Abstract
Antigen binding to the B cell antigen receptor (BCR) initiates an array of signaling events. These include endocytosis of ligand-receptor complexes via clathrin-coated pits, trafficking of the internalized ligand to lysosomes, degradation of the associated proteins to peptides, and peptide presentation on nascent major histocompatibility complex class II to T cells. The signal transduction events supporting BCR internalization are not well understood. We have identified a pathway supporting BCR internalization that includes the Vav1 and/or Vav3 isoforms and the GTPase dynamin. Vav1 and -3 are not required for B cell development and maturation, nor for a variety of BCR-induced signaling events nor for BCR signaling leading to major histocompatibility complex class II and CD80 expression, but Vav1 and/or -3 are absolutely required for BCR endocytosis and BCR-induced Rac-GTP loading. This is the first demonstration of a link between Vav and Rac in BCR internalization leading to antigen presentation to T cells.
Signal transduction events induced by antigen-triggered clustering of the B cell antigen receptor (BCR)2 include the activation of Src family and Syk tyrosine kinases and the tyrosine phosphorylation of the BCR-associated proteins Igα/β. These very early events stimulate multiple adapter proteins (BLNK, Bam32, Grb2, LAB, and Gab2) and effector enzymes (Vav, phospholipase C, and phosphatidylinositol 3-kinase (PI3K)). Once active, the effector enzymes generate small molecule second messengers, including inositol trisphosphate, diacylglycerol, increased cytoplasmic Ca2+, and 3-phosphoinositide lipids. These mediators activate additional effector enzymes (protein kinase C, Tec family kinases, Ras/Raf, MAP kinase, other small GTPases) to amplify the signal from the BCR. The amplified signal culminates in transcription factor activation, causing a resting B cell to become capable of presenting antigen to T cells and of entering the cell cycle. The early signal transduction events that drive B cell responses in health and disease have been well studied for the past 20 years (1–3).
However, none of these events is sufficient to drive a humoral immune response to a vaccine. The production of high affinity, class switched antibodies and long lived memory depends on physical contact between helper T cells to receive the essential CD40 signal (4, 5). Because T-B contact is driven by antigen, the B cells must present antigen to the helper T cells for responses to vaccines. Therefore, one of the primary functions of BCR signal transduction is to cause the antigen-bound receptor to internalize the BCR and associated antigen, and to process and present the antigenic peptide to helper T cells. There have been numerous studies of signal transduction events elicited by the antigen-bound BCR, linking signal transduction events to entry into the cell cycle and new gene expression. Very few studies have shown a connection between those signaling events and BCR internalization following antigen binding.
It may be that antigen is internalized through BCR in the complete absence of a biochemical signal. The issue was addressed several years ago using anti-Ig reagents that do or do not cross-link the BCR (6). Thus, although monovalent reagents could internalize and be presented to T cells, reagents that cross-link the BCR to induce signal transduction were 10-fold more efficient in delivering BCR (and thus bound antigen) to lysosomal and MHC class II peptide loading compartments (6). A second study (7) found that any mutation in the BCR transmembrane domain that prevented increased cytoplasmic Ca2+ likewise prevented antigen presentation by B cells expressing the mutant BCR. These findings are consistent with a need for BCR signaling to support BCR internalization and antigen presentation.
Despite the lack of a clear and defined signal transduction pathway supporting BCR internalization, some events are known to occur. Antigen binding causes the BCR to co-localize with clathrin, and clathrin is phosphorylated by a member of the Src family of protein-tyrosine kinases (8). Recent studies show that antigen-induced BCR internalization requires actin reorganization in a Btk-dependent manner (9). B cells lacking actin-binding protein 1 (10) or the adapter proteins Bam32 (11) or Linker of Activation of B cells (LAB) (12) fail to internalize their BCR. However, it is not known how these proteins are activated nor what lies up- or downstream of their activation. Indeed, despite the importance of BCR internalization to humoral immune responses (6, 7), there is very little mechanistic information available.
Our published work (13) shows an important role for the small GTPase Rac in antigen-induced BCR internalization. If a protein contributes to that process, it follows that animals lacking that protein should have defects in vaccine responses. The issue has been difficult to address in vivo because Rac1−/− mice are not viable (14). Rac2-deficient mice show a slightly impaired vaccine response (15), but the humoral immunity may be due to redundancy of Rac1 for Rac2 function (16).
In lymphocytes, the Vav isoforms (Vav1,2,3) catalyze the GTP/GDP exchange of guanine nucleotides for Rac (17). Vav-deficient animals have been described (18–22), but many of these mice show lymphopoiesis defects, and thus vaccine responses are difficult to measure. Most combinations of Vav deficiencies (Vav1−/−, Vav1,2−/−, Vav1,3−/−, Vav2,3−/−, and Vav1,2,3−/−) show gross deficiencies in T cell development and a lack of mature T cells in the periphery (20). B cell development in these animals shows some impairment in the ratio of peripheral subsets (20) and Vav1,2,3−/− mice fail to respond to vaccination by Ig production (20, 23).
The receptor internalization process has been well studied for receptor tyrosine kinases, which in many ways are similar to lymphocyte antigen receptors. Epidermal growth factor receptor is internalized through clathrin-coated pits to early endosomes where it either recycles back to the plasma membrane or moves to late endosome/lysosomes for degradation. Internalization through clathrin requires dynamin (24), a GTPase that assembles into a collar-like structure at the neck of coated pits (reviewed in Ref. 25). Mutations of dynamin that render its GTPase domain inactive (K44A dynamin) block growth factor receptor internalization (26).
We explored the role of Vav and dynamin in BCR internalization and antigen presentation to T cells. Using dominant-negative mutants or RNA interference strategies, we find both proteins are required for BCR endocytosis and for formation of a class II-peptide complex. The role of Vav was studied in more detail. We found that curiously mice having only Vav2 (Vav1,3−/− mice) show normal B cell development and Vav1,3-deficient B cells show normal BCR-induced signaling events. Vav1,3−/− B cells likewise respond to anti-BCR stimulation by undergoing a variety of signal transduction events and up-regulation of MHC class II and of CD80 in a way indistinguishable from wild-type B cells. Despite the apparently normal stimulation of known BCR signaling events, the Vav1,3-deficient B cells fail to internalize their antigen receptors. The data suggest that BCR signaling leading to activation of transcription factors, new protein expression, and other preparations for B-T interaction are distinct from those supporting BCR endocytosis.
EXPERIMENTAL PROCEDURES
Mice
Animals were purchased from The Jackson Laboratories (Bar Harbor, ME) or Taconic Farms (Germantown, NY) and bred at our facility. The Vav1,3−/− mice (20) were bred at our facility, and the animals were genotyped at day 10 after birth. Littermate wild-type C57BL/6 mice were used as controls for Vav1,3−/−. All experiments have been approved by the Institutional Animal Care and Use Committee at this institution.
Cell Culture
The murine B cell lymphoma M12g4Rd and parental M12.4 were maintained at 37 °C in RPMI 1640 medium containing 10% fetal bovine serum, 50 μm 2-mercaptoethanol, and penicillin/streptomycin.
Materials
F(ab′)2 antibodies to BCR were from Pierce and Zymed Laboratories Inc.. Antibodies to Rac, phosphorylated Akt, and MAP kinases were purchased from Cell Signaling (Danvers, MA). FITC-conjugated F(ab′)2 fragment of Cy5 rabbit anti-mouse, FITC donkey anti-rabbit IgG, FITC and PE donkey anti-mouse IgG, and the Cy3-donkey anti-goat IgG were purchased from Jackson ImmunoResearch (West Grove, PA). Phosphocholine (PC)-labeled keyhole limpet hemocyanin (KLH) and ovalbumin (Ova) were obtained from Biosearch Technologies, Inc. (Novato, CA). PC-KLH was conjugated with Cy5 using Fluorolink Cy5-reactive dye from Amersham Biosciences. Ova peptide 323–339 was purchased from GenScript Corp. (Piscataway, NJ). The Rac activation kit and anti-phosphotyrosine antibodies were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). Antibodies to actin, dynamin-1 and -2, Vav, phosphorylated Vav (pVav), and normal rabbit IgG were bought from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). (R)-Phycoerythrin-conjugated goat anti-rabbit IgG was from Southern Biotech (Birmingham, AL). Guinea pig complement was obtained from Sigma or Calbiochem. Recombinant protein A/G-agarose was from Invitrogen. Mouse IL-2 enzyme-linked immunosorbent assay kit and fluorochrome-conjugated antibodies to B220, CD19, CD3, IgM, IgD, MHC class II, CD80, and CD86 were bought from BD Biosciences.
Cell Purification and Stimulations and Lysates
Splenic B cells were prepared from the spleens of 5–8-week-old BALB/c mice as described earlier (13). In brief, erythrocytes were removed by osmotic lysis, and T cells were removed by incubation with anti-Thy 1.2 followed by guinea pig complement. The resulting population was 85–95% pure B cells. For immunoprecipitations and whole cell lysates, 0.5–1 × 106 cells were washed and resuspended in 100 μl of phosphate-buffered saline and stimulated at 37 °C with 15–20 μg/ml F(ab′)2 rabbit anti-mouse IgG or IgM or the protein antigens described below. Cells were lysed in TN-1 buffer (50 mm Tris-HCl, pH 8.0, 125 mm NaCl, 10 mm EDTA, 1% Triton, 10 mm NaF, 3 mm Na3VO4, 10 mm Na4P2O7, 10 μg/ml aprotinin, 10 μg/ml leupeptin) and centrifuged at 16,000 × g for 10 min at 4 °C. The cleared lysate was assessed for protein level and placed directly in SDS sample buffer (2.3% SDS, 50 mm Tris, pH 6.8, 10% glycerol). The proteins were separated by SDS-PAGE. The proteins were electrophoretically transferred to nitrocellulose membranes, probed with the indicated antibodies, and visualized by an enhanced chemiluminescence. The bands were quantitated by measuring individual chemiluminescence by using a Lumi-Imager F1 work station (Roche Applied Science). In most cases, the signal present in the unstimulated lane was defined as 1.0, and the signal present in the stimulated lane is the amount of signal relative to the unstimulated level. For MAP kinase and Akt, the fold stimulation was determined from the ratio of the phosphorylated to total amount of each kinase within an individual experiment.
BCR Internalization Assay (See Fig. 1)
FIGURE 1.
Assay for BCR internalization. A, primary B cells or the M12g4Rd cell line was incubated with anti-BCR or PC-KLH, respectively. In primary cells, the BCR forms a cap within 2 min, and the cells can be fixed and stained for microscopic analysis. The remaining surface BCR after internalization can be detected with a fluorochrome-labeled anti-BCR antibody. B, BCR internalization in primary B cells. Unstained B cells (solid line) show autofluorescence only; anti-BCR-stained B cells kept at 4 °C (gray peak) show a high level of fluorescence; anti-BCR-stained B cells transferred to 37 °C (dotted line) show an intermediate fluorescence, indicating loss of surface BCR. These data are representative of more than 50 similar measurements. C, BCR internalization in M12g4Rd B cells. Unstained B cells (solid line) show autofluorescence only; PC-KLH-treated B cells kept at 4 °C (gray peak) show a high level of fluorescence; PC-KLH-treated B cells transferred to 37 °C (dotted line) show decreased BCR fluorescence. These data are representative of more than 20 similar measurements. D, B cells were stained with FITC-anti-BCR at 4 °C and transferred to 37 °C followed by staining with PE-anti-BCR. B cells were analyzed for fluorescence at the indicated times after 37 °C incubation. These results are representative of three separate experiments.
Splenic B cells, M12.4, or M12g4Rd B cells were incubated with primary anti-Ig, PC-Ova, or PC-KLH for 30 min at 4 °C and washed to remove unbound antibody or antigen. The cells were then incubated at 37 °C for the indicated time. The reaction was stopped by moving samples to 4 °C, and the receptors remaining on the surface were detected by a fluorescently labeled anti-Ig. Mean fluorescence intensity of the samples was then analyzed using FACScan. As controls, samples were either stained with secondary antibody alone, or the samples were maintained at 4 °C to prevent BCR internalization. Percent internalized BCR was obtained by the mean fluorescence intensity (MFI) of the sample and the control in the following calculation: % internalization = ((MFI no internalization, 4 °C sample control) − (MFI sample) ÷ (MFI no internalization, 4 °C sample control)) × 100. Unless otherwise indicated, the data were taken 30 min after anti-BCR or antigen stimulation.
Fluorescence Confocal Microscopy
The methods have been described previously in more detail (13). Briefly, 2 × 106 cells were stimulated with Cy5-labeled anti-Ig for 2–5 min. The cells were then fixed with 2% paraformaldehyde and permeabilized with 0.05% saponin-containing buffer. For intracellular staining, fixed cells were incubated with a neutralizing antibody (2.4G2) overnight at 4 °C to block Fc receptors. The cells were then stained with antibody to dynamin-2 and pVav for 1 h at 4 °C, followed by staining with fluorescently labeled secondary antibodies for 30 min at 4 °C. The cells were then washed twice and added to poly-l-lysine (0.1%, w/v; Sigma)-coated coverslips. The slips were mounted to slides using antifade solution and viewed under a Zeiss LSM 510 laser scanning microscope. Co-localization of proteins was calculated based on their correlation coefficient after scanning the perimeter of the cell, as described (13). For quantitation, we report correlation coefficients of a minimum of 10 cells.
Transfections
For transfections, plasmids containing Δ6AVav (27) and full-length Vav were cloned into the expression vector pLEGFP-C1, which co-expresses green fluorescent protein (GFP) along with the gene of interest. WT K44A dynamin was cloned into the expression vector MIGR1, which also has a GFP expression cassette. A previously described 17-nucleotide sequence was used to target mouse Dyn2 mRNA (5′-GGACCAGGCAGAGAATG-3′ (28)). The nucleotide was cloned into the RNAi-ready pSIREN-RetroQ vector from Clontech following the manufacturer's protocol. M12g4Rd cells were transfected by electroporation as follows: 1 × 107 cells were electroporated with the indicated plasmid DNA in a 4-mm gap cuvette containing warm RPMI medium, using a Bio-Rad GenePulser at 340 V and 960 microfarads. The cells were resuspended in 8–10 ml of warm growth medium and cultured for 36–72 h before analysis. Following culture, the cells were sorted for equal expression of GFP.
Ca2+ Measurements
Ca2+ was monitored as described previously (29). Splenic B cells were loaded with INDO-1AM (Sigma) at 3 μm for 45–60 min at 37 °C, washed in PBS containing 1 mm CaCl2, and loaded into a 1-mm cuvette in a luminescence spectrometer (LS50B, PerkinElmer Life Sciences), using FLWinLab software. INDO-1 was excited at 355 nm, and fluorescence was monitored at 380 nm.
Flow Cytometry
Cells were stained as described above under “Fluorescence Confocal Microscopy.” Briefly, cells were stimulated with anti-Ig, fixed, and stained for cell surface markers using fluorescent-labeled antibodies specific to the proteins under study. Cells were then washed and analyzed by flow cytometry; a minimum of 10,000 events were analyzed. The mean fluorescence intensity was calculated for every sample and was plotted as an average of three independent experiments unless otherwise stated. The experiment in Fig. 4A used primary splenocytes from the indicated strain and gated on CD19+ cells.
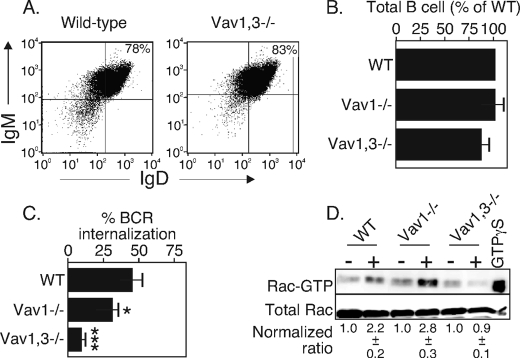
FIGURE 4.
Vav1 and/or -3 is required for BCR internalization. A, flow cytometry analysis of IgM/IgD staining of splenocytes from WT or Vav1,3−/− mice, gated on CD19-expressing B cells. The data are representative of three separate animals. B, total splenic B cells of WT, Vav1−/−, or Vav1,3−/− mice. Totals are calculated from total splenocytes and percent CD19+ from three (WT) or four animals (Vav1−/−, Vav1,3−/−). C, BCR internalization of B cells from the above strains using the assay in Fig. 1. *, p < 0.05; ***, p < 0.001. The data are representative of three experiments. D, Rac-GTP loading of unstimulated (−) or BCR-stimulated for 4 min (+) splenic B cells from the above strains. The assay was performed and quantitated as described in Fig. 3. The data are representative of three separate experiments.
Rac Assay
The protocol is based on a Rac-GTP pulldown method using GST-p21-activated kinase as described (30). Briefly, cells were stimulated and lysed as above, and the lysates were incubated with GST-PAK pre-bound to glutathione-agarose beads. After washing, the beads were incubated with Laemmli SDS-PAGE sample buffer, and the eluted proteins were subjected to SDS-PAGE. After the transfer of proteins to a nitrocellulose membrane, Western blotting was performed using a pan-monoclonal antibody to Rac isoforms. The amount of GTP-loaded Rac was quantitated by measuring the mass of GTP-Rac in comparison with the mass of total Rac. The value was then normalized to the GTP-Rac/total Rac ratio of unstimulated cells.
Antigen Presentation Assay
M12g4Rd or parental M12.4 B cells were cultured in RPMI 1640 medium in 24-well plates at 200,000/well. M12g4Rd B cells were stimulated with PC-Ova or PC-KLH (5 μg/ml or the indicated doses) to generate class II MHC loaded with Ova peptide on their surface. T cell hybridomas (DO.11.10 (31)) were then added to the wells containing B cells at a concentration of 100,000/well, and the co-culture was maintained at 37 °C for 24 h. Following incubation, the plates were frozen at −70 °C and thawed. Plates were centrifuged 2,000 × g, and supernatants were harvested. The supernatant was then assayed for IL-2. In the indicated experiments, B cells were co-cultured with DO.11.10 or B04 (32) in the presence of an Ova peptide.
Statistics
All experiments were done in triplicate and repeated multiple times, as indicated in the figure legends. The data are reported as the average and standard error of the triplicate samples from within each experiment.
RESULTS
Internalization Assay
Our internalization assay uses two models. First, we used primary B cells stimulated with F(ab′)2 fragments of anti-immunoglobulin to test BCR internalization of wild-type or knock-out animals, measured by flow cytometry or fluorescence confocal microscopy. Second, we used a B cell line (M12g4Rd) transfected with a BCR specific for the hapten PC (33). The M12g4Rd B cell line is used to introduce mutated forms of genes or RNA knockdown strategies and stimulated with PC-labeled KLH (PC-KLH) but does not form a distinctive BCR cap and thus is less useful for fluorescence microscopy. Primary B cells form an obvious cap but cannot be transfected. Hence the two models are used for distinct purposes but both readily internalize their BCR upon contact with antigen (in the case of M12g4Rd) or anti-BCR (in both cases).
We quantitate internalization by flow cytometry, as shown conceptually in Fig. 1A and as raw data in Fig. 1, B and C. Primary cells (Fig. 1B) or the M12g4Rd cell line (Fig. 1C) was incubated with unlabeled anti-BCR antibodies (B) or with PC-KLH (C). The BCRs of primary cells form caps within 120 s of stimulation, a point at which the cells can be fixed and stained for fluorescence microscopy. We move some samples to 37 °C and incubate for various periods of time to permit internalization (Fig. 1, B and C, dotted line). All the samples were then stained for the amount of remaining surface BCR using fluorochrome-labeled antibodies. There is no binding of this labeled antibody in the absence of anti-BCR (Fig. 1, B and C, solid line). One control sample was treated with isotype antibody, and a second was treated with unlabeled anti-BCR or antigen and kept at 4 °C to prevent internalization. The BCR fluorescence level of the 4 °C sample defines “no internalization” or complete external BCR receptor (Fig. 1, B and C, gray peak). The data were reported as percent internalization, using the calculation under “Experimental Procedures.” Using similar strategies, we have previously documented that the BCR appears within an endosome in the B cell (13).
The decrease of anti-BCR fluorescence could potentially be due to receptor shedding rather than BCR internalization. To test for receptor shedding, we labeled B cells at 4 °C with FITC-rabbit anti-BCR and washed away unbound antibodies. After incubation at 4 or 37 °C, we labeled the remaining rabbit anti-BCR on the B cell surface with PE donkey anti-rabbit. We found (Fig. 1D) that the PE anti-rabbit Ig was able to detect progressively less surface material as the B cells were incubated at 37 °C, showing either BCR internalization or BCR shedding. However, the amount of FITC rabbit anti-BCR remained constant, indicating the receptor is not lost from the surface and either remains at the surface or is internalized. Thus, our measurements reflect BCR internalization and not receptor shedding.
Dynamin Is Required for BCR Internalization
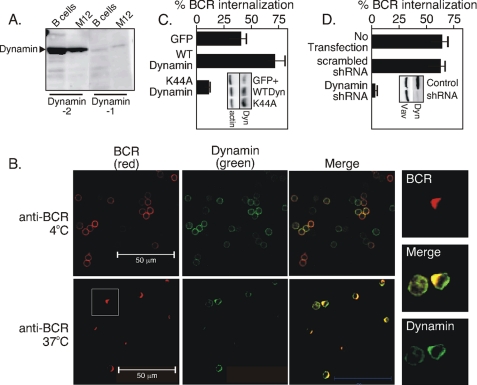
Earlier studies established a role for clathrin in BCR internalization (8). Dynamin is a GTPase that assists internalization of clathrin-coated pits in many receptor tyrosine kinases. Lysates of splenic B cells or from the M12g4Rd B cell line showed (Fig. 2A) that dynamin-2 is the predominant isoform present in both B cell models. We then asked if dynamin co-caps with the BCR following stimulation of cells with an anti-Ig reagent. Here, splenic B cells were incubated with Cy5-labeled anti-BCR antibodies at 4 °C (Fig. 2B, top panels) or moved to 37 °C (Fig. 2B, bottom panels). The cells were then permeabilized with saponin and stained with antibodies to dynamin-2. We found (Fig. 2B) that dynamin-2 co-localized with the BCR cap after 2 min at 37 °C. To make the point more clear, we show in Fig. 2B a set of images to the right that are digitally magnified anti-BCR-stimulated cells within the white square. The co-localization was quantitated in 10–20 cells, as we earlier described (13), by determining the correlation coefficient. In this method, perfectly co-localized fluorescent signals have a correlation coefficient of 1.0. We found that dynamin and BCR had correlation coefficients of 0.49 ± 0.01 in the unstimulated state, which rose to 0.91 ± 0.01 after 2 min of treatment with anti-BCR antibodies, indicating a high degree of co-localization.
FIGURE 2.
Dynamin co-caps with the BCR and is required for BCR internalization. A, immunoblot of B cell lysates with anti-dynamin-1 and -2. B, confocal fluorescence microscopy analysis of dynamin subcellular location using Cy5-labeled BCR antibodies (red), goat anti-dynamin-2 and FITC-labeled anti-goat Ig antibodies (green), and the merge of red and green fluorescence (yellow) before (anti-BCR, 4 °C) and after BCR stimulation (anti-BCR, 37 °C). Data represent 7–8 identical experiments. C, BCR internalization assay at 30 min after anti-BCR of M12g4Rd cells transfected with GFP alone, wild-type dynamin (WT dynamin), or the GTPase-deficient K44A dynamin mutant (K44A dynamin). The inset shows levels of the endogenous (GFP+) or transfected proteins (WTDyn, K44A). The data represent four similar trials. D, BCR internalization assay of M12g4Rd cells at 30 min after anti-BCR. Cells were transfected with nothing (no transfection), scrambled shRNA (scrambled shRNA), or shRNA targeting dynamin-2 (dynamin shRNA). The inset shows an immunoblot of control cells (control) transfected with scrambled shRNA or M12g4Rd transfected with shRNA to dynamin-2 (shRNA), immunoblotted with antibodies to dynamin or Vav. The data are representative of three separate experiments.
To test whether dynamin has a role in BCR internalization, we transfected the mouse B cell line M12g4Rd with either empty vector or vector containing wild-type dynamin or dominant-negative dynamin (K44A dynamin) lacking a functional GTPase domain. The transfectants were sorted for GFP expression to collect the cells expressing the transfected gene and gated to ensure similar levels of transfection. Analysis of BCR internalization in the sorted populations revealed that cells transfected with wild-type dynamin (Fig. 2C) showed an increase in receptor internalization as compared with cells transfected with control vector. In contrast, overexpression of the GTPase dynamin mutant (K44A dynamin) blocked BCR internalization. The inset in Fig. 2C shows the dynamin was indeed overexpressed in the cells transfected with wild-type dynamin (WTDyn; 2.9 ± 0.2-fold over GFP+; average and standard error of four separate transfections) or the K44A mutant of dynamin (K44A; 3.0 ± 0.4) relative to the endogenous dynamin in vector-only transfected cells. The data suggest dynamin is needed for BCR endocytosis and is limiting to the process because its overexpression improves BCR endocytosis. In an additional approach, M12g4Rd was co-transfected with shRNA targeting dynamin or with scrambled shRNA and with a vector encoding GFP. The GFP-expressing cells were sorted and applied to BCR internalization assay. We found (Fig. 2D, inset) that the shRNA targeting dynamin reduced dynamin protein expression by ∼80% within 72 h of transfection. Likewise, BCR internalization was completely blocked in cells expressing the shRNA targeting dynamin, but not in cells expressing scrambled shRNA (Fig. 2D). Together, these findings show that dynamin is required for BCR internalization.
Vav Is Required for BCR Internalization
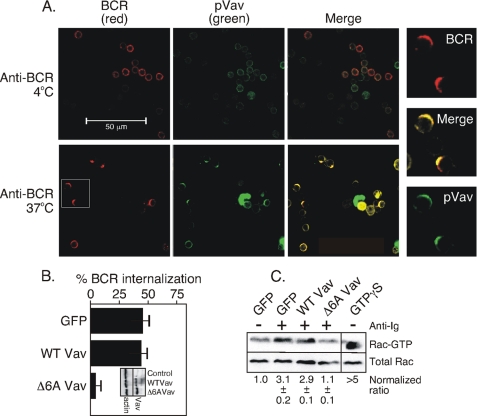
BCR internalization requires Rac (13), and Rac activation in lymphocytes might be accomplished by Vav (reviewed in Ref. 17). To understand the contribution of Vav to BCR internalization, we first examined Vav localization with respect to BCR in B cells using confocal microscopy (Fig. 3A) with an antibody to tyrosine-phosphorylated Vav. We found that pVav co-capped with the BCR following the receptor stimulation. As in Fig. 2, we show a set of images that are digitally magnified and obtained from the anti-BCR-stimulated cells in the white square. The BCR and pVav showed a correlation coefficient of 0.50 ± 0.01 in B cells held at 4 °C and 0.92 ± 0.02 in B cells at 37 °C, 2 min after BCR stimulation, again showing a high degree of co-localization. To test the role of Vav catalytic activity, we transfected the M12g4Rd B cells with either an empty vector, a vector containing wild-type Vav, or a dominant-negative Vav mutant lacking six essential residues within the catalytic domain (Δ6A Vav) (27). Following transfection, cells were sorted for equal amounts of GFP expression and then evaluated for BCR internalization. As above, we performed Western blots of an equal number of sorted GFP+ cells to test for protein expression levels. We found that WTVav and Δ6AVav were overexpressed 2.8 ± 0.2- and 2.9 ± 0.3-fold (the average and standard error of four separate transfections), respectively, relative to the endogenous Vav (Fig. 3B, inset). Regarding BCR endocytosis, we found (Fig. 3B) that receptor internalization was blocked in cells expressing the dominant-negative Δ6AVav mutant relative to cells transfected with the control vector containing GFP alone. Unlike the dynamin result, BCR endocytosis was not improved upon overexpression of Vav, suggesting Vav is not limiting to the process.
FIGURE 3.
Vav is required for BCR internalization. A, confocal microscopy of pVav (green), BCR (red), and the merge of red and green fluorescence (yellow) before (anti-BCR, 4 °C) and after BCR stimulation (anti-BCR, 37 °C). The data are representative of eight separate experiments. B, BCR internalization measured for 30 min with anti-BCR in M12g4Rd cells overexpressing wild-type Vav or Δ6AVav. The inset shows the levels of the endogenous (Control) or transfected (WTVav, Δ6AVav) proteins. The data are representative of four separate experiments. C, Rac-GTP loading of unstimulated (−) or BCR-stimulated for 4 min (+) B cells from the above transfectants as obtained using GST-PAK and as described previously (30). The material bound to GST-PAK was separated on 15% SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to pan-Rac. In a separate lane, nonhydrolyzable GTPγS was added to a lysate of the GFP+ cells as a positive control for Rac-GTP. The lanes between this control and anti-BCR-stimulated cells expressing the Δ6AVav mutant have been removed. The ratio of Rac-GTP to total Rac and normalized to the unstimulated sample is shown below the blot and was determined by quantitation of the chemiluminescent signal from the indicated proteins. The data are representative of four separate experiments.
If Vav is the sole Rac activator in B cells, the activation of Rac should likewise be impaired in B cells expressing the dominant-negative Δ6AVav mutant. We tested this possibility using a GST-PAK pulldown assay for GTP-bound Rac, as described (30). We found (Fig. 3C) that the amount of Rac-GTP was elevated ∼3-fold over background in anti-BCR-stimulated vector-transfected cells and in anti-BCR-stimulated cells overexpressing wild-type Vav. In contrast, there was no increase in Rac-GTP in B cells expressing the Δ6AVav mutant. These findings show that Vav is required for BCR internalization and that Vav acts proximal to Rac activation.
We used B cells from Vav1,3−/− mice as a second test of the role of Vav in BCR internalization. T cell development in these animals is markedly affected (20), but the mice have normal B cell development (Fig. 4, A and B) (20). Using our BCR internalization assay shown in Fig. 1, we found that splenic B cells lacking both the Vav1 and -3 isoforms failed to internalize their receptors (Fig. 4C) and failed to activate Rac (Fig. 4D). These findings are consistent with studies using the transfected Vav mutants and again show the importance of Vav1,3 to BCR-induced Rac activation and to BCR internalization.
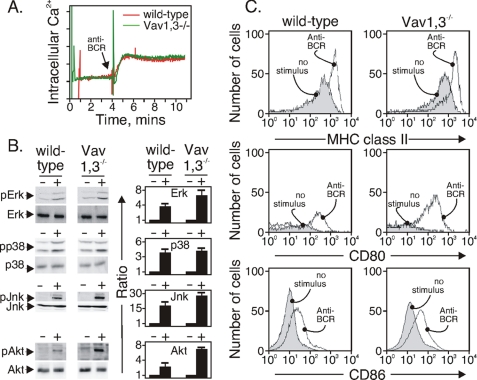
It is possible that the B cells from Vav1,3−/− mice exhibit other signaling defects that could negatively influence BCR endocytosis. We tested the ability of the BCR of Vav1,3-deficient B cells to stimulate Ca2+ influx, an event that requires the coordinated action of BLNK, phospholipase Cγ2, Btk, and inositol trisphosphate formation (34). We also tested BCR-induced activation of MAP kinase modules ERK, Jun kinase, and p38 and the survival kinase Akt. These pathways require the activation of numerous adapter proteins, primary (PI3K and phospholipase γ2) and secondary enzymes (Raf and other MAPKKKs) (35). We found that, relative to wild-type B cells, Vav1,3-deficient B cells displayed normal changes in cytoplasmic Ca2+ (Fig. 5A) and normal or elevated activation of all three MAP kinase modules (Fig. 5B). The activation of Akt in B cells of Vav1,3−/− mice, revealed by Akt phosphorylation and dependent on PI3K (36), was likewise elevated relative to wild-type B cells (Fig. 5B). The relative fold increase of the activation of each kinase is shown in Fig. 5B in the bar graph immediately to the right of each blot and is the average and standard error of four separate experiments. Thus, Vav1,3-deficient B cells are capable of BCR signaling, but they fail to internalize their BCR.
FIGURE 5.
Vav1,3 is not required for BCR-induced signaling events. A, anti-BCR-stimulated Ca2+ influx (arrow) of INDO-1-loaded B cells from the indicated strains. The data are representative of three separate experiments. B, anti-BCR-stimulated (+) activation of MAP kinase modules and pAkt in B cells from the indicated strains. Equal amounts of protein were loaded on the gel and quantitated as described under “Experimental Procedures.” The data are representative of four separate experiments. C, expression of MHC class II, CD80, and CD86 in unstimulated or anti-BCR-stimulated B cells from the indicated strains. Surface proteins were measured after 24 h with anti-BCR, and the data are representative of two separate experiments.
BCR stimulation causes B cells to up-regulate MHC class II and CD80/86 in preparation for their essential interaction with T cells. We tested whether these events were intact in the Vav1,3-deficient B cells by comparing with splenic B cells of wild-type mice and staining the cells with antibodies to MHC class II and CD80 and CD86 24 h after BCR stimulation. We found (Fig. 5C) that B cells from both wild-type and Vav1,3−/− mice up-regulated MHC class II and CD80 and CD86 following BCR stimulation. As shown earlier (37), CD80 expression was very low or absent on unstimulated B cells of both strains but rose to a higher level, and all cells became positive after BCR stimulation. Thus, B cells lacking these Vav isoforms are capable of up-regulating the most appropriate molecules for B-T interaction. However, because of the defect in BCR endocytosis, we hypothesized that B cells lacking a functional Vav or dynamin molecule would be defective in displaying MHC class II containing antigenic peptide.
Impaired BCR Endocytosis Reduces MHC Class II-mediated Antigen Presentation
It was difficult to test the appearance of antigenic peptides in a polyclonal system, because the individual BCRs expressed on primary B cells each have a unique protein specificity; hence, a population of primary B cells has many unique protein specificities. As a model for the process, we used the Balb/c-derived parent B cell line M12.4 and its progeny, M12g4Rd. These lines differ only by virtue of the transfection and expression in M12.4 of an antigen receptor specific for the hapten PC to create the M12g4Rd B cell line (33). The BCR-transfected M12g4Rd is able to present antigenic peptides derived from PC-labeled proteins to T helper cells (33).
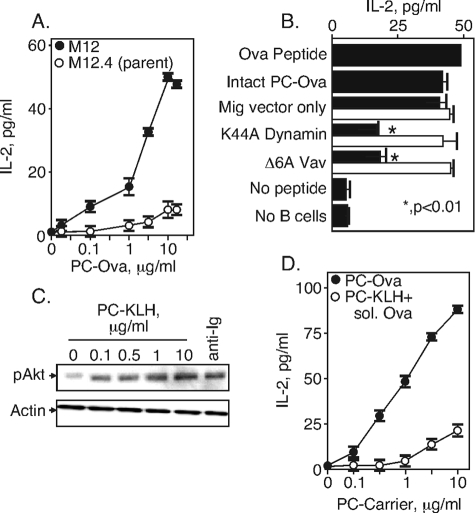
For these studies, we used PC-Ova and the Ova/I-Ad-specific DO.11.10 T cell line (31) as a tool to measure the appearance of MHC class II peptide on the surface of the B cell. The BCR-transfected M12g4Rd B cells express MHC class II and CD80/86, and the levels do not change upon treatment with PC-Ova or upon transfection (data not shown). The B cells were pulsed with the indicated concentration of PC-Ova for 4 h and then washed free of antigen. The antigen-pulsed B cells were then co-incubated for 24 h with DO.11.10 T cells, used here as a reporter for the presence of class II-Ova peptide on the antigen-pulsed B cells. At 24 h, the supernatant was collected and assayed for IL-2 production as a read-out for T cell recognition of class II-Ova peptide. We found (Fig. 5A) that the BCR-transfected M12g4Rd B cell line presented PC-Ova to DO.11.10 at doses about 2 logs lower than did the parental cell line. These data indicate that IL-2 responses at these antigen doses using this model are dependent on expression of an antigen-specific BCR. We used 5 μg/ml PC-Ova for subsequent experiments because this amount was on the linear portion of the curve and induced an easily detectable amount of IL-2 from DO.11.10.
To test the contribution of Vav and dynamin to BCR-mediated antigen internalization and presentation, M12g4Rd B cells were transfected with vector only or with the catalytically inactive dynamin (K44A dynamin) or Vav mutant (Δ6AVav) mentioned above. The GFP+- transfected cells were sorted, pulsed with 5 μg/ml PC-Ova for 4 h, and incubated for 24 h with DO.11.10. As an additional control, we added the processed synthetic Ova-(323–339) 17-mer peptide (38) to the transfected M12g4Rd B cells. We found (Fig. 6B, black bars) that M12g4Rd B cells expressing either the mutant dynamin or the mutant Vav showed a deficiency in antigen presentation, consistent with their deficiencies in BCR internalization. However, IL-2 responses were essentially restored when the processed, synthetic Ova peptide was applied to B cells expressing these mutant proteins (Fig. 6, white bars). Therefore, the defect in cells expressing dominant-negative Vav or dynamin is not because of the capacity of these cells to display MHC class II-peptide complexes that trigger T cells and is consistent with BCR internalization being the key event. We conclude that defects in BCR signal transduction altering BCR internalization similarly alter the formation of class II-peptide complexes, thus forming the T cell antigen receptor ligands to support B-T interaction and T cell activation.
FIGURE 6.
Antigen presentation to Ova-specific T cells requires dynamin and Vav. A, M12g4Rd or parental M12.4 were incubated with the indicated doses of PC-Ova for 4 h, washed and incubated with DO.11.10. IL-2 was measured after 24 h of incubation with the B cells. B, M12g4Rd B cells were transfected with dominant-negative mutants of dynamin or Vav as indicated. The GFP+-transfected cells were sorted and pulsed with PC-ovalbumin protein (black bars) or were provided the Ova peptide (ISQAVHAAHAEINEAGR; white bars). Antigen-pulsed M12g4Rd cells were washed and incubated with DO.11.10 Ova-specific T cells for 24 h. IL-2 production by the T cells was measured by enzyme-linked immunosorbent assay. *, p < 0.01. The data are representative of four separate experiments. C, induction of pAkt in M12g4Rd B cells by PC-KLH. M12g4Rd were incubated for 5 min with the indicated dose of PC-KLH or with anti-Ig (10 μg/ml). Whole cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies specific for pAkt. D, inefficient antigen presentation by pinocytosis. B cells were pulsed with the indicated amounts of PC-Ova or of PC-KLH containing 10 μg/ml soluble Ova. After 4 h, the B cells were washed and incubated with DO.11.10 before IL-2 measurements as above. The data are representative of two identical experiments.
B cells can present antigen that has been internalized by pinocytosis (39) as well as through the BCR. We have interpreted the findings in Fig. 6B as indicating that disrupting the BCR endocytic signaling apparatus likewise disrupts antigen presentation. However, an alternative possibility to BCR-mediated endocytosis is that BCR stimulation by PC-Ova elevates pinocytic activity, and a fraction of the ovalbumin protein is taken up in this nonspecific way. To test this possibility, we stimulated the B cells with various doses of an unrelated PC-labeled protein, KLH. PC-KLH caused phosphorylation of Akt in the M12g4Rd cell line, which was maximal at 1 μg/ml, consistent with the anti-hapten specificity of the BCR (Fig. 5C). To test the effect of BCR stimulation on antigen pinocytosis, we co-incubated the M12g4Rd cells with various doses of PC-KLH and a fixed amount of 10 μg/ml unconjugated, soluble ovalbumin. We found (Fig. 6D) that the DO.11.10 cells responded efficiently to PC-Ova and less efficiently to PC-KLH presenting soluble Ova. These data show that presentation of ovalbumin peptides by these B cells must be derived from BCR-internalized antigen and not from pinocytosed antigens.
DISCUSSION
Mature B cells capture antigen through their clonally restricted BCR. Antigen-induced BCR clustering triggers the induction of genes necessary for entry into the cell cycle by production of cyclins and cyclin-dependent kinases. BCR signaling also initiates a sequence of intracellular biochemical events that elicit increased class II and CD80/86 expression (40, 41). Increased expression of both receptors promotes the interaction with antigen-specific T cells to produce the humoral immune response. However, B-T interaction is limited by and requires a ligand for the T cell antigen receptor, antigenic peptide presented by B cell MHC class II molecules. The formation of the T cell ligand requires that BCR internalize, process, and present peptides from the antigenic protein. It was not clear whether the same set of kinases (e.g. Src and Tec family kinases, Syk), the abundant adapter proteins (e.g. BLNK, Bam32, Grb2, LAB, and Gab2), and the numerous enzymes (e.g. phospholipase C, PI3K, Vav, Raf, and MAP kinases) that activate transcription factors and genes regulating cell cycle transitions are likewise involved in BCR internalization.
Here we report some of the early signal transduction events that govern BCR internalization. We show that the internalization process requires the GTPase dynamin and the guanine nucleotide exchange factor Vav. The requirement for dynamin was expected, given the involvement of clathrin (8). Although our earlier report showed a need for the small GTPase Rac in BCR endocytosis (13), and Vav is a major GEF for Rac (17), the requirement for Vav was not immediately apparent because there are more than 30 known GEFs acting on Rac isoforms, identified by genetic or biochemical searches (42). Furthermore, the role of Vav in BCR-induced Rac activation was difficult to study because Vav-deficient animals often show dramatic deficiencies in mature lymphocytes.
Mice lacking all three Vav isoforms do not develop mature B or T cells (20). Mice lacking two of three Vav isoforms (the Vav1,3−/− mice) develop B cells normally (20) (Fig. 3), and the B cells appear to be able to signal normally via their BCR (Fig. 5). Indeed, some of the BCR signaling events were improved in Vav1,3-deficient B cells, and particularly those dependent on PI3K (formation of pAkt, pJNK, and pERK). Likewise, the Vav1,3-deficient cells were able to up-regulate MHC class II, CD80, and CD86 in response to BCR stimulation. Nevertheless, Vav1,3−/− B cells fail to internalize their BCR upon BCR stimulation (Fig. 4), a Rac-dependent event (13). These latter findings suggest Vav2, the only isoform present in Vav1,3-deficient B cells, is sufficient for BCR signal transduction to support all B cell developmental functions and at least some transcription factor activation. However, Vav2 is insufficient for BCR endocytosis. The data further support the notion that BCR signaling leading to activation of transcription factors and BCR signaling leading to receptor internalization sometimes use a distinct set of proteins.
It is interesting that the Vav1-deficient B cells showed a partial defect in BCR endocytosis but no defect in Rac activation. The observation suggests Vav might make contributions beyond activation of Rac. The hypothesis is possible because Vav contains a number of protein interaction domains and could be involved in recruitment of other proteins essential to BCR endocytosis, a function additional to its GEF activity.
It is not clear how Vav is activated in this system. In other cellular systems, Vav is activated by tyrosine phosphorylation at Tyr-174 (43), which relieves internal steric hindrance of the N-terminal calponin homology on the catalytic Dbl homology domain (44). A recent report suggests Vav might be phosphorylated by Btk (9). Vav phosphorylation is achieved by Vav recruitment to the site of tyrosine kinases associated with the receptors. Vav binds to the scaffolding protein LAT in T cells (45), to phosphorylated CD19 in B cells (46), to DAP10 in NK cells (47), and to PI3K-generated lipids in many cytokine receptors (27, 48, 49). CD19 is tyrosine-phosphorylated upon BCR stimulation (50), and PI3K is activated to cause formation of 3-phosphoinositide lipids (51). However, recent studies have identified a LAT homolog in B cells, termed LAB (52), expressing nine cytoplasmic tyrosine residues. It is not known if LAB is able to bind Vav, as does the T cell homolog LAT (53). However, LAB-deficient B cells show a defect in BCR endocytosis (12), consistent with the possibility that LAB recruits Vav to support Vav activation.
The actin cytoskeleton plays an important role in BCR internalization (54), which is regulated by Rho family GTPases (reviewed in Ref. 55). Our earlier studies indicated a role for Rac in BCR internalization (13), but there are many known GEFs acting on Rac isoforms (42). We were particularly interested in Vav for two reasons. First, relative to other Rac GEFs, Vav is the predominant Rac GEF expressed in B cells (17). Second, Vav-deficient animals show defects in BCR-dependent events, especially B cell development (18–20). Our data show not only that Vav1 and/or -3 is needed for BCR internalization, but also that Vav acts as the exclusive GEF for Rac activation in B cells because BCR-induced Rac activation was completely blocked in Vav-deficient B cells.
Vaccines are a powerful means to avoid infectious disease, but the responses to vaccination are variable in efficacy. Primary vaccination failure occurs in a heritable way in 2–10% of individuals receiving the measles vaccine (56) and 5–20% of hepatitis B vaccine (57, 58). Besides HLA, heritable features identified include the Killer cell inhibitory receptors (59) that recruit inhibitory phosphatases and block signaling by activating receptors. Genes encoding CD40 ligand (60) and TAP (61), a transport protein involved in antigen presentation, are other identified genetic components. Understanding the link between BCR signal transduction and internalization leading to antigen presentation is important for optimizing the response to vaccines where humoral immunity is the goal. This study identifies dynamin and Vav1 and/or -3 as important regulators of the humoral immune response, because they are involved in BCR signaling that supports BCR endocytosis, leading to processing and presenting antigen to T cells. Finding other heritable features that permit antibody responses in humans will help in understanding how vaccines work, but might also lead to better vaccines and a better understanding of adverse responses to existing vaccines.
Acknowledgments
We are grateful to David Parker, Lou Justement, and Paul Kincade for critical review of the manuscript.
Footnotes
- BCR
- B cell antigen receptor
- GEF
- guanine nucleotide exchange factor
- GFP
- green fluorescent protein
- KLH
- keyhole limpet hemocyanin
- LAB
- linker of activation of B cells
- MFI
- mean fluorescence intensity
- PC
- phosphorylcholine
- PI3K
- phosphatidylinositol 3-kinase
- Ova
- ovalbumin
- WT
- wild type
- FITC
- fluorescein isothiocyanate
- MHC
- major histocompatibility complex
- MAP
- mitogen-activated protein
- IL
- interleukin
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- ERK
- extracellular signal-regulated kinase
- pVav
- phosphorylated Vav
- shRNA
- short hairpin RNA
- PE
- phycoerythrin.
REFERENCES
- 1.Harwood N. E., Batista F. D. (2008) Immunity 28, 609–619 [DOI] [PubMed] [Google Scholar]
- 2.Gupta N., DeFranco A. L. (2007) Semin. Cell Dev. Biol. 18, 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambier J. C., Gauld S. B., Merrell K. T., Vilen B. J. (2007) Nat. Rev. Immunol. 7, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall L. S., Aruffo A., Ledbetter J. A., Noelle R. J. (1993) J. Clin. Immunol. 13, 165–174 [DOI] [PubMed] [Google Scholar]
- 5.Foy T. M., Shepherd D. M., Durie F. H., Aruffo A., Ledbetter J. A., Noelle R. J. (1993) J. Exp. Med. 178, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W., Cho H., Cheng P., Pierce S. K. (1995) J. Immunol. 155, 4255–4263 [PubMed] [Google Scholar]
- 7.Shaw A. C., Mitchell R. N., Weaver Y. K., Campos-Torres J., Abbas A. K., Leder P. (1990) Cell 63, 381–392 [DOI] [PubMed] [Google Scholar]
- 8.Stoddart A., Dykstra M. L., Brown B. K., Song W., Pierce S. K., Brodsky F. M. (2002) Immunity 17, 451–462 [DOI] [PubMed] [Google Scholar]
- 9.Sharma S., Orlowski G., Song W. (2009) J. Immunol. 182, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onabajo O. O., Seeley M. K., Kale A., Qualmann B., Kessels M., Han J., Tan T. H., Song W. (2008) J. Immunol. 180, 6685–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niiro H., Allam A., Stoddart A., Brodsky F. M., Marshall A. J., Clark E. A. (2004) J. Immunol. 173, 5601–5609 [DOI] [PubMed] [Google Scholar]
- 12.Mutch C. M., Sanyal R., Unruh T. L., Grigoriou L., Zhu M., Zhang W., Deans J. P. (2007) Int. Immunol. 19, 19–30 [DOI] [PubMed] [Google Scholar]
- 13.Phee H., Rodgers W., Coggeshall K. M. (2001) Mol. Cell. Biol. 21, 8615–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugihara K., Nakatsuji N., Nakamura K., Nakao K., Hashimoto R., Otani H., Sakagami H., Kondo H., Nozawa S., Aiba A., Katsuki M. (1998) Oncogene 17, 3427–3433 [DOI] [PubMed] [Google Scholar]
- 15.Croker B. A., Tarlinton D. M., Cluse L. A., Tuxen A. J., Light A., Yang F. C., Williams D. A., Roberts A. W. (2002) J. Immunol. 168, 3376–3386 [DOI] [PubMed] [Google Scholar]
- 16.Abo A., Pick E., Hall A., Totty N., Teahan C. G., Segal A. W. (1991) Nature 353, 668–670 [DOI] [PubMed] [Google Scholar]
- 17.Turner M., Billadeau D. D. (2002) Nat. Rev. Immunol. 2, 476–486 [DOI] [PubMed] [Google Scholar]
- 18.Zhang R., Alt F. W., Davidson L., Orkin S. H., Swat W. (1995) Nature 374, 470–473 [DOI] [PubMed] [Google Scholar]
- 19.Holsinger L. J., Graef I. A., Swat W., Chi T., Bautista D. M., Davidson L., Lewis R. S., Alt F. W., Crabtree G. R. (1998) Curr. Biol. 8, 563–572 [DOI] [PubMed] [Google Scholar]
- 20.Fujikawa K., Miletic A. V., Alt F. W., Faccio R., Brown T., Hoog J., Fredericks J., Nishi S., Mildiner S., Moores S. L., Brugge J., Rosen F. S., Swat W. (2003) J. Exp. Med. 198, 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swat W., Xavier R., Mizoguchi A., Mizoguchi E., Fredericks J., Fujikawa K., Bhan A. K., Alt F. W. (2003) Int. Immunol. 15, 215–221 [DOI] [PubMed] [Google Scholar]
- 22.Charvet C., Canonigo A. J., Bécart S., Maurer U., Miletic A. V., Swat W., Deckert M., Altman A. (2006) J. Immunol. 177, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 23.Stephenson L. M., Miletic A. V., Kloeppel T., Kusin S., Swat W. (2006) J. Immunol. 177, 8620–8625 [DOI] [PubMed] [Google Scholar]
- 24.Vieira A. V., Lamaze C., Schmid S. L. (1996) Science 274, 2086–2089 [DOI] [PubMed] [Google Scholar]
- 25.Song B. D., Schmid S. L. (2003) Biochemistry 42, 1369–1376 [DOI] [PubMed] [Google Scholar]
- 26.He Y. Y., Huang J. L., Chignell C. F. (2006) Oncogene 25, 1521–1531 [DOI] [PubMed] [Google Scholar]
- 27.Vedham V., Phee H., Coggeshall K. M. (2005) Mol. Cell. Biol. 25, 4211–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez T. S., Hamann M. J., McCarney S., Savoy D. N., Lubking C. M., Heldebrant M. P., Labno C. M., McKean D. J., McNiven M. A., Burkhardt J. K., Billadeau D. D. (2005) Nat. Immunol. 6, 261–270 [DOI] [PubMed] [Google Scholar]
- 29.Cooney D. S., Phee H., Jacob A., Coggeshall K. M. (2001) J. Immunol. 167, 844–854 [DOI] [PubMed] [Google Scholar]
- 30.Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 31.Kappler J. W., Skidmore B., White J., Marrack P. (1981) J. Exp. Med. 153, 1198–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shastri N., Gammon G., Horvath S., Miller A., Sercarz E. E. (1986) J. Immunol. 137, 911–915 [PubMed] [Google Scholar]
- 33.Webb C. F., Das C., Coffman R. L., Tucker P. W. (1989) J. Immunol. 143, 3934–3939 [PubMed] [Google Scholar]
- 34.Kurosaki T., Tsukada S. (2000) Immunity 12, 1–5 [DOI] [PubMed] [Google Scholar]
- 35.Gauld S. B., Dal Porto J. M., Cambier J. C. (2002) Science 296, 1641–1642 [DOI] [PubMed] [Google Scholar]
- 36.Jacob A., Cooney D., Tridandapani S., Kelley T., Coggeshall K. M. (1999) J. Biol. Chem. 274, 13704–13710 [DOI] [PubMed] [Google Scholar]
- 37.Greenwald R. J., Freeman G. J., Sharpe A. H. (2005) Annu. Rev. Immunol. 23, 515–548 [DOI] [PubMed] [Google Scholar]
- 38.Buus S., Sette A., Colon S. M., Miles C., Grey H. M. (1987) Science 235, 1353–1358 [DOI] [PubMed] [Google Scholar]
- 39.Arai C., Ichijo T., Tanaka Y., Okada Y., Umeda M., Uchida T., Kiniwa M., Kakiuchi T. (2003) Eur. J. Immunol. 33, 1806–1815 [DOI] [PubMed] [Google Scholar]
- 40.Natarajan K., Sahoo N. C., Rao K. V. (2001) J. Immunol. 167, 114–122 [DOI] [PubMed] [Google Scholar]
- 41.Lenschow D. J., Sperling A. I., Cooke M. P., Freeman G., Rhee L., Decker D. C., Gray G., Nadler L. M., Goodnow C. C., Bluestone J. A. (1994) J. Immunol. 153, 1990–1997 [PubMed] [Google Scholar]
- 42.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 43.Miletic A. V., Sakata-Sogawa K., Hiroshima M., Hamann M. J., Gomez T. S., Ota N., Kloeppel T., Kanagawa O., Tokunaga M., Billadeau D. D., Swat W. (2006) J. Biol. Chem. 281, 38257–38265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aghazadeh B., Lowry W. E., Huang X. Y., Rosen M. K. (2000) Cell 102, 625–633 [DOI] [PubMed] [Google Scholar]
- 45.Zhang W., Irvin B. J., Trible R. P., Abraham R. T., Samelson L. E. (1999) Int. Immunol. 11, 943–950 [DOI] [PubMed] [Google Scholar]
- 46.O'Rourke L. M., Tooze R., Turner M., Sandoval D. M., Carter R. H., Tybulewicz V. L., Fearon D. T. (1998) Immunity 8, 635–645 [DOI] [PubMed] [Google Scholar]
- 47.Graham D. B., Cella M., Giurisato E., Fujikawa K., Miletic A. V., Kloeppel T., Brim K., Takai T., Shaw A. S., Colonna M., Swat W. (2006) J. Immunol. 177, 2349–2355 [DOI] [PubMed] [Google Scholar]
- 48.Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R. D., Krishna U. M., Falck J. R., White M. A., Broek D. (1998) Science 279, 558–560 [DOI] [PubMed] [Google Scholar]
- 49.Tamás P., Solti Z., Bauer P., Illés A., Sipeki S., Bauer A., Faragó A., Downward J., Buday L. (2003) J. Biol. Chem. 278, 5163–5171 [DOI] [PubMed] [Google Scholar]
- 50.Chalupny N. J., Kanner S. B., Schieven G. L., Wee S. F., Gilliland L. K., Aruffo A., Ledbetter J. A. (1993) EMBO J. 12, 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brauweiler A., Tamir I., Dal Porto J., Benschop R. J., Helgason C. D., Humphries R. K., Freed J. H., Cambier J. C. (2000) J. Exp. Med. 191, 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen E., Zhu M., Zhang W., Koonpaew S., Zhang W. (2003) Nat. Immunol. 4, 117–123 [DOI] [PubMed] [Google Scholar]
- 53.Paz P. E., Wang S., Clarke H., Lu X., Stokoe D., Abo A. (2001) Biochem. J. 356, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown B. K., Song W. (2001) Traffic 2, 414–427 [DOI] [PubMed] [Google Scholar]
- 55.Heasman S. J., Ridley A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 56.Poland G. A., Ovsyannikova I. G., Jacobson R. M. (2008) Vaccine 26, 6183–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruskall M. S., Alper C. A., Awdeh Z., Yunis E. J., Marcus-Bagley D. (1992) J. Exp. Med. 175, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuckerman J. N. (1996) J. Med. Virol. 50, 283–288 [DOI] [PubMed] [Google Scholar]
- 59.Neumann-Haefelin C., Thimme R. (2007) Genes Immun. 8, 181–192 [DOI] [PubMed] [Google Scholar]
- 60.Goncalves L., Albarran B., Salmen S., Borges L., Fields H., Montes H., Soyano A., Diaz Y., Berrueta L. (2004) Virology 326, 20–28 [DOI] [PubMed] [Google Scholar]
- 61.Hayney M. S., Poland G. A., Dimanlig P., Schaid D. J., Jacobson R. M., Lipsky J. J. (1997) Vaccine 15, 3–6 [DOI] [PubMed] [Google Scholar]