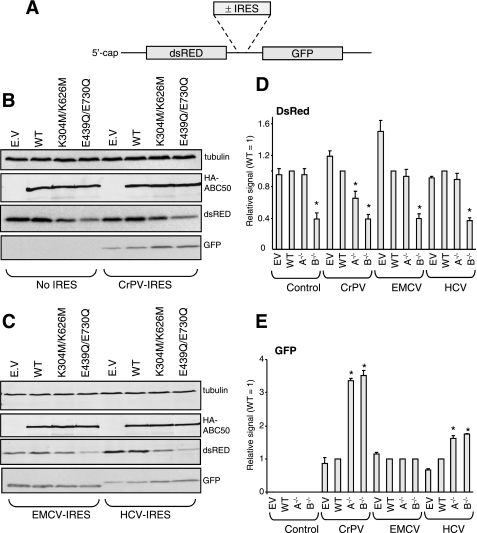
Differential Effects of ABC50 Mutants on IRES-driven Reporters
Certain viral mRNAs contain IRES elements that permit efficient translation of the downstream cistron without the full complement of initiation factors that is normally required for cap-dependent translation. An extreme example of this is the CrPV IRES, which does not even require eIF2 (34, 35), since it initiates translation from a non-AUG codon and does not require methionyl-tRNA. Other IRES elements tested here are reported to show no requirement for eIF2 (HCV; at least under certain conditions (36)) or a relatively low requirement for this factor (EMCV (37)).
To further characterize the effects of the dominant interfering ABC50 mutants on IRES-driven translation, we made use of bicistronic vectors containing the coding region for dsRED, translated in a cap-dependent way, and the coding region for GFP, in some cases preceded by a viral IRES (depicted in Fig. 7A). Cells were transfected with vectors containing no IRES or the IRES elements from CrPV, HCV, or EMCV, together with vectors for HA-tagged ABC50, the two Walker box mutants, or, as a control, the empty vector. Forty hours later, cells were lysed, and samples of lysate were analyzed by Western blot for HA-ABC50, dsRED, and GFP as well as for tubulin as a “loading control.”
FIGURE 7.
Effects of dominant-interfering mutants of ABC50 on cap- and IRES-driven translation. HEK293 cells were transfected with vectors containing the cistrons encoding dsRED and GFP plus, as indicated in A, IRES elements (derived from the CrPV, HCV, or EMCV RNAs). B and C, cells were co-transfected with wild type or mutant ABC50 or, as a control, the corresponding empty vector (EV). Forty hours later, cells were lysed, and samples were analyzed for the indicated proteins by Western blot. All data are representative of at least three replicate experiments in which essentially identical data were obtained. The expression levels of HA-ABC50 were similar across 12 replicate experiments. D and E, quantitation of the data, from three independent experiments ± S.E. for the expression of dsRED or GFP. *, p < 0.05 versus WT control. In D and E, A−/− indicates the double Walker box A mutant (K304M/K626M) and B−/− indicates the double Walker box B mutant (E439Q/E730Q).
As shown in Fig. 7B, the three forms of ABC50 expressed at very similar levels. The K304M/K626M mutants slightly inhibited dsRED production, whereas the E439Q/E730Q exerted a more pronounced inhibitory effect, consistent with other findings in this study (Fig. 7, B and C; quantitation in Fig. 7D). When the vector lacking an IRES was used, no GFP was detected (Fig. 7B), confirming that translation of this downstream cistron requires an active IRES element to be present. Introducing the CrPV IRES allowed expression of GFP, and strikingly, this was not inhibited by either ABC50 mutant (Fig. 7, B and E). In fact, both mutants actually markedly enhanced GFP synthesis from this reporter (Fig. 7, B, C, and E). This effect was consistently observed in three independent experiments. A similar, but more modest, stimulation was observed for the HCV construct (Fig. 7, C and E); due to the way this vector was constructed, the GFP polypeptide generated here is slightly larger than for the other vectors). This indicates that translation driven by the CrPV and HCV IRES does not have a strong requirement, if any, for ABC50. The enhanced expression of GFP seen in the presence of the ABC50 mutants when the CrPV and HCV IRES-containing vectors are used could be a consequence of the general repression of cap-dependent translation making more (e.g. elongation) factors and ribosomes available for translation of cistrons driven by these IRESs. In the case of the vector containing the EMCV IRES, neither inhibition nor stimulation was observed (Fig. 7, C and E) when the ABC50 mutants were co-expressed. This suggests that the EMCV IRES does not display a strong requirement for functional NBDs in ABC50.
Importantly, these data reveal that ABC50 exerts differing effects on the expression of different cistrons, depending on their mode of translation initiation. This supports the earlier conclusion that ABC50 plays a role in the initiation of translation and indicates that ABC50 mutants do not interfere with translation elongation, since, if this were so, the expression of all cistrons would be impaired.
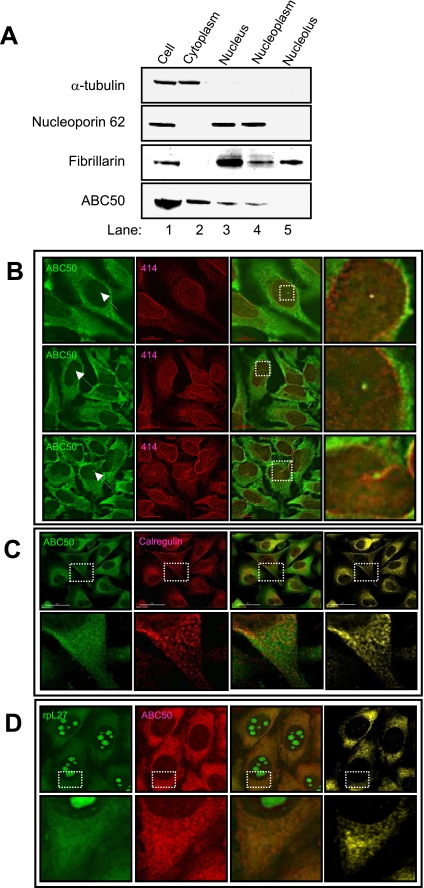
ABC50 Localizes Primarily to the Cytoplasm
The above data indicate that the main function of ABC50 is in mRNA translation rather than ribosome biogenesis (where ABCE1/RLI1 plays a role). We would therefore anticipate that endogenous ABC50 would mainly be located in the cytosol rather than the nucleolus, where ribosome assembly occurs. To study the subcellular localization of endogenous ABC50, HeLa cells were fractionated into cytoplasmic and nuclear fractions (20). As expected, α-tubulin and the nuclear pore protein nucleoporin p62 were detected in the cytoplasmic and nucleoplasmic fractions, respectively (Fig. 8A). Fibrillarin was detected in both the nucleoplasmic and nucleolar fractions, also in agreement with the roles of this protein in both of these nuclear compartments (39). ABC50 was predominantly located in the cytoplasmic fraction, whereas a smaller amount of ABC50 was also found in the nuclear fraction (Fig. 8A, lanes 2 and 3). When the isolated nuclei were further fractionated into nucleoplasmic and nucleolar fractions, ABC50 was detected only in the nucleoplasm (Fig. 8A, lanes 4 and 5). This suggests that ABC50 is unlikely to play a direct role in the early steps of ribosome biogenesis (as RLI1 does) (15) (which occur in the nucleolus).
FIGURE 8.
ABC50 is found in the cytoplasmic and nuclear compartments of HEK293 cells. A, subcellular fractionation of HeLa cells. HeLa cells were separated into cytoplasmic and nuclear fractions, and the latter was further fractionated into nucleoplasmic and nucleolar fractions. The fractions were probed with antibodies against (from top to bottom) α-tubulin, nucleoporin p62, fibrillarin, and ABC50. B, immunofluorescence of ABC50 (green) confirms that the majority of ABC50 was located in the cytoplasm, with a weak, finely punctate, nuclear staining. In some cells (fewer than 5%), ABC50 was also seen in one bright spot in the nucleoplasm. These ABC50 spots were colocalized with nucleoporin p62 (red). C, ABC50 co-localizes with the ER. HeLa cells were immunostained with anti-ABC50 (green) and anti-calregulin (red) (marker for endoplasmic reticulum). The right panel (yellow) shows co-localization sites for both proteins. D, ABC50 colocalizes with ribosomal proteins in the cytoplasm but not in the nucleus. HeLa cells stably expressing YFP-tagged L27 (green) were immunostained with anti-ABC50 (red). The right panel (yellow) shows co-localization sites for both proteins. Broken squares denote the areas shown enlarged on the extreme right (B) or below (C and D).
We considered it important to try to confirm these observations by immunofluorescence experiments. Anti-ABC50 predominantly labeled the cytoplasm of the cells, with a weak punctate nuclear staining (Fig. 8B). In a small percentage of the cells (<5%), ABC50 was also found in a single spot in the nucleus (Fig. 8B, arrows). Double immunofluorescence with anti-nucleoporin p62 revealed that these nuclear spots labeled by anti-ABC50 colocalized with the nuclear envelope and are likely to be invaginations of this structure (40).
Since proteins on the endoplasmic reticulum (ER; an important site of mRNA translation) are also detected in these nuclear invaginations, we asked if the ABC50 in HeLa cells was located on the ER by performing double immunofluorescence using anti-ABC50 and ER marker anti-calregulin (Fig. 8C). Anti-calregulin labeled a punctate pattern in the cytoplasm, which consistently overlapped with ABC50 in essentially all of the cells visualized. In addition to this punctuate staining, there was also a diffuse component in the ABC50 staining that was not labeled by anti-calregulin, suggesting that although a ABC50 is found on the ER, there is also a diffuse, non-ER cytoplasmic population of ABC50. ABC50 is thus found in the two major loci where active translation occurs.
We then asked how the distribution pattern of ABC50 was compared with that of ribosomal subunits. To visualize ribosomal subunits without using antibodies against ribosomal proteins, which are prone to fixation artifacts (41), we used a HeLa cell line stably expressing ribosomal protein L27-YFP (19). The YFP-tagged L27 was detected in isolated large ribosomal subunits and polyribosomes (data not shown). As shown in Fig. 8D, L27-YFP was predominantly located in the nucleolus, with additional weak cytoplasmic signals. Like ABC50, the cytoplasmic signal for L27-YFP was composed of a punctate pattern with a diffuse background. Both the punctate and diffuse components of the L27-YFP signal overlapped with ABC50 (Fig. 8D), although ABC50 was more concentrated in the punctate regions.
ABC50 colocalized with L27 in the cytoplasm but not in the nucleolus. This suggests that ABC50 binds to mature, functional ribosomes but not the preribosomes that are found in the nucleolus.
DISCUSSION
This study provides the first detailed information on ABC50, a member of the ABCF family of ABC proteins that lack membrane-spanning domains. Earlier work showed that Gcn20p and eEF3 (also ABCF family members) play key roles in translation and in its control (3, 4, 8, 42), whereas recent work has revealed a role for ABCE1 (also called RLI1) in ribosome preinitiation complex assembly and in ribosome biogenesis (12, 15, 16). Recent work has shown that the related Drosophila protein “pixie” associates with 40 S ribosomal subunits and is required for translation initiation (17). Less was known about ABC50 than its relatives; the present investigation substantially advances our understanding of this protein, its interactions, and its role in mRNA translation.
Our previous work (2, 21) showed that ABC50 interacts with eIF2 and suggested that ABC50 promoted the association of eIF2 with initiator methionyl-tRNA in vitro. This implied that ABC50 probably played a role in mRNA translation, probably at the initiation stage. The present report provides the first direct data supporting this; two types of experiments lead us to conclude that ABC50 does indeed play an essential role in translation initiation. First, the RNA interference experiments show that knocking down the level of expression of ABC50 leads to a severe inhibition of reporter proteins from the vector that was co-transfected with the siRNAs. Indeed, both cap-dependent and EMCV IRES-driven translation were impaired, demonstrating that the role of ABC50 is not restricted to events involved in cap-dependent translation, such as the function of the eIF4F complex. This is consistent with the finding that ABC50 promotes the activity of eIF2 (2), which is required for both modes of translation initiation. Second, the expression of specific point mutants of ABC50 in which both Walker box motifs were disrupted led to a reduction in polyribosome levels. This is again fully consistent with a role in initiation, the process that leads to formation of polyribosomes. ABC50 does apparently associate with elongating ribosomes; however, if ABC50 functioned primarily in elongation, the expression of interfering mutants with impaired function would be expected to result in the build-up of polyribosomes, rather than their loss.
Our recent data showed that the N terminus of ABC50 was sufficient for its interaction with eIF2 (21). In contrast, the N terminus of ABC50 is not sufficient to mediate association with ribosomes, either NBD1 or NBD2 also being required (21). However, mutations within the NBDs (Fig. 2), in various ways, individually or in combination, did not prevent this interaction. Nonetheless, variants of ABC50 with mutations in both Walker box A or B motifs exert a dominant-interfering phenotype with respect to cap-dependent and impaired polyribosome assembly. These data clearly show that the Walker box motifs play key roles in the functions of ABC50 and that the roles of the A and B boxes in terms of the function of ABC50 in translation are quite distinct. The latter conclusion is exemplified by the data (Fig. 6B) showing that the polysomal distribution of the Walker box A and B mutants differs markedly. Our earlier data (2) indicated that the binding of ABC50 to ribosomes was enhanced by ATP. The observation that the K304M/K626M mutant (which does not bind ATP) actually shows increased association with polysomes is therefore puzzling. The present findings suggest that ATP binding is not required for the basal level of interaction of ABC50 with ribosomes, but it is possible that ATP may somehow stimulate binding. Alternatively, it is possible, in principle, that the effect of ATP on the binding of endogenous ABC50 to ribosomes (2) is mediated through another ATP-binding protein, not via ABC50 itself.
The data for IRES-driven translation provide important information about the function of ABC50. First, a marked difference was observed between the effects of siRNA-mediated knockdown of ABC50 and those of the Walker box mutants. Although depleting cells of ABC50 strongly impaired EMCV IRES-driven translation (Fig. 4C), neither Walker box mutants expressed in cells exerted much effect on this (Fig. 7C). Indeed, a reporter cistron driven by either the HCV (36) or CrPV (34) IRES actually showed increased expression in cells expressing these mutants (Fig. 7, B and C); this may reflect increased availability, under these conditions where general translation initiation is inhibited, of ribosomes and translation factors for translation of these IRESs that show decreased factor requirements (34, 43–45). These observations imply that these IRESs have limited or no dependence on functional Walker boxes in ABC50, consistent with their low requirement (HCV (36) and EMCV (37)) or lack of requirement (CrPV (34)) for the ABC50 partner protein, eIF2. Indeed, CrPV-driven translation is actually enhanced when eIF2B activity is impaired (35). These findings are all consistent with the idea, based on our earlier work (2), that ABC50 normally acts to promote eIF2 activity. Further, the data for EMCV imply that feature(s) of ABC50 other than functional Walker boxes are needed for the function of the EMCV IRES. Further work is needed to establish what these features are; it is worth noting that the extreme N terminus of Gcn20p is sufficient to complement disruption of the Gcn20 gene in yeast (4), showing that Gcn20p has a function(s) that does not require the NBDs.
Although mutating the A or B Walker boxes in ABC50 creates mutants that interfere to similar extents with overall protein synthesis, the consequences of these mutations are quite different. Expressing a variant in which the A boxes are mutated (which prevents ATP binding) does not have a marked effect on formation of polysomes (Fig. 5E), but the mutant ABC50 shows a marked accumulation in polysomes (Fig. 6B). In contrast, the Walker box B mutant, which, by analogy with other ABC proteins, is expected to bind ATP but fail to hydrolyze it, causes marked loss of polysomes and greater inhibition of cap-dependent translation (Fig. 7B). However, this mutant does not build up in polysomes. These data are consistent with our finding that intact Walker boxes are not required for ABC50 to bind to ribosomes (Fig. 2). They suggest that ATP binding may be needed for release of ABC50 from ribosomes and that ATP hydrolysis by ABC50 may be necessary for ribosomes to complete initiation or enter elongation.
The present data distinguish ABC50 from the other ABC family proteins that are known to play roles linked to mRNA translation. Unlike Gcn20p, ABC50 binds to eIF2 and to both initiating and elongating ribosomes. Our data also suggest that ABC50 does not play a role in ribosome biogenesis, thus differentiating it from RLI1/ABCE1. First, although some ABC50 is found in the nucleus, it localizes to the nucleoplasm, not to the nucleolus, where rDNA transcription and ribosome assembly occur. Second, knocking down ABC50 had no apparent effect upon the levels of either ribosomal RNA (18 and 28 S) or ribosomal proteins.
Thus, ABC50 plays a positive role in translation initiation. It represents a fourth distinct functionally defined class of the non-membrane ABC proteins that play roles in the biogenesis, function, or control of the translational machinery of eukaryotic cells (Table 1).
TABLE 1.
Functions of ABC proteins involved in mRNA translation
| Protein | Known functions | Reference(s) |
|---|---|---|
| eEF3 | Required for binding of eEF1A/GTP/amino acyl-tRNA to the ribosomal A-site in certain fungi | Refs. 38 and 42 |
| Rli1 (ABCE1) | Interacts with eIF2, eIF3, and eIF5 | Refs. 12, 14, and 15 |
| Role in translation initiation | ||
| Role in ribosome biogenesis | ||
| Gcn20 | Interacts with Gcn1 and elongating ribosomes | Refs. 3, 4, and 46 |
| N-terminal region required for activation of the eIF2α kinase Gcn2 in response to accumulation of uncharged tRNA | ||
| ABC50 (ABCF1) | Interacts with eIF2 and initiating but not elongating ribosomes | Refs. 2 and 21; this work |
| Required for translation initiation |
Acknowledgments
We thank other former members of the Proud laboratory, especially Drs. Maria Buxad|$$|Aae, Josep Parra, and Andrew Tee, for technical help and advice and Dr. Ian Kerr (University of Nottingham) for helpful discussions.
This work was supported by a project grant from the UK Biotechnology and Biological Sciences Research Council (to C. G. P.) and by funding from the University of British Columbia.
- eIF2
- eukaryotic initiation factor 2
- ABC
- ATP-binding cassette
- DMEM
- Dulbecco's modified Eagle's medium
- NBD
- nucleotide-binding domain
- BES
- N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid
- siRNA
- small interfering RNA
- PBS
- phosphate-buffered saline
- GFP
- green fluorescent protein
- CAT
- chloramphenicol acetyltransferase
- IRES
- internal ribosome entry segment
- CrPV
- cricket paralysis virus
- HCV
- hepatitis C virus
- EMCV
- encephalomyocarditis virus
- HA
- hemagglutinin
- UTR
- untranslated region
- WT
- wild type
- ER
- endoplasmic reticulum
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Richard M., Drouin R., Beaulieu A. D. (1998) Genomics 53, 137–145 [DOI] [PubMed] [Google Scholar]
- 2.Tyzack J. K., Wang X., Belsham G. J., Proud C. G. (2000) J. Biol. Chem. 275, 34131–34139 [DOI] [PubMed] [Google Scholar]
- 3.Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G. (1995) EMBO J. 14, 3184–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G. (1997) Mol. Cell. Biol. 17, 4474–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins C. F., Linton K. J. (2004) Nat. Struct. Mol. Biol. 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 6.Locher K. P. (2004) Curr. Opin. Struct. Biol. 14, 426–431 [DOI] [PubMed] [Google Scholar]
- 7.Kerr I. D. (2004) Biochem. Biophys. Res. Commun. 315, 166–173 [DOI] [PubMed] [Google Scholar]
- 8.Triana-Alonso F. J., Chakraburtty K., Nierhaus K. H. (1995) J. Biol. Chem. 270, 20473–20478 [DOI] [PubMed] [Google Scholar]
- 9.Sandbaken M. G., Lupisella J. A., DiDomenico B., Chakraburtty K. (1990) J. Biol. Chem. 265, 15838–15844 [PubMed] [Google Scholar]
- 10.Kozak L., Gopal G., Yoon J. H., Sauna Z. E., Ambudkar S. V., Thakurta A. G., Dhar R. (2002) J. Biol. Chem. 277, 33580–33589 [DOI] [PubMed] [Google Scholar]
- 11.Bisbal C., Martinand C., Silhol M., Lebleu B., Salehzada T. (1995) J. Biol. Chem. 270, 13308–13317 [DOI] [PubMed] [Google Scholar]
- 12.Dong J., Lai R., Nielsen K., Fekete C. A., Qiu H., Hinnebusch A. G. (2004) J. Biol. Chem. 279, 42157–42168 [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z., Fang L. L., Johnsen R., Baillie D. L. (2004) Biochem. Biophys. Res. Commun. 323, 104–111 [DOI] [PubMed] [Google Scholar]
- 14.Chen Z. Q., Dong J., Ishimura A., Daar I., Hinnebusch A. G., Dean M. (2006) J. Biol. Chem. 281, 7452–7457 [DOI] [PubMed] [Google Scholar]
- 15.Kispal G., Sipos K., Lange H., Fekete Z., Bedekovics T., Janáky T., Bassler J., Aguilar Netz D. J., Balk J., Rotte C., Lill R. (2005) EMBO J. 24, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarunin A., Panse V. G., Petfalski E., Dez C., Tollervey D., Hurt E. C. (2005) EMBO J. 24, 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen D. S., Leevers S. J. (2007) J. Biol. Chem. 282, 14752–14760 [DOI] [PubMed] [Google Scholar]
- 18.Hall-Jackson C. A., Cross D. A., Morrice N., Smythe C. (1999) Oncogene 18, 6707–6713 [DOI] [PubMed] [Google Scholar]
- 19.Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. (2004) J. Cell Biol. 166, 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutherland H. G., Lam Y. W., Briers S., Lamond A. I., Bickmore W. A. (2004) Exp. Cell Res. 294, 94–105 [DOI] [PubMed] [Google Scholar]
- 21.Paytubi S., Morrice N. A., Boudeau J., Proud C. G. (2008) Biochem. J. 409, 223–231 [DOI] [PubMed] [Google Scholar]
- 22.Reimer G., Rose K. M., Scheer U., Tan E. M. (1987) J. Clin. Invest. 79, 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann A., Roeder R. G. (1991) Nucleic Acids Res. 19, 6337–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trinkle-Mulcahy L., Andrews P. D., Wickramasinghe S., Sleeman J., Prescott A., Lam Y. W., Lyon C., Swedlow J. R., Lamond A. I. (2003) Mol. Biol. Cell 14, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26.Tee A. R., Proud C. G. (2002) Mol. Cell. Biol. 22, 1674–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szakács G., Ozvegy C., Bakos E., Sarkadi B., Váradi A. (2001) Biochem. J. 356, 71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaitseva J., Jenewein S., Wiedenmann A., Benabdelhak H., Holland I. B., Schmitt L. (2005) Biochemistry 44, 9680–9690 [DOI] [PubMed] [Google Scholar]
- 29.Urbatsch I. L., Gimi K., Wilke-Mounts S., Senior A. E. (2000) Biochemistry 39, 11921–11927 [DOI] [PubMed] [Google Scholar]
- 30.Tombline G., Bartholomew L. A., Urbatsch I. L., Senior A. E. (2004) J. Biol. Chem. 279, 31212–31220 [DOI] [PubMed] [Google Scholar]
- 31.Moody J. E., Millen L., Binns D., Hunt J. F., Thomas P. J. (2002) J. Biol. Chem. 277, 21111–21114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Does D. C., Presenti C., Schulze K., Dinkelaker S., Tampé R. (2006) J. Biol. Chem. 281, 5694–5701 [DOI] [PubMed] [Google Scholar]
- 33.Yang H., Hamada K., Terashima H., Izuta M., Yamaguchi-Sihta E., Kondoh O., Satoh H., Miyazaki M., Arisawa M., Miyamoto C., Kitada K. (1996) Biochim. Biophys. Acta 1310, 303–308 [DOI] [PubMed] [Google Scholar]
- 34.Wilson J. E., Pestova T. V., Hellen C. U., Sarnow P. (2000) Cell 102, 511–520 [DOI] [PubMed] [Google Scholar]
- 35.Deniz N., Lenarcic E. M., Landry D. M., Thompson S. R. (2009) RNA 15, 932–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster A. M., Jan E., Sarnow P. (2006) RNA 12, 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005) Science 309, 1577–1581 [DOI] [PubMed] [Google Scholar]
- 38.Andersen C. B., Becker T., Blau M., Anand M., Halic M., Balar B., Mielke T., Boesen T., Pedersen J. S., Spahn C. M., Kinzy T. G., Andersen G. R., Beckmann R. (2006) Nature 443, 663–668 [DOI] [PubMed] [Google Scholar]
- 39.Chen D., Huang S. (2001) J. Cell Biol. 153, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fricker M., Hollinshead M., White N., Vaux D. (1997) J. Cell Biol. 136, 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas F., Kutay U. (2003) J. Cell Sci. 116, 2409–2419 [DOI] [PubMed] [Google Scholar]
- 42.Belfield G. P., Ross-Smith N. J., Tuite M. F. (1995) J. Mol. Evol. 41, 376–387 [PubMed] [Google Scholar]
- 43.Robert F., Kapp L. D., Khan S. N., Acker M. G., Kolitz S., Kazemi S., Kaufman R. J., Merrick W. C., Koromilas A. E., Lorsch J. R., Pelletier J. (2006) Mol. Biol. Cell 17, 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivas-Estilla A. M., Svitkin Y., Lopez Lastra M., Hatzoglou M., Sherker A., Koromilas A. E. (2002) J. Virol. 76, 10637–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez J., Yaman I., Merrick W. C., Koromilas A., Wek R. C., Sood R., Hensold J., Hatzoglou M. (2002) J. Biol. Chem. 277, 2050–2058 [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Barrio M., Dong J., Ufano S., Hinnebusch A. G. (2000) EMBO J. 19, 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]