Abstract
The thrombin mutant W215A/E217A (WE) is a potent anticoagulant both in vitro and in vivo. Previous x-ray structural studies have shown that WE assumes a partially collapsed conformation that is similar to the inactive E* form, which explains its drastically reduced activity toward substrate. Whether this collapsed conformation is genuine, rather than the result of crystal packing or the mutation introduced in the critical 215–217 β-strand, and whether binding of thrombomodulin to exosite I can allosterically shift the E* form to the active E form to restore activity toward protein C are issues of considerable mechanistic importance to improve the design of an anticoagulant thrombin mutant for therapeutic applications. Here we present four crystal structures of WE in the human and murine forms that confirm the collapsed conformation reported previously under different experimental conditions and crystal packing. We also present structures of human and murine WE bound to exosite I with a fragment of the platelet receptor PAR1, which is unable to shift WE to the E form. These structural findings, along with kinetic and calorimetry data, indicate that WE is strongly stabilized in the E* form and explain why binding of ligands to exosite I has only a modest effect on the E*-E equilibrium for this mutant. The E* → E transition requires the combined binding of thrombomodulin and protein C and restores activity of the mutant WE in the anticoagulant pathway.
Thrombin is the pivotal protease of blood coagulation and is endowed with both procoagulant and anticoagulant roles in vivo (1). Thrombin acts as a procoagulant when it converts fibrinogen into an insoluble fibrin clot, activates clotting factors V, VIII, XI, and XIII, and cleaves PAR12 and PAR4 on the surface of human platelets thereby promoting platelet aggregation (2). Upon binding to thrombomodulin, a receptor present on the membrane of endothelial cells, thrombin becomes unable to interact with fibrinogen and PAR1 but increases >1,000-fold its activity toward the zymogen protein C (3). Activated protein C generated from the thrombin-thrombomodulin complex down-regulates both the amplification and progression of the coagulation cascade (3) and acts as a potent cytoprotective agent upon engagement of EPCR and PAR1 (4).
The dual nature of thrombin has long motivated interest in dissociating its procoagulant and anticoagulant activities (5–12). Thrombin mutants with anticoagulant activity help rationalize the bleeding phenotypes of several naturally occurring mutations and could eventually provide new tools for pharmacological intervention (13) by exploiting the natural protein C pathway (3, 14, 15). Previous mutagenesis studies have led to the identification of the E217A and E217K mutations that significantly shift thrombin specificity from fibrinogen toward protein C relative to the wild type (10–12). Both constructs were found to display anticoagulant activity in vivo (10, 12). The subsequent discovery of the role of Trp-215 in controlling the balance between pro- and anti-coagulant activities of thrombin (16) made it possible to construct the double mutant W215A/E217A (WE) featuring >19,000-fold reduced activity toward fibrinogen but only 7-fold loss of activity toward protein C (7). These properties make WE the most potent anticoagulant thrombin mutant engineered to date and a prototype for a new class of anticoagulants (13). In vivo studies have revealed an extraordinary potency, efficacy, and safety profile of WE when compared with direct administration of activated protein C or heparin (17–19). Importantly, WE elicits cytoprotective effects (20) and acts as an antithrombotic by antagonizing the platelet receptor GpIb in its interaction with von Willebrand factor (21).
What is the molecular mechanism underscoring the remarkable functional properties of WE? The mutant features very low activity toward synthetic and physiological substrates, including protein C. However, in the presence of thrombomodulin, protein C is activated efficiently (7). A possible explanation is that WE assumes an inactive conformation when free but is converted into an active form in the presence of thrombomodulin. The ability of WE to switch from inactive to active forms is consistent with recent kinetic (22) and structural (23, 24) evidence of the significant plasticity of the trypsin fold. The active form of the protease, E, coexists with an inactive form, E*, that is distinct from the zymogen conformation (25). Biological activity of the protease depends on the equilibrium distribution of E* and E, which is obviously different for different proteases depending on their physiological role and environmental conditions (25). The E* form features a collapse of the 215–217 β-strand into the active site and a flip of the peptide bond between residues Glu-192 and Gly-193, which disrupts the oxyanion hole. These changes have been documented crystallographically in thrombin and other trypsin-like proteases such as αI-tryptase (26), the high temperature requirement-like protease (27), complement factor D (28), granzyme K (29), hepatocyte growth factor activator (30), prostate kallikrein (31), prostasin (32, 33), complement factor B (34), and the arterivirus protease nsp4 (35). Hence, the questions that arise about the molecular mechanism of WE function are whether the mutant is indeed stabilized in the inactive E* form and whether it can be converted to the active E form upon thrombomodulin binding.
Structural studies of the anticoagulant mutants E217K (36) and WE (37) show a partial collapse of the 215–217 β-strand into the active site that abrogates substrate binding. The collapse is similar to, but less pronounced than, that observed in the structure of the inactive E* form of thrombin where Trp-215 relinquishes its hydrophobic interaction with Phe-227 to engage the catalytic His-57 and residues of the 60-loop after a 10 Å shift in its position (24). These more substantial changes have been observed recently in the structure of the anticoagulant mutant Δ146–149e (38), which has proved that stabilization of E* is indeed a molecular mechanism capable of switching thrombin into an anticoagulant. It would be simple to assume that both E217K and WE, like Δ146–149e, are stabilized in the E* form. However, unlike Δ146–149e, both E217K and WE carry substitutions in the critical 215–217 β-strand that could result into additional functional effects overlapping with or mimicking a perturbation of the E*-E equilibrium. A significant concern is that both structures suffer from crystal packing interactions that may have biased the conformation of side chains and loops near the active site (24). The collapsed structures of E217K and WE may be artifactual unless validated by additional structural studies where crystal packing is substantially different.
To address the second question, kinetic measurements of chromogenic substrate hydrolysis by WE in the presence of saturating amounts of thrombomodulin have been carried out (37), but these show only a modest improvement of the kcat/Km as opposed to >57,000-fold increase observed when protein C is used as a substrate (7, 37). The modest effect of thrombomodulin on the hydrolysis of chromogenic substrates is practically identical to that seen upon binding of hirugen to exosite I (37) and echoes the results obtained with the wild type (39) and other anticoagulant thrombin mutants (7, 9, 10, 12, 38). That argues against the ability of thrombomodulin alone to significantly shift the E*-E equilibrium in favor of the E form. Binding of a fragment of the platelet receptor PAR1 to exosite I in the D102N mutant stabilized in the E* form (24) does trigger the transition to the E form (23), but evidence that a similar long-range effect exists for the E217K or WE mutants has not been presented.
In this study we have addressed the two unresolved questions about the mechanism of action of the anticoagulant thrombin mutant WE. Here we present new structures of the mutant in its human and murine versions, free and bound to a fragment of the thrombin receptor PAR1 at exosite I. The structures are complemented by direct energetic assessment of the binding of ligands to exosite I and its effect on the E*-E equilibrium.
MATERIALS AND METHODS
The human and murine thrombin versions of the mutant WE (hWE and mWE, respectively) were expressed, purified, and validated as described previously (7, 40). The soluble fragment of human PAR1, 42SFLLRNPNDKYEPFWEDEEKN62, corresponding to the sequence downstream from the cleavage site at Arg-41 and containing the hirudin-like motif 52YEPFWE57 for binding exosite I, was synthesized by solid phase, purified to homogeneity by high pressure liquid chromatography (HPLC), and tested for purity by mass spectrometry. hWE was concentrated to 8.0 mg/ml in 50 mm choline chloride, 20 mm Tris, pH 7.4. mWE was concentrated to 9.8 mg/ml in 50 mm choline chloride, 20 mm Tris, pH 7.4. Crystallization was achieved at 22 °C by the vapor diffusion technique, with each crystallization reservoir containing 500 μl of solution. Equal volumes of the protein sample and reservoir solution (1 μl each) were mixed to prepare the hanging drops. The reservoir solutions are provided in Table 1. Diffraction quality crystals grew in 1–3 weeks and were cryoprotected in a solution similar to the reservoir solution but containing 15% glycerol prior to flash freezing. X-ray diffraction data for mWE-3, mWE-PAR1, hWE, and hWE-PAR1 (Table 1) were collected at the Advanced Photon Source (Beamline Biocars 14-BMC, Argonne National Laboratory), and those for mWE-1 and mWE-2 were collected with a home source RAXIS IV detector (Rigaku/MSC, The Woodlands, TX). The data were indexed, integrated, and scaled with the HKL2000 software package (41). The structures were solved by molecular replacement using MOLREP from the CCP4 suite (42) and Protein Data Bank accession code 1TQ0 (37) as a search model. Refinement and electron density generation were performed with the Crystallography and NMR System (43) for mWE-3, mWE-PAR1, hWE, and hWE-PAR1 or with REFMAC (44) for mWE-1 and mWE-2; 5% of the reflections were randomly selected as a test set for cross-validation. Ramachandran plots were calculated using PROCHECK (45). Statistics for data collection and refinement are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession codes: 3HK3 for mWE-1, 3HK6 for mWE-2, 3EDX for mWE-3, 3HKI for mWE-PAR1, 3EE0 for hWE, and 3HKJ for hWE-PAR1).
TABLE 1.
Crystallographic data for the thrombin mutant W215A/E217A (human and murine) free and bound to a fragment of PAR1
| mWE-1 | mWE-2 | mWE-3 | mWE-PAR1 | hWE | hWE-PAR1 | |
|---|---|---|---|---|---|---|
| Buffer/salt | 0.2 m NH4Cl | 0.2 m NH4H2PO4 | 0.2 m Na2SO4 | 100 mm Tris | 100 mm MES | 100 mm HEPES |
| pH | 6.3 | 4.6 | 6.6 | 8.5 | 6.5 | 7.5 |
| Polyethylene glycol | 3,350 (20%) | 3,350 (14%) | 3,350 (20%) | 10,000 (20%) | 400 (30%) | 10,000 (20%) |
| PDBa ID | 3HK3 | 3HK6 | 3EDX | 3HKI | 3EE0 | 3HKJ |
| Data collection | ||||||
| Wavelength (Å) | 1.54 | 1.54 | 0.9 | 0.9 | 0.9 | 0.9 |
| Space group | P212121 | P41212 | P212121 | P21212 | P212121 | P1 |
| Unit cell dimensions (Å) | a = 48.6 | a = 70.4 | a = 97.7 | a = 231.4 | a = 40.1 | a = 53.1, α = 98.9° |
| b = 63.9 | b = 70.4 | b = 105.8 | b = 51.0 | b = 60.0 | b = 61.8, β = 110.3° | |
| c = 95.0 | c = 293.1 | c = 176.8 | c = 80.5 | c = 120.0 | c = 67.8, γ = 92.9° | |
| Molecules/asymmetric unit | 1 | 2 | 3 | 2 | 1 | 2 |
| Resolution range (Å) | 40–1.94 | 40–3.2 | 40–2.4 | 40–2.2 | 40–2.75 | 40–2.6 |
| Observations | 124,234 | 80,314 | 380,536 | 311,467 | 43,454 | 69,365 |
| Unique observations | 21,296 | 12,201 | 67,622 | 45,052 | 7,810 | 22,864 |
| Completeness (%) | 94.9 (87.0) | 93.3 (72.8) | 94.5 (79.9) | 90.9 (80.9) | 97.5 (85.3) | 93.4 (79.3) |
| Rsym (%) | 8.1 (28.0) | 10.5 (37.8) | 5.9 (46.1) | 5.1 (36.5) | 9.0 (32.3) | 5.2 (19.9) |
| I/σ(I) | 17.2 (3.3) | 14.6 (3.5) | 21.2 (2.5) | 28.6 (4.9) | 15.4 (2.9) | 18.5 (4.2) |
| Refinement | ||||||
| Resolution (Å) | 40-1.94 | 40-3.2 | 40-2.4 | 40-2.2 | 40-2.75 | 40-2.6 |
| |F|σ(|F|) | >0 | >0 | >0 | >0 | >0 | >0 |
| Rcryst, Rfree | 0.181, 0.235 | 0.222, 0.314 | 0.214, 0.244 | 0.227, 0.261 | 0.239, 0.323 | 0.195, 0.234 |
| Reflections (working/test) | 20,160/1,098 | 11,572/583 | 62,795/3,371 | 40,411/3,592 | 7,683/418 | 21,050/1,079 |
| Protein atoms | 2,289 | 4,882 | 7,353 | 5,041 | 2,218 | 4,714 |
| Solvent molecules | 240 | 0 | 227 | 217 | 0 | 154 |
| r.m.s.d. bond lengthsb (Å) | 0.013 | 0.019 | 0.007 | 0.011 | 0.008 | 0.007 |
| r.m.s.d. anglesc (°) | 1.4 | 1.9 | 1.3 | 1.8 | 1.3 | 1.5 |
| r.m.s.d. ΔB (Å2) (mm/ms/ss)c | 0.76/0.55/2.17 | 0.74/0.37/1.72 | 3.45/4.18/5.55 | 2.32/2.63/3.22 | 1.57/1.76/2.01 | 1.24/1.19/1.60 |
| 〈B〉 protein (Å2) | 25.6 | 44.9 | 54.6 | 41.2 | 38.4 | 40.6 |
| 〈B〉 solvent (Å2) | 34.8 | 48.9 | 40.4 | 37.7 | ||
| Ramachandran plot | ||||||
| Most favored (%) | 98.8 | 97.1 | 97.4 | 98.5 | 99.2 | 99.0 |
| Generously allowed (%) | 1.2 | 2.4 | 1.8 | 1.3 | 0.9 | 0.2 |
| Disallowed (%) | 0.0 | 0.5 | 0.8 | 0.2 | 0.0 | 0.8 |
a PDB, Protein Data Bank.
b Root-mean-squared deviation (r.m.s.d.) from ideal bond lengths and angles and in B-factors of bonded atoms.
c mm, main chain-main chain; ms, main chain-side chain; ss, side chain-side chain.
Binding of the exosite I ligand hirugen was studied directly by isothermal titration calorimetry under experimental conditions of 5 mm Tris, 0.1% polyethylene glycol 8000, and 200 mm NaCl, pH 8.0 at 25 °C using an iTC200 calorimeter (MicroCal Inc., Northampton, MA) with the sample cell containing thrombin and the syringe injecting hirugen. Details of the experimental procedures have been presented elsewhere (38). Thermodynamic parameters were extracted from a curve fit to the data using software (Origin 7.0) provided by MicroCal that was consistent with a one-site binding model. Experiments were performed in triplicate with excellent reproducibility (<10% variation in thermodynamic parameters). The effect of ligand binding to exosite I on the E*-E equilibrium can be understood in terms of the properties of the linkage scheme (Scheme 1).
The scheme involves the four possible intermediates for an enzyme existing in two states, E* and E, binding an allosteric effector, L, such as hirugen or thrombomodulin with an equilibrium association constant, K* or K. The equilibrium distribution of E* and E is reflected by the constant r = [E*]/[E] in the absence of L and rL = [E*L]/[EL] in the presence of L. Conservation of energy in Scheme 1 requires (46, 47)
 |
which implies that the original distribution of E* and E forms quantified by the equilibrium constant r can only be affected to an extent r/rL equal to the ratio K/K*. If L binds to E with an affinity x-fold higher than that of E*, then the relative population of E* relative to E will decrease x-fold in the presence of L. In other words, the transition of E* to E is never complete upon binding of an allosteric effector (unless K/K* → ∞) but obeys in magnitude the constraints imposed by Equation 1.
SCHEME 1.

RESULTS
Six crystal structures of hWE and mWE were obtained, four in the absence of any ligands and two bound to the extracellular fragment of human PAR1. The six models vary in number of molecules per asymmetric unit, space group, and crystallization conditions including pH (Table 1), as well as in a number of critical residues that confer murine thrombin high catalytic activity in the absence of Na+ activation (40). Yet the structures present a common active site architecture differing significantly from the wild type in the E form (48). Each of the 11 hWE or mWE protein monomers shows collapse of the 215–217 β-strand into the active site, and the majority show disruption of the oxyanion hole (Fig. 1). Binding of PAR1 to exosite I does not correct the collapsed conformation.
FIGURE 1.
Cα traces of engineered proteases hWE and mWE compared with native thrombin. Left, new hWE structure with only one molecule in the asymmetric unit (wheat) is nearly identical to the previous structure, 1TQ0 (light green), and differs from the active E form (1SGT (red)) (48) for the collapse of the 215–217 β-strand (arrow) into the active site. Right, mWE-1 (light blue), mWE-2 (wheat), and mWE-3 (light green) differ from wild-type murine thrombin (2OCV (red)) (51) at both the 215–217 β-strand (arrow) and the oxyanion hole.
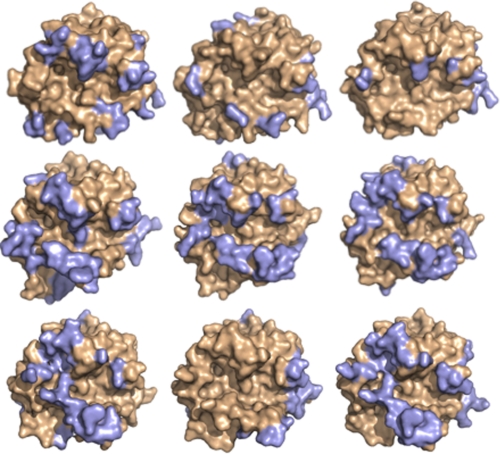
Only one molecule is present in the asymmetric unit of the new structure of hWE, unlike the previous structure, 1TQ0 (37). The P212121 space group differs from the previous P213 lattice and has smaller cell dimensions. In the new hWE structure, the 215–217 β-strand collapses into the S1 pocket (Fig. 1) and is accompanied by complete disorder of the 186- and 220-loops responsible for Na+ binding. Although the electron density does not enable definition of the autolysis loop and the 220-loop in hWE, the conformational state of what can be observed suggests solvent exposure of the Na+ binding site and increased solvent accessibility of the normally buried 187RGD189 sequence. Such a conformation bears similarity to the K+-bound form of thrombin (49, 50) as well as to the structure of the thrombin mutant E217K (36). The new structure confirms the collapsed conformation under different crystallization conditions, space groups, and crystal packing (Fig. 2) lending support to the validity of the three-dimensional architecture of the mutant WE first observed in the structure 1TQ0 (37).
FIGURE 2.
Surface representation of the nine free monomers of hWE and mWE (wheat). The wide variety of crystal contacts <4 Å (light blue) documented in the data sets proves that the collapse of the 215–217 β-strand (arrow) is not an artifact of crystal packing. Top row, hWE, 1TQ0 molecule A, 1TQ0 molecule B. Middle row, mWE-1, mWE-2 molecule A, mWE-2 molecule B. Bottom row, mWE-3 molecules A, B, and C.
Crystals of mWE in the free form were obtained with one, two, and three molecules in the asymmetric unit (mWE-1, mWE-2, and mWE-3). As in the hWE, the 215–217 β-strand collapses into the active site (Fig. 1) although the structures were solved under different crystallization conditions, space groups, and crystal packing (Fig. 2). The oxyanion hole is disrupted in all monomers because of a flip of the Glu-192—Gly-193 peptide bond (Fig. 3). The ψ angle of Glu-192 decreases by 75°, and the ϕ angle of Gly-193 decreases by 180°. In this altered conformation, the carbonyl O atom of Glu-192 is placed within H-bonding distance to both the OH and backbone N atoms of the catalytic Ser-195, resulting in an architecture of the triad and oxyanion hole that is incompatible with stabilization of the catalytic transition state (Fig. 3). In turn, the stretch of residues from Glu-192 to Asp-194 assumes a short helical geometry. Overall, the conformational change is less drastic than that observed in the E* form documented in the D102N (24) and Δ146–149e (38) mutants of human thrombin, although it goes in the same direction.
FIGURE 3.
Comparison of the oxyanion hole in hWE and mWE in the presence and absence of PAR1. Left, PAR1 binding in hWE (light green) induces rotation of the Glu-192—Gly-193 peptide bond such that the backbone O atom is in H-bonding distance of both the OH and backbone N atom of the catalytic Ser-195. Right, in all mWE structures the Glu-192—Gly-193 peptide bond is flipped whether free (wheat) or bound to the PAR1 peptide (light green).
All six monomers of mWE are highly similar to one another throughout their entire structures with the exception of the surface exposed 145-, 186-, and 220-loops. All monomers show the 186-loop flipped toward the active site with only slight variations, which exposes the Na+ binding site to solvent. Molecule B of hWE-3 is unique in presenting a more extended 145-loop because of packing between adjacent molecules (Fig. 2). Unlike the hWE crystal structure and 1TQ0, the Na+-binding loops are well defined due in part to the recruitment of Arg-187 as an intramolecular cation mimic and favorable electrostatics due to the presence of the critical Lys-222 in the murine enzyme. In wild-type murine thrombin, and unlike human thrombin, Lys-222 buries into the Na+ binding site and effectively mimics the Na+ activation mechanism (40, 51). In all mWE structures, Lys-222 is dislodged from its position between the 186- and 220- loops and rotates upward to become stabilized by Glu-146 from the autolysis loop, albeit in slightly different orientations in each monomer. In all three monomers of mWE-3, Arg-187 substitutes into the Na+ site and recruits polar contacts from the backbone O atoms of Phe-184a, Lys-185, and Lys-224. Arg-187 assumes a similar buried conformation in the D102N mutant of thrombin (23, 24). In molecule B of mWE-3, the backbone O atom of Lys-222 is also recruited into the ligand shell of Arg-187. In contrast, the other three monomers from mWE-1 and mWE-2 present Arg-187 in an orientation more similar to wild-type murine thrombin, leaving the Na+ binding site exposed to solvent. All three structures of mWE support the conclusion that the disrupted active site architecture is the dominant conformation in solution that is generated by the double Ala substitution at residues 215 and 217.
To test whether the structural defects of the WE conformation are corrected upon ligand binding to exosite I, as documented recently for the E* form (23), both hWE and mWE were crystallized in complex with an extracellular fragment of PAR1. In the hWE-PAR1 complex, eight residues of the peptide are visible bound to molecule A, whereas seven residues are visible bound to molecule B (Fig. 4). In the mWE-PAR1 complex, the peptide bound to molecule A is defined as being over 15 residues in length, whereas in the peptide bound to molecule B only 9 residues are visible in the electron density (Fig. 4). In both structures the PAR1 fragment engages exosite I of thrombin through a number of polar and hydrophobic interactions. Residue Tyr-52 and Phe-55 of PAR1 interlock with Phe-34 of thrombin, and this complementary hydrophobic packing is assisted by a cation-π interaction between Arg-67 in thrombin and the aromatic ring of Phe-55 of PAR1. Additional H-bond contacts link Asp-50, Tyr-52, and Thr-54 of PAR1 to Leu-40, Arg-73, and Tyr-76 of thrombin. Lack of distinct contacts in the C-terminal portion of PAR1 is unlike that found in hirudin (52) or PAR3 (53) binding, which present additional C-terminal acidic moieties to extend the reach of these peptides farther back into exosite I. The paucity of contacts by the C-terminal residues of PAR1 forces the peptide to adopt two different binding poses to pack Trp-56 against the surface of thrombin. These peptide conformations are similar to that observed in the previously determined complex with the D102N mutant of thrombin (23). However, PAR1 binding to hWE or mWE restores neither the collapse of the 215–217 β-strand (Fig. 4) nor the disruption of the oxyanion hole (Fig. 3). The PAR1-bound structures are nearly identical to those in the free state.
FIGURE 4.
PAR1 binding to hWE and mWE does not restore active site architecture. Left, hWE free (wheat) and hWE bound (light green) to the PAR1 peptide (gold) are nearly identical with the exception of the oxyanion hole (see legend for Fig. 3). Right, mWE free (wheat) is nearly identical to the PAR1 (gold) bound state (light green).
The integrity of exosite I in the WE mutant is supported by calorimetric measurements of the binding of hirugen (Fig. 5). Hirugen binding to exosite I in the wild type elicits changes in activity toward chromogenic substrates that are practically identical to those elicited by thrombomodulin (39). Calorimetry provides a direct and highly informative method of studying binding interactions that, in the case of ligands like hirugen, have thus far relied on indirect kinetic assays (37, 39, 54) or fluorescence probes added to the ligand or the enzyme (55). The binding of hirugen to the wild type is characterized by an equilibrium association constant of K = 7.4 106 m−1 and an enthalpy of −14.5 kcal/mol, underscoring a significant entropy loss in the interaction. The binding of hirugen to the mutant WE features an equilibrium association constant K = 2.1 106 m−1, which is <4-fold lower than that of the wild type. The enthalpy change associated with binding is −14.0 kcal/mol, practically identical to that of the wild type. These results support the conclusion that exosite I is affected modestly by the WE mutation or the collapse in the 215–217 β-strand. It follows from linkage Scheme 1 that binding of ligands to exosite I of the mutant WE should have only a modest effect on the E*-E equilibrium. If the mutant WE is stabilized in the E* form, binding of ligands such as thrombomodulin and hirugen to exosite I are expected to have only a modest effect on the catalytic activity of the enzyme toward small chromogenic substrates, as indeed was found when measured experimentally (37).
FIGURE 5.
Hirugen binding to thrombin wild type (left) and WE (right) measured by isothermal titration calorimetry. The top panel shows the heat exchanged in each individual titration for the thrombin sample (lower trace) and the hirugen buffer control (upper trace, left). The bottom panel is the integration of the data to yield the overall heat exchanged as a function of the ligand/protein molar ratio. Experimental conditions are: 5 mm Tris, 0.1% polyethylene glycol 8000, 200 mm NaCl, pH 8.0, 25 °C. The enzyme and hirugen concentrations are: 10 and 105 μm (thrombin wild type); 12 and 220 μm (WE). Titration curves were fit using the Origin software of the iTC200, with best-fit parameter values: thrombin wild type, K = 7.4 ± 0.1 106 m−1, ΔG = −9.4 ± 0.1 kcal/mol, ΔH = −14.5 ± 0.1 kcal/mol, TΔS = −5.1 ± 0.1 kcal/mol; WE, K = 2.1 ± 0.5 106 m−1, ΔG = −8.6 ± 0.1 kcal/mol, ΔH = −14.0 ± 0.1 kcal/mol, TΔS = −5.4 ± 0.1 kcal/mol. The value of the stoichiometric constant, N, was 0.97 ± 0.01 for thrombin wild type and 1.02 ± 0.01 for the WE mutant.
DISCUSSION
The anticoagulant thrombin mutant WE has little activity toward substrate but features significant activity toward the anticoagulant protein C in the presence of the cofactor thrombomodulin (7). Because of its remarkable success in preclinical studies (17–19, 21), elucidation of the molecular mechanism underlying the function of WE is necessary to eventually remove any residual activity toward fibrinogen and PAR1 in future thrombin variants. A new paradigm, which emerged from the analysis of recent crystal structures of trypsin-like proteases (23, 24, 26–35), offers a context in which to interpret the peculiar properties of WE and other anticoagulant thrombin mutants (9, 10, 12, 38). Trypsin-like proteases such as thrombin exists in equilibrium between two forms, E* (inactive) and E (active) (25). The biological activity of the protease is determined by the exact partitioning between the inactive E* and active E forms. The allosteric equilibrium may be shifted using ad hoc cofactors or small molecules as well as site-directed mutations. The mutant Δ146–149e carrying a deletion of nine residues in the autolysis loop of thrombin has been shown recently to adopt the E* conformation (38), and its anticoagulant profile has been explained in terms of stabilization of E* until binding of protein C and thrombomodulin restore activity by triggering the conversion to the E form. The mutant WE features a collapse of the 215–217 β-strand and disruption of the oxyanion hole, which resembles the conformation of E* (24). This collapse is not artifactual, as it is observed in structures of the human and murine enzymes obtained under different experimental conditions, space groups, and crystal packing (Fig. 2) lending support to the validity of the conformation first observed in the structure 1TQ0 (37). It is likely that the collapsed conformation of WE is indeed the E* form biased by the effect of removing the indole of Trp-215. The side chain of Trp-215 is critical in maintaining the open conformation of the active site in the E form through a strong hydrophobic interaction with Phe-227 (48). Likewise, in the E* form, the hydrophobic interaction of Trp-215 with His-57 and Tyr-60a in the 60-loop produces the collapsed conformation that obliterates substrate access to the active site (24). When the side chain of Trp-215 is eliminated with the W215A substitution, the 215–217 β-strand loses a key structural element to stabilize either E or E*. Hence, it is not surprising that the conformation of the 215–217 β-strand in the WE mutant occupies a position that is intermediate to the two limiting positions in the E and E* forms. The collapse is sufficient to compromise substrate binding to the active site even though it does not move the 215–217 β-strand fully to the position seen in the E* form.
The crystal structures of human and murine thrombin mutant WE bound to a fragment of PAR1 in exosite I show no conversion to the E form and no change in the collapsed 215–217 β-strand as observed in the free form. These observations seem to contradict recent crystallographic evidence that the D102N mutant assumes the E* form when free and the E form when bound to a fragment of PAR1 at exosite I (23). However, attention to the principles embodied by scheme 1 shows that the results are not in contradiction. Linkage Scheme 1 establishes that an allosteric effector can only shift the E*-E equilibrium in favor of E by an amount equal to the ratio of affinities of the two forms. Because exosite I, unlike the active site, is similarly accessible in the E* and E forms (1, 23, 24, 48), binding of thrombomodulin, hirugen, or fragments of PAR1 to the two forms is unlikely to result in extreme perturbations of the E*-E equilibrium. In fact, binding of hirugen to exosite I in the WE mutant takes place with an affinity that is <4-fold lower than that of the wild type, confirming that the collapsed conformation of the mutant that compromises access to the active site has little effect on the architecture of exosite I. The available structures of D102N do not imply an all-or-none distribution of E* and E in solution. Kinetic studies in which the E*-E equilibrium distribution can be measured directly (22) prove that r = 1.1 for D102N free in solution (24). Therefore, it would have been equally likely for this mutant to crystallize in the E form when free, but E* predominated under the crystallization conditions explored thus far (23, 24). If binding to exosite I takes place with 4-fold higher affinity in the E form, as suggested by the hirugen binding data reported in this study (Fig. 5), then a value of rL = 0.28 is obtained for the mutant D102N bound to exosite I (see Equation 1). Under these conditions, the fraction of D102N in the active E form would be 78% of the total. It is not surprising, then that the structure of D102N bound to a fragment of PAR1 to exosite I has revealed the active E form (23). However, future crystallization studies may trap the D102N-PAR1 complex with the enzyme in the E* form because of the significant fraction (22%) of this inactive conformation still present in solution. The value of r for the WE mutant cannot be estimated directly from kinetic studies because Trp-215 is a major fluorophore reporting the E*-E interconversion (22). However, a lower boundary of r = 200 can be inferred from the functional properties of the mutant Δ146–149e that crystallizes in the E* form when free (38) and is a less potent anticoagulant compared with WE. Binding of ligands to exosite I of WE should give a value of rL = 50 (see Equation 1), which translates into 2% of the enzyme in the E form when bound. Under these conditions, it would be very difficult to trap the WE-PAR1 complex with the enzyme in the E form, and the E* form documented in the present study reflects the conformation of 98% of the molecules in solution.
The foregoing argument also explains why thrombomodulin and hirugen have little effect on the hydrolysis of chromogenic substrates by the mutants WE (37) and Δ146–149e (38) as well as why physiological substrates like fibrinogen and PAR1 are not cleaved efficiently by the mutants WE and Δ146–149e (7, 38). Assuming that these substrates bind to exosite I before contacting the active site, the interaction with exosite I would be insufficient to energetically move the E*-E equilibrium completely in favor of the active E form. On the other hand, when protein C and thrombomodulin are present in solution, then most of the activity of the mutant WE is restored. The thrombomodulin-protein C complex acting as a substrate may have a more profound effect on E*-E equilibrium by accessing additional regions of the thrombin surface beyond exosite I (56). In that case, the change in affinity between the E* and E forms shown in Scheme 1 may be substantial and could ensure significant repopulation of the E form that is not possible with either protein C or thrombomodulin alone.
We conclude that the mutant WE is stabilized in the E* form, but the complete collapse of the 215–217 β-strand is impeded by the absence of the side chain of Trp-215. Binding to exosite I fails to shift substantially the E*-E equilibrium in favor of the E form unless protein C and thrombomodulin act in combination. The result of this mechanism is a mutant that acts efficiently in the anticoagulant pathway and retains minimal activity toward the procoagulant substrate fibrinogen and the prothrombotic substrate, PAR1. The intriguing functional properties of the mutant WE (20, 21), its well established potency as an anticoagulant in non-human primates (17, 19), and the current elucidation of the role of E* in switching thrombin into an anticoagulant (38) should make it possible to engineer a thrombin mutant with exclusive activity toward protein C for therapeutic applications.
This work was supported, in whole or in part, by National Institutes of Health Research Grants HL49413, HL58141, and HL73813 (to E. D. C.).
- PAR
- protease-activated receptor
- WE
- W215A/E217A
- hWE
- human thrombin version of WE
- mWE
- murine thrombin version of WE.
REFERENCES
- 1.Di Cera E. (2008) Mol. Aspects Med. 29, 203–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davie E. W., Kulman J. D. (2006) Semin. Thromb. Hemost. 32, Suppl. 1, 3–15 [DOI] [PubMed] [Google Scholar]
- 3.Esmon C. T. (2003) Chest 124, 26S–32S [DOI] [PubMed] [Google Scholar]
- 4.Riewald M., Petrovan R. J., Donner A., Mueller B. M., Ruf W. (2002) Science 296, 1880–1882 [DOI] [PubMed] [Google Scholar]
- 5.Le Bonniec B. F., Esmon C. T. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7371–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Q. Y., Sheehan J. P., Tsiang M., Lentz S. R., Birktoft J. J., Sadler J. E. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6775–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantwell A. M., Di Cera E. (2000) J. Biol. Chem. 275, 39827–39830 [DOI] [PubMed] [Google Scholar]
- 8.Dang Q. D., Guinto E. R., Di Cera E. (1997) Nat. Biotechnol. 15, 146–149 [DOI] [PubMed] [Google Scholar]
- 9.Dang Q. D., Sabetta M., Di Cera E. (1997) J. Biol. Chem. 272, 19649–19651 [DOI] [PubMed] [Google Scholar]
- 10.Gibbs C. S., Coutré S. E., Tsiang M., Li W. X., Jain A. K., Dunn K. E., Law V. S., Mao C. T., Matsumura S. Y., Mejza S. J., Paborsky L. R., Leung L. L. (1995) Nature 378, 413–416 [DOI] [PubMed] [Google Scholar]
- 11.Tsiang M., Jain A. K., Dunn K. E., Rojas M. E., Leung L. L., Gibbs C. S. (1995) J. Biol. Chem. 270, 16854–16863 [DOI] [PubMed] [Google Scholar]
- 12.Tsiang M., Paborsky L. R., Li W. X., Jain A. K., Mao C. T., Dunn K. E., Lee D. W., Matsumura S. Y., Matteucci M. D., Coutré S. E., Leung L. L., Gibbs C. S. (1996) Biochemistry 35, 16449–16457 [DOI] [PubMed] [Google Scholar]
- 13.Bates S. M., Weitz J. I. (2006) Br. J. Haematol. 134, 3–19 [DOI] [PubMed] [Google Scholar]
- 14.Di Cera E. (2007) J. Thromb. Haemost. 5, 196–202 [DOI] [PubMed] [Google Scholar]
- 15.Griffin J. H. (1995) Nature 378, 337–338 [DOI] [PubMed] [Google Scholar]
- 16.Arosio D., Ayala Y. M., Di Cera E. (2000) Biochemistry 39, 8095–8101 [DOI] [PubMed] [Google Scholar]
- 17.Gruber A., Cantwell A. M., Di Cera E., Hanson S. R. (2002) J. Biol. Chem. 277, 27581–27584 [DOI] [PubMed] [Google Scholar]
- 18.Gruber A., Fernández J. A., Bush L., Marzec U., Griffin J. H., Hanson S. R., Di Cera E. (2006) J. Thromb. Haemost. 4, 392–397 [DOI] [PubMed] [Google Scholar]
- 19.Gruber A., Marzec U. M., Bush L., Di Cera E., Fernández J. A., Berny M. A., Tucker E. I., McCarty O. J., Griffin J. H., Hanson S. R. (2007) Blood 109, 3733–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feistritzer C., Schuepbach R. A., Mosnier L. O., Bush L. A., Di Cera E., Griffin J. H., Riewald M. (2006) J. Biol. Chem. 281, 20077–20084 [DOI] [PubMed] [Google Scholar]
- 21.Berny M. A., White T. C., Tucker E. I., Bush-Pelc L. A., Di Cera E., Gruber A., McCarty O. J. (2008) Arterioscler. Thromb. Vasc. Biol. 18, 329–334 [DOI] [PubMed] [Google Scholar]
- 22.Bah A., Garvey L. C., Ge J., Di Cera E. (2006) J. Biol. Chem. 281, 40049–40056 [DOI] [PubMed] [Google Scholar]
- 23.Gandhi P. S., Chen Z., Mathews F. S., Di Cera E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pineda A. O., Chen Z. W., Bah A., Garvey L. C., Mathews F. S., Di Cera E. (2006) J. Biol. Chem. 281, 32922–32928 [DOI] [PubMed] [Google Scholar]
- 25.Di Cera E. (2009) IUBMB Life 61, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohr K. B., Selwood T., Marquardt U., Huber R., Schechter N. M., Bode W., Than M. E. (2006) J. Mol. Biol. 357, 195–209 [DOI] [PubMed] [Google Scholar]
- 27.Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. (2002) Nature 416, 455–459 [DOI] [PubMed] [Google Scholar]
- 28.Jing H., Babu Y. S., Moore D., Kilpatrick J. M., Liu X. Y., Volanakis J. E., Narayana S. V. (1998) J. Mol. Biol. 282, 1061–1081 [DOI] [PubMed] [Google Scholar]
- 29.Hink-Schauer C., Estébanez-Perpiñá E., Wilharm E., Fuentes-Prior P., Klinkert W., Bode W., Jenne D. E. (2002) J. Biol. Chem. 277, 50923–50933 [DOI] [PubMed] [Google Scholar]
- 30.Shia S., Stamos J., Kirchhofer D., Fan B., Wu J., Corpuz R. T., Santell L., Lazarus R. A., Eigenbrot C. (2005) J. Mol. Biol. 346, 1335–1349 [DOI] [PubMed] [Google Scholar]
- 31.Carvalho A. L., Sanz L., Barettino D., Romero A., Calvete J. J., Romão M. J. (2002) J. Mol. Biol. 322, 325–337 [DOI] [PubMed] [Google Scholar]
- 32.Rickert K. W., Kelley P., Byrne N. J., Diehl R. E., Hall D. L., Montalvo A. M., Reid J. C., Shipman J. M., Thomas B. W., Munshi S. K., Darke P. L., Su H. P. (2008) J. Biol. Chem. 283, 34864–34872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spraggon G., Hornsby M., Shipway A., Tully D. C., Bursulaya B., Danahay H., Harris J. L., Lesley S. A. (2009) Protein Sci. 18, 1081–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponnuraj K., Xu Y., Macon K., Moore D., Volanakis J. E., Narayana S. V. (2004) Mol. Cell 14, 17–28 [DOI] [PubMed] [Google Scholar]
- 35.Barrette-Ng I. H., Ng K. K., Mark B. L., Van Aken D., Cherney M. M., Garen C., Kolodenko Y., Gorbalenya A. E., Snijder E. J., James M. N. (2002) J. Biol. Chem. 277, 39960–39966 [DOI] [PubMed] [Google Scholar]
- 36.Carter W. J., Myles T., Gibbs C. S., Leung L. L., Huntington J. A. (2004) J. Biol. Chem. 279, 26387–26394 [DOI] [PubMed] [Google Scholar]
- 37.Pineda A. O., Chen Z. W., Caccia S., Cantwell A. M., Savvides S. N., Waksman G., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 39824–39828 [DOI] [PubMed] [Google Scholar]
- 38.Bah A., Carrell C. J., Chen Z., Gandhi P. S., Di Cera E. (2009) J. Biol. Chem. 284, 20034–20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vindigni A., White C. E., Komives E. A., Di Cera E. (1997) Biochemistry 36, 6674–6681 [DOI] [PubMed] [Google Scholar]
- 40.Bush L. A., Nelson R. W., Di Cera E. (2006) J. Biol. Chem. 281, 7183–7188 [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 42.Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 43.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 45.Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 46.Di Cera E. (1995) Thermodynamic Theory of Site-specific Binding Processes in Biological Macromolecules, Cambridge University Press, Cambridge, UK [Google Scholar]
- 47.Wyman J., Gill S. J. (1990) Binding and Linkage, University Science Books, Mill Valley, CA [Google Scholar]
- 48.Pineda A. O., Carrell C. J., Bush L. A., Prasad S., Caccia S., Chen Z. W., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 31842–31853 [DOI] [PubMed] [Google Scholar]
- 49.Carrell C. J., Bush L. A., Mathews F. S., Di Cera E. (2006) Biophys. Chem. 121, 177–184 [DOI] [PubMed] [Google Scholar]
- 50.Papaconstantinou M. E., Carrell C. J., Pineda A. O., Bobofchak K. M., Mathews F. S., Flordellis C. S., Maragoudakis M. E., Tsopanoglou N. E., Di Cera E. (2005) J. Biol. Chem. 280, 29393–29396 [DOI] [PubMed] [Google Scholar]
- 51.Marino F., Chen Z. W., Ergenekan C., Bush-Pelc L. A., Mathews F. S., Di Cera E. (2007) J. Biol. Chem. 282, 16355–16361 [DOI] [PubMed] [Google Scholar]
- 52.Rydel T. J., Tulinsky A., Bode W., Huber R. (1991) J. Mol. Biol. 221, 583–601 [DOI] [PubMed] [Google Scholar]
- 53.Bah A., Chen Z., Bush-Pelc L. A., Mathews F. S., Di Cera E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11603–11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayala Y., Di Cera E. (1994) J. Mol. Biol. 235, 733–746 [DOI] [PubMed] [Google Scholar]
- 55.Kroh H. K., Tans G., Nicolaes G. A., Rosing J., Bock P. E. (2007) J. Biol. Chem. 282, 16095–16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu H., Bush L. A., Pineda A. O., Caccia S., Di Cera E. (2005) J. Biol. Chem. 280, 7956–7961 [DOI] [PubMed] [Google Scholar]