Abstract
IgY is the principal serum antibody in birds and reptiles, and an IgY-like molecule was the evolutionary precursor of both mammalian IgG and IgE. A receptor for IgY on chicken monocytes, chicken leukocyte receptor AB1 (CHIR-AB1), lies in the avian leukocyte receptor cluster rather than the classical Fc receptor cluster where the genes for mammalian IgE and IgG receptors are found. IgG and IgE receptors bind to the lower hinge region of their respective antibodies with 1:1 stoichiometry, whereas the myeloid receptor for IgA, FcαRI, and the IgG homeostasis receptor, FcRn, which are found in the mammalian leukocyte receptor cluster, bind with 2:1 stoichiometry between the heavy chain constant domains 2 and 3 of each heavy chain. In this paper, the extracellular domain of CHIR-AB1 was expressed in a soluble form and shown to be a monomer that binds to IgY-Fc with 2:1 stoichiometry. The two binding sites have similar affinities: Ka1 = 7.22 ± 0.22 × 105 m−1 and Ka2 = 3.63 ± 1.03 × 106 m−1 (comparable with the values reported for IgA binding to its receptor). The affinity constants for IgY and IgY-Fc binding to immobilized CHIR-AB1 are 9.07 ± 0.07 × 107 and 6.11 ± 0.02 × 108 m−1, respectively, in agreement with values obtained for IgY binding to chicken monocyte cells and comparable with reported values for human IgA binding to neutrophils. Although the binding site for CHIR-AB1 on IgY is not known, the data reported here with a monomeric receptor binding to IgY at two sites with low affinity suggest an IgA-like interaction.
Fc receptors link the specificity of the adaptive immune system with the effector mechanisms of innate immune cells. In birds and reptiles, IgY is the principal serum antibody, and both mammalian IgG and IgE have evolved from an IgY-like ancestor, so studies of IgY offer insights into their origins (1). The historical contribution of chicken immunology to a wider understanding of the subject has been considerable (2), and recently several chicken IgY-Fc receptors have been identified. In this paper, the chicken antibody, IgY, is shown to bind to a chicken leukocyte receptor, CHIR-AB1,4 in a different manner from that of its mammalian orthologues, IgG and IgE, to their respective Fc receptors.
Phagocytosis, mediated in mammals by IgG, and passive cutaneous anaphylaxis, mediated by both IgG and IgE in mammals, have been observed in chickens (3, 4), presumably both effected by IgY. In vitro, IgY binds to monocyte cell lines (5, 6), and an IgY receptor (CHIR-AB1) has been identified that is able to mediate the influx of calcium into cells (5).
The genes for the mammalian high affinity IgE receptor, and several IgG receptors, are located in the classical Fc receptor cluster, whereas in chickens, this cluster is represented by a single gene, the product of which has been expressed and found not to bind IgY (7). Intriguingly, the first IgY leukocyte receptor, CHIR-AB1, was found to be a member of the chicken leukocyte receptor cluster (LRC) (5), adjacent to over 100 genes with high intersequence homology (8). This finding, together with phylogenetic analysis of the orthologous Fc receptor gene clusters (7, 9), implies that during the evolution of the IgY-like ancestor of both IgG and IgE, antibody-Fc binding function migrated from proteins expressed in the LRC to those in the classical Fc receptor cluster. The human LRC is the site of FcαRI, the leukocyte receptor for IgA (an antibody involved in mucosal immunity), the fetal IgG receptor (FcRn, involved in adult IgG homeostasis), and also a number of natural killer cell receptors including the HLA-G ligand, KIR2DL4 (10). A further leukocyte receptor for chicken IgY, also related to LRC receptors, was identified recently, on chromosome 20 (11), and remains to be characterized.
Typically, the stoichiometry of the receptor-antibody complex differs for receptors located in the classical Fc receptor cluster and the LRC. Crystal structures of IgG complexes with FcγRIII and of IgE with FcϵRI show 1:1 receptor:antibody stoichiometry, with the receptor binding across both heavy chains in the lower hinge (12). In contrast, the crystal structure of FcαRI complexed with IgA shows 2:1 stoichiometry (13) as does that of FcRn with IgG (14), with the two receptors binding between the heavy chain constant domains 2 and 3 on each heavy chain. The IgY/receptor interaction could have either stoichiometry; on the one hand, IgY is an orthologue of IgG and IgE, which can both show 1:1 stoichiometry, but on the other hand, the location of the IgY receptor, CHIR-AB1, in the same gene cluster as the IgA and FcRn receptors suggests the possibility of a 2:1 stoichiometry. Consistent with either of these binding modes, the crystal structure of IgY-Fc reveals that many of the residues located in the receptor-binding sites in human IgE, IgG, and IgA are present and accessible in IgY (15).
The single extracellular domain of the chicken leukocyte IgY receptor, CHIR-AB1, has been expressed in insect cells by Arnon et al. (16), who showed that this preparation consists of a mixture of soluble monomer and dimer. Because of the heterogeneity of the protein, it was not possible to ascertain whether the observed 2:1 stoichiometry of receptor binding to antibody involved two monomers or a single dimer binding to IgY. Thus, it was not possible to answer the question of whether the antibody-receptor complex most resembles that of human IgA or of IgG and IgE. We have expressed the extracellular domain of CHIR-AB1 in human HEK cells. It is a monomer, and we report here that it binds to IgY and IgY-Fc with 2:1 stoichiometry.
EXPERIMENTAL PROCEDURES
Cloning of smCHIR-AB1
Soluble monomeric CHIR-AB1 (smCHIR-AB1) was prepared by changing the sequence of the soluble dimeric CHIR-AB1-IgGFc fusion construct (here named sfpCHIR-AB1) used by Viertlboeck et al. (5) to introduce a TEV protease cleavage site flanked by stuffer sequences (17) at the C terminus of the CHIR-AB1 extracellular domain. The following sequence includes codons for the last two amino acids of the soluble extracellular domain of CHIR-AB1 and the first two amino acids of IgG-Fc: AGCCACGAATTCTATGATATTCCAACTACTGCTAGCGAGAATTTGTATTTTCAGGGTGAGCTCAAAACCGGATCCGCCGAGCCC.
The mutated fusion construct was cloned into pCEP4 (Invitrogen) and expressed from HEK293E cells (18). The supernatant was bound to a protein A-Sepharose column (GE Healthcare) and eluted using 0.1 m glycine, pH 2.5, into 2 m Tris, pH 8.6, to effect neutralization. It was dialyzed into 50 mm Tris, pH 8.0, 0.5 mm EDTA, 50 mm sodium chloride and cleaved using a His6-tagged TEV protease (a gift from Dr. M. Conte in this laboratory).
Cleaved IgG-Fc was removed from the preparation by a second passage through the protein A column, TEV protease was removed by passage through a His-Trap HP column (GE Healthcare), and the smCHIR-AB1 was further purified on a Superdex 200 column (GE Healthcare).
Preparation and Characterization of Proteins
Fcυ2–4 was prepared as described previously (6). sfpCHIR-AB1 (5) was expressed in HEK293E cells and purified on a protein A-Sepharose column. Size exclusion chromatography was performed on a Gilson HPLC system using a Superdex 200 column. Serum IgY (sIgY; Stratech Scientific) was purified by HPLC.
The concentration of protein solutions was determined by measuring their absorbance at 280 nm using extinction coefficients calculated from the amino acid sequence using ProtParam (19).
The percentage of carbohydrate in serum IgY, Fcυ2–4 and smCHIR-AB1 was estimated using a kit from Pierce (23260). The measurement was performed twice for smCHIR-AB1.
Deglycosylation under Denaturing Conditions
Purified serum IgY and smCHIR-AB1 were deglycosylated using PNGase F (New England Biolabs, P0704S), as per the manufacturer's instructions. The samples were assayed for removal of carbohydrate by SDS-PAGE using the method of Laemmli (20) and stained with Coomassie Brilliant Blue R250 (British Drug House) for protein and periodic acid-Schiff for carbohydrate (Pierce).
Sedimentation Equilibrium
Sedimentation equilibrium experiments were performed on smCHIR-AB1, Fcυ2–4, and mixtures of the two using a Beckman Optima XL-A analytical ultracentrifuge as described previously (21). The samples were dialyzed into 0.125 m sodium chloride, 50 mm Tris, pH 7.4, 0.05% sodium azide (TBSa), and the data were acquired as an average of 25 absorbance measurements at a wavelength of 280 nm and a radial spacing of 0.001 cm. Sedimentation equilibrium experiments were performed at 4 °C and rotor speeds of 19,000, 16,000, and 13000 rpm for smCHIR-AB1 alone and 8,500, 10,000, and 12000 rpm for both Fcυ2–4 alone and Fcυ2–4 mixed with smCHIR-AB1. For the interaction study, the two proteins were mixed in 1:1, 2:1, and 1:2 molar ratios, with the overall protein loading concentrations corresponding to a measured A280 of ∼0.6, and preincubated for 1 h at 25 °C. The base lines were measured following overspeed runs at 42,000 rpm for 5 h, which resulted in meniscus depletion, and the base lines were then fixed for all curve fits. The data for smCHIR-AB1 and Fcυ2–4 alone at equilibrium were analyzed in terms of a single ideal solute to obtain the buoyant molecular mass M(1 − V¯ρ) as previously described (22). The molecular masses were calculated using the previously determined solvent density (22). Because both the Fcυ2–4 and smCHIR-AB1 are expressed in mammalian cells, they are N-glycosylated, and this affects the estimate of V̄. The carbohydrate content of smCHIR-AB1 was 41%, and that of Fcυ2–4 was 9.5% (see “Results”). The glycans on smCHIR-AB1, produced in HEK cells, were modeled as a monofucosylated bi-antennary complex carbohydrate. The glycans on Fcυ2–4 expressed from NS0 cells were modeled as high mannose structures. The carbohydrates led to a V̄ at 4 °C of 0.692 for smCHIR-AB1 and 0.715 for Fcυ2–4 calculated using SEDNTERP (23). The equilibrium data for the 1:1, 2:1, and 1:2 mixtures of smCHIR-AB1 and Fcυ2–4 were simultaneously fitted to a range of models using the experimental buoyant molecular masses, M(1 − V̄ρ), determined for the individual components as described previously (21). By using the buoyant molecular masses, no errors associated with estimating density or partial specific volume are included in the fit. The models used included 1:1, 2:1, 1:2, and two-component noninteracting models.
Equilibrium Gel Filtration Experiments
Equilibrium gel filtration experiments (24) to determine the smCHIR-AB1-Fcυ2–4 complex stoichiometry were performed on an AKTA fast protein liquid chromatography system (GE Healthcare) with a Superdex 200 column. The column buffer was TBSa with 5.38 μm Fcυ2–4. 10 μm smCHIR-AB1 was mixed with Fcυ2–4 in the following molar ratios: 4:1, 3:1, 2:1, 1:1, and 0.5:1 in column buffer and equilibrated at 25 °C for 1 h before injection onto the column.
Surface Plasmon Resonance (SPR) Measurement
The interactions between smCHIR-AB1 or sfpCHIR-AB1 and serum IgY, yolk IgY, or Fcυ2–4 were measured using a Biacore 3000 instrument with Biacore global analysis software 4.0.1 (GE Healthcare). Equilibrium data were analyzed using Microcal Origin v6.0. To minimize signals resulting from aggregated proteins, the samples were purified by gel filtration chromatography on a Superdex 200 column prior to use. After immobilization, increasing concentrations of analyte were injected at a flow rate of 30 μl/min, and association and dissociation phases were monitored for 3 min each. The measurements were performed at 25 °C in 10 mm Hepes, pH 7.4, 150 mm NaCl, and 0.005% surfactant P20.
To bind smCHIR-AB1 to oriented IgY, 8000 resonance units (RU) of protein A (Sigma; P6031) were coupled directly to a CM5 chip using standard amine chemistry (Biacore manual, 2001). 426, 854, and 1304 RU of polyclonal yolk anti-protein A IgY (Norwegian Antibodies; nonimmune IgY does not bind to Protein A) were captured onto flow cells 2, 3, and 4, respectively, and the protein A on flow cell 1 used as a blank subtraction. Serial dilutions of smCHIR-AB1 at concentrations from 2500 nm down to 39 nm were used as analyte.
In other experiments, 2000 RU of sIgY were coupled directly to a CM5 sensor chip, and flow cell 1 was coupled with 2000 RU of human IgG-Fc (Bethyl Laboratories; P80-104). The analyte was smCHIR-AB1 at concentrations from 3700 to 29 nm. With IgY bound to protein A or coupled directly to the sensor chip, the signal from smCHIR-AB1 returned to base line without using a regenerant.
For a comparison with cell binding (6), sfpCHIR-AB1 was coupled directly to a CM5 sensor chip at levels of 66, 99, and 147 RU with 60 RU of human IgG-Fc as control, and the analyte was sIgY at concentrations from 687 to 2.8 nm. To compare IgY with Fcυ2–4, concentrations of Fcυ2–4 from 100 to 3.1 nm were used as analyte. With sfpCHIR-AB1 as ligand, regeneration after IgY binding was effected by two injections of 0.2 m glycine, pH 2.25.
Detection of IgY Binding to MQ-NCSU Cells Using Flow Cytometry (FACS)
Chicken MQ-NCSU monocyte cells were grown and prepared as described previously (6). Incubations and washes used PBS with 1% bovine serum albumin, and 10 nm sIgY and 33 nm anti-IgY FITC (Promega) were used for labeling. To investigate the effect of smCHIR-AB1 on IgY binding to cells, sIgY was preincubated with 1.2 μm smCHIR-AB1 for 1 h before adding the mixture to cells. The cells were analyzed using a FACSCalibur instrument (BD Biosciences).
RESULTS
Characterization of smCHIR-AB1
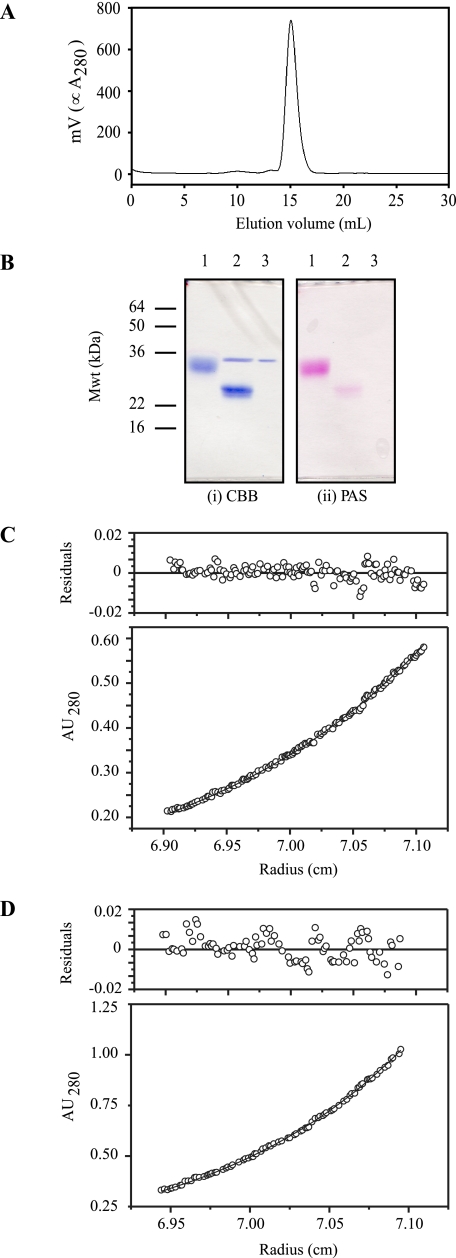
Gel filtration of 10 μm smCHIR-AB1, the highest concentration used in any of the experiments in this paper, shows a single peak (Fig. 1A), which remained after storage of the protein at 4 °C for 4 weeks in TBSa. The molecular mass calculated from the amino acid sequence is 13.5 kDa, and there are two potential glycosylation sites. Measurement of the carbohydrate content of smCHIR-AB1 gave a value of 41% for smCHIR-AB1 and 9.5% for Fcυ2–4. The value for serum IgY was 6% and was used to calculate the stoichiometry determined by SPR. The gel in Fig. 1B shows that when most of the carbohydrate is removed from smCHIR-AB1 (Fig. 1B, panel ii, lane 2) by PNGase F, there is a considerable change in migration from that of the untreated protein (Fig. 1B, panel i, lanes 1 and 2). Both the smCHIR-AB1 (Fig. 1C) and the Fcυ2–4 (Fig. 1D) data from sedimentation equilibrium in the analytical ultracentrifuge fit a single-species model, and the buoyant molecular masses were 5970 ± 32 and 23107 ± 109 Da for smCHIR-AB1 and Fcυ2–4, respectively. This corresponds to a molecular mass of 82,580 Da for Fcυ2–4, which is thus a two-chain molecule as expected. The experimental molecular mass of smCHIR-AB1, 19,702 Da, is consistent with that of a monomer with two glycosylated sites. An unglycosylated dimer would have a minimum molecular mass of 27 kDa, outside the error limit of the value for the molecular mass of smCHIR-AB1 calculated from the analytical ultracentrifuge data. Thus, there is no evidence for dimerization of smCHIR-AB1.
FIGURE 1.
Characterization of soluble monomeric CHIR-AB1. A, HPLC Superdex 200 gel filtration profile of smCHIR-AB1. The single peak shows that it is homogeneous. B, SDS-PAGE gel stained for protein (panel i) and carbohydrate (panel ii). Lane 1, smCHIR-AB1. Lane 2, smCHIR-AB1 treated with PNGase. Lane 3, PNGase alone. C, a representative set of sedimentation equilibrium data and residuals for smCHIR-AB1, collected at 16,000 RPM and fitted to a single ideal species model. The residuals are randomly distributed around zero, and the buoyant molecular mass obtained from simultaneously fitting three sets of data is 5970 ± 32. D, a representative set of sedimentation equilibrium data and residuals for Fcυ2–4, collected at 10,000 RPM and fitted to a single ideal species model. The residuals are randomly distributed around zero, and the buoyant molecular mass obtained from simultaneously fitting six sets of data is 23107 ± 109.
Stoichiometry of the Monomeric smCHIR-AB1/Fcυ2–4 Interaction
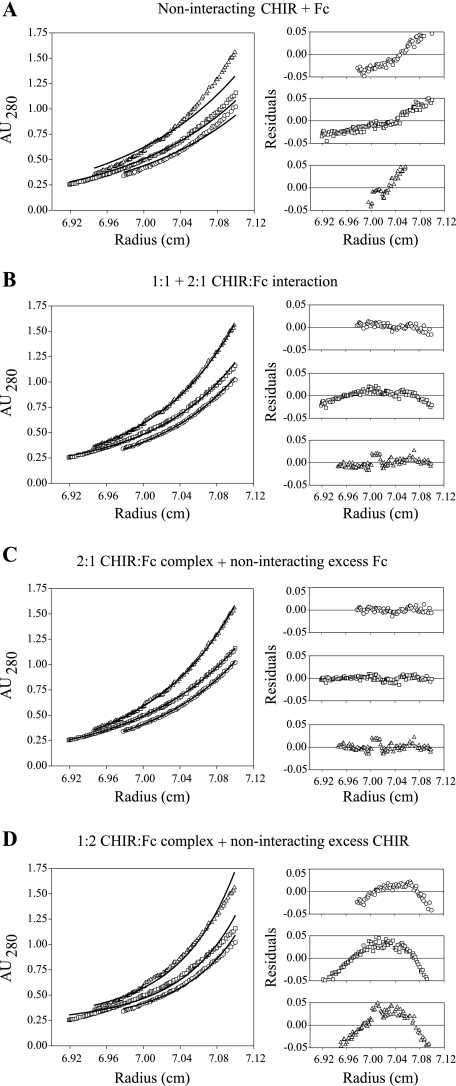
smCHIR-AB1 and Fcυ2–4 were mixed in different ratios and analyzed by sedimentation equilibrium in the analytical ultracentrifuge. The panels in Fig. 2 show the residuals resulting from a selection of the models used to analyze the interaction between the two proteins. The data were fitted best by a model in which there were two noninteracting species: a complex of two smCHIR-AB1 bound to one Fcυ2–4 and excess Fcυ2–4 alone (Fig. 2C). The data are not well described by an equilibrium model where smCHIR-AB1 and Fcυ2–4 interact to form 1:1 and 2:1 complexes (Fig. 2B), indicating that the 2:1 complex is of sufficient affinity not to be in equilibrium with uncomplexed material at the concentrations used for the experiment.
FIGURE 2.
Sedimentation equilibrium of monomeric CHIR-AB1 and IgY-Fc. Sedimentation equilibrium analyses of mixtures of smCHIR-AB1 and Fcυ2–4; the data acquired at 10,000 rpm are shown fitted to a selection of models. The three cells containing different ratios of Fcυ2–4:smCHIR-AB1 were fitted simultaneously to each model using Sigmaplot as described previously (27), and the three sets of residuals are shown for each fit (1:1, 2:1, and 1:2 Fc:CHIR mixtures are shown as circles, squares, and triangles, respectively). The models represent the following: A, a noninteracting mix of smCHIR-AB1 and Fcυ2–4; B, a 1:1 and 2:1 equilibrium between smCHIR-AB1 and Fcυ2–4; C, a mixture of 2:1 smCHIR-AB1-Fcυ2–4 complex and excess noninteracting Fcυ2–4; and D, a mixture of 1:2 smCHIR-AB1-Fcυ2–4 complex and excess noninteracting smCHIR-AB1. It is clear that only C shows residuals randomly distributed around zero, and it was found to be the only model that fitted the data well. The data at 8,500 and 12,000 rpm were also found to fit only to this model (not shown).
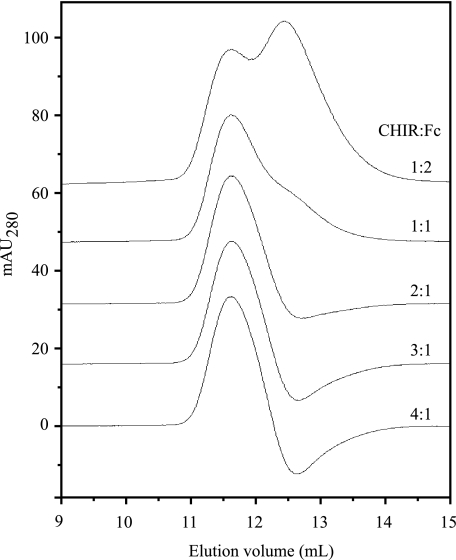
The interaction between smCHIR-AB1 and Fcυ2–4 was also investigated by gel filtration. Complexes can dissociate over the time course of an experiment, but in the method of Hummel and Dreyer (24), the column is equilibrated with one of the proteins at a concentration above the Kd of the interaction (the higher the concentration, the more precise the result), and mixtures of the two interacting proteins were injected. When there is an excess of the protein equilibrant, in this case Fcυ2–4, a peak of free equilibrant is observed as well as complex, but when the second protein (smCHIR-AB1) is in excess, the concentration of uncomplexed equilibrant can be lower than that used to equilibrate the column, leading to a trough at the retention time of the equilibrant. The molar ratio of the two proteins under conditions where neither a peak nor a trough is observed yields the stoichiometry of the interaction. Fcυ2–4, rather than whole serum IgY, was used in this experiment to ensure resolution of the peaks corresponding to complexed and free Fcυ2–4. The column was equilibrated with 5.38 μm Fcυ2–4 in TBSa buffer and mixtures of 10 μm smCHIR-AB1 and 20 (0.5:1), 10 (1:1), 5 (2:1), 3.3 (3:1), and 2.5 (4:1) μm Fcυ2–4 were injected in column buffer (containing Fcυ2–4) in separate experiments. The elution profiles are shown in Fig. 3. At an injected Fcυ2–4 concentration of 10 μm, a peak of free Fcυ2–4 is observed, whereas with 3.3 μm of injected Fcυ2–4, there is a trough. The absence of either peak or trough occurs close to a 2:1 molar ratio of smCHIR-AB1:Fcυ2–4 (Fig. 3), showing that two molecules of monomeric CHIR-AB1 are able to bind one molecule of Fcυ2–4.
FIGURE 3.
Equilibrium gel filtration of monomeric CHIR-AB1 and IgY-Fc. Gel filtration elution peaks from a Superdex 200 column resulting from a series of injections of mixtures of 10 μm smCHIR-AB1 and different concentrations of Fcυ2–4 to give molar ratios of 0.5:1, 1:1, 2:1, 3:1, and 4:1 are shown. The column was pre-equilibrated in buffer containing 5.38 μm Fcυ2–4. Absorbance at 280 nm (in mAU) is shown for the 4:1 result, with the other traces offset vertically by 16 mAU for display purposes.
SPR Measurements of smCHIR and sfpCHIR-AB1 Binding to IgY
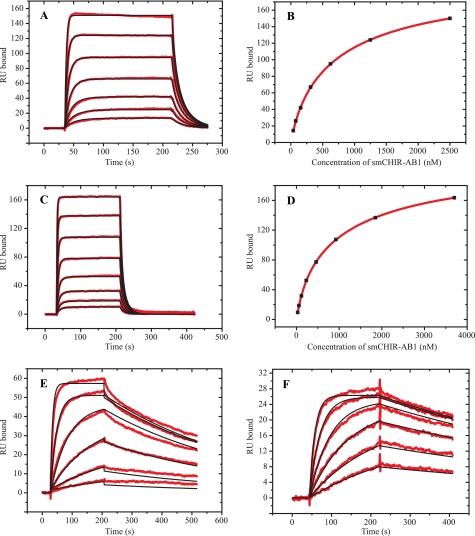
To maximize accessibility of smCHIR-AB1 to the two binding sites in IgY-Fc, protein A coupled to a CM5 chip was used to capture anti-protein A yolk IgY. Unlike that of human IgG, the Fc region of IgY does not bind protein A so that, in this experiment, IgY binds to the immobilized protein A and is therefore captured on the sensor chip by means of the Fab region only. The best fit for sensorgrams resulting from injections of smCHIR-AB1 was obtained with the heterogeneous ligand model, yielding two separate sites each with its own association constant of 7.22 ± 0.22 × 105 and 3.63 ± 1.03 × 106 m−1, respectively (Fig. 4A). An equilibrium binding curve returned Ka values of 9.06 ± 3.22 × 105 and 4.57 ± 1.73 × 106 m−1 (Fig. 4B). The Rmax value for the two sites together was 218 RU. Using 180 and 150 g/liter, respectively, for the refractive index increment for protein and carbohydrate (25), the expected value for Rmax can be calculated for 1:1 and 2:1 stoichiometry of smCHIR-AB1 binding to IgY. A value of 168 kDa (26) was used for the molecular weight of IgY, and 13.5 kDa was the molecular weight calculated for smCHIR-AB1. The measured percentages of carbohydrate were 6 and 41%, respectively. The resultant Rmax was 163 RU for 1:1 and 326 RU for 2:1 stoichiometry. These calculated values assume 100% active IgY, and thus any value above 1:1 implies the presence of two sites. The nonintegral value obtained may be due to the presence of some inactive IgY or to the partial inaccessibility of one of the sites when IgY is bound to protein A. These Ka values for oriented yolk IgY were similar to those determined in an equivalent experiment using serum IgY: Ka = 3.49 ± 0.01 × 105 and 1.91 ± 0.01 × 106 m−1. Nonimmune serum IgY was captured by monoclonal mouse anti-chicken IgY-Fabs (Biodesign International), and the analyte was smCHIR-AB1 (sensorgrams not shown).
FIGURE 4.
SPR measurement of CHIR-AB1 binding to IgY and IgY-Fc. A, an SPR sensorgram and fit, using a heterogeneous ligand model, of 39–2500 nm smCHIR-AB1 binding to anti-protein A yolk IgY captured on protein A immobilized to a CM5 sensor chip. B, binding curve using Req in RU from A for bound smCHIR-AB1 and nanomolar concentration of analyte for free smCHIR-AB1. C, sensorgram and fit, using a heterogeneous ligand model, of 32–4000 nm smCHIR-AB1 binding to serum IgY immobilized on a CM5 sensor chip. D, binding curve using Req in RU from C for bound smCHIR-AB1 and nanomolar concentration of analyte for free smCHIR-AB1. E, sensorgram and fit, using a 1:1 binding model, of 2.8–687 nm serum IgY binding to sfpCHIR-AB1 immobilized on a CM5 sensor chip. F, sensorgram and fit, using a 1:1 binding model, of 3–100 nm Fcυ2–4 binding to sfpCHIR-AB1 immobilized on a CM5 sensor chip.
Serum IgY was also coupled directly to a CM5 chip (2000 RU) with smCHIR-AB1 as analyte (Fig. 4C). Analysis of the sensorgrams using the heterogeneous ligand model gave a fit with Ka values of 5.17 ± 0.12 × 105 and 3.40 ± 0.05 × 106 m−1 (Fig. 4C). Analysis of the equilibrium binding curve (Fig. 4D) showed that χ2 was 34 times lower using a two-site than a one-site model, giving Ka values of 5.57 ± 1.26 × 105 and 3.61 ± 0.76 × 106 m−1 and Rmax = 212 RU (Fig. 4D), which agree well with the values derived from the kinetic analysis and also with the values derived with oriented yolk IgY.
Difficulties were experienced in the reverse experiment: coupling smCHIR-AB1 to the biosensor surface and using IgY as analyte. smCHIR-AB1 at pH 3.5, one pH unit below its iso-electric point calculated from the amino acid sequence (19), did not preconcentrate when injected onto a CM5 chip, probably because of the extensive glycosylation of the protein. Biotinylated smCHIR-AB1 was coupled to a streptavidin-coated chip, with serum IgY as analyte, but very slow association rates were observed, suggesting that biotinylation of smCHIR-AB1 interferes with the CHIR/IgY interaction. However, CHIR-AB1 can be coupled to a CM5 chip as a fusion protein with IgG-Fc (5), sfpCHIR-AB1, which displays two domains of soluble extracellular receptor on the flexible hinge of IgG-Fc. With IgY as analyte, a 1:1 model yields a Ka value of 9.07 ± 0.07 × 107 m−1 (Fig. 4E). This higher value must be an avidity effect caused by the presence of two separate binding sites for CHIR-AB1 on IgY interacting with either two receptor molecules on a CHIR-AB1 fusion protein or adjacent molecules in the highly flexible dextran matrix on the biosensor surface.
Because whole IgY was used for the SPR measurements, but Fcυ2–4 for gel filtration and analytical ultracentrifugation, a comparison by SPR of IgY and Fcυ2–4 binding to immobilized sfpCHIR-AB1 was made. With Fcυ2–4 as analyte, the fit obtained using a 1:1 model gave a Ka value of 6.11 ± 0.02 × 108 m−1 (Fig. 4F). The association rate constant for Fcυ2–4 (7.21 × 105 m−1 s−1) was slightly faster than that for whole IgY (1.96 × 105 m−1 s−1), and the dissociation rate constant (1.18 × 10−3 s−1) was slightly slower than that for IgY (2.16 × 10−3 s−1). These two small effects are additive, so that the overall Ka for Fcυ2–4 is approximately six times higher than for IgY. The higher equilibrium association constants for Fc fragments compared with intact antibodies are also characteristic of IgE (27) and IgG (28).
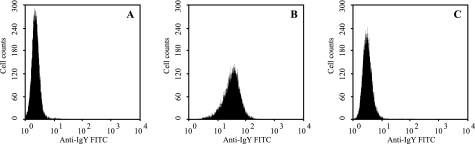
To ascertain whether the Fc receptor for IgY on the monocyte cell line MQ-NCSU (6) is CHIR-AB1, MQ-NCSU cells were assayed for IgY binding in the presence of the soluble receptor. A 100-fold excess of smCHIR-AB1 almost totally inhibited IgY binding (Fig. 5).
FIGURE 5.
Effect of soluble CHIR-AB1 on IgY binding to chicken monocytes. FACS histograms of sIgY binding to MQ-NCSU cells, detected by anti-IgY FITC, are shown. A, buffer alone followed by anti-IgY FITC. B, 10 nm sIgY followed by anti-IgY FITC. C, 10 nm sIgY + 1.2 μm smCHIR-AB1 followed by anti-IgY FITC.
DISCUSSION
This is the first study of the interaction between homogeneous soluble receptor CHIR-AB1 and chicken IgY and its Fc fragment; CHIR-AB1 prepared by Arnon et al. (16) was an equilibrium mixture of monomer and dimer. Thus, it was not possible to establish the competency of monomeric receptor to bind to IgY, nor whether the observed 2:1 receptor:IgY stoichiometry was due to one receptor dimer or two monomers binding to IgY. There are several explanations for the difference between the two preparations of CHIR-AB1. The receptor reported here, smCHIR-AB1, was expressed in HEK293 cells and has a carbohydrate content of ∼40%. In contrast, data for soluble CHIR-AB1 (16), which was expressed from insect cells, imply a 22% carbohydrate content. Moreover, one of the two predicted glycosylation sites on CHIR-AB1, Asn83, lies close to the interface between the two monomers in the crystal structure of the dimer, and no carbohydrate residues are visible at this position (16). It is possible that a longer carbohydrate chain on the more highly glycosylated smCHIR-AB1 sterically hinders formation of the salt bridge between Arg72 and Asp44 in the dimer. Second, the weak periodic acid-Schiff staining of smCHIR-AB1 after PNGase treatment could be due to O-glycosylation of Thr40, Ser77, or Thr81, which might prevent dimer formation. Lastly, we cannot exclude the possibility that the additional amino acids at the C terminus of smCHIR (seven residues of the stalk that connects the Ig and transmembrane domains in the full CHIR-AB1 sequence and part of the TEV protease site remaining after cleavage) may bend back and interfere with the dimer interface. Regardless of cause, the data reported here show that smCHIR-AB1, expressed in mammalian cells, is entirely monomeric. Using three techniques, analytical ultracentrifugation, gel filtration, and SPR, the data presented here show that monomeric smCHIR-AB1 forms a complex with Fcυ2–4 with a stoichiometry of 2:1.
In SPR experiments reported here, smCHIR-AB1 binds to oriented IgY-anti-protein A, immobilized on a protein A surface. Analysis of equilibrium binding using a two-site interaction yields Ka values of 9.06 ± 3.22 × 105 and 4.57 ± 1.73 × 106 m−1, and with kinetic analysis using the heterogeneous ligand model (Biacore software 4.01), Ka = 7.22 ± 0.32 × 105 and 3.63 ± 1.03 × 106 m−1. These values agree well with one another and are almost within the error of the corresponding values obtained by coupling serum IgY directly to a CM5 chip: 5.17 ± 0.12 × 105 and 3.40 ± 0.05 × 106 m−1 (equilibrium analysis), and 5.57 ± 1.26 × 105 and 3.61 ± 0.60 × 106 m−1 (kinetic analysis). The Ka values for the two sites differ by a factor of approximately six, whereas those for FcαRI binding to IgA-Fc, which also shows 2:1 stoichiometry, differ only by a factor of approximately two (29). In the crystal structure of the IgA-Fc-receptor complex, IgA-Fc has 2-fold rotational symmetry (13), and the two sites are therefore structurally identical. The crystal structure of uncomplexed Fcυ3–4 (15) shows an asymmetry between the two heavy chains, and if this asymmetry persists in solution and upon complex formation, it could explain the more pronounced difference in affinity between the two receptor-binding sites on IgY-Fc than on IgA-Fc.
The greater than 25-fold difference in affinity between the interaction of a single receptor (mean Ka = 6.1 × 105 and 3.4 × 106 m−1, when IgY is immobilized on a sensor chip) and the apparent availability of two receptors when sfpCHIR-AB1 is immobilized (Ka = 9.1 × 107 m−1) can be explained in terms of an avidity effect with two smCHIR-AB1 molecules binding at independent sites on an IgY molecule.
This higher Ka value, as a result of avidity, agrees well with the result obtained in cell binding studies, Ka = 5 × 107 m−1 (6). The apparent concentration of the receptor is raised by its confinement to the two dimensions of the plasma membrane so that even a small number of receptors/cell is capable of engaging IgY with 2:1 stoichiometry. In the IgA system, there is the same agreement between the Ka of 1.1 × 108 m−1 for IgA binding to immobilized FcαRI and the Ka of 0.48–1.7 × 108 m−1 obtained for IgA binding to cells (29). Although another IgY-Fc receptor, ggFcR, has been described on chicken monocytes (11), in the FACS assay reported here, almost all IgY binding to cells was inhibited by an excess of smCHIR-AB1. Either the affinity of ggFcR and any other receptors for IgY are so low that they dissociate within the time scale of the experiment, the receptors are present in much lower numbers than CHIR-AB1, or they bind to an overlapping region on IgY-Fc, and the binding can thus be blocked by smCHIR-AB1.
The 2:1 stoichiometry for CHIR-AB1 binding to IgY and the value of the affinity constant for each binding site pose the question of whether the binding site for CHIR-AB1 on IgY is homologous to that of FcαRI on IgA. In support of this, there are a surprising number of amino acid identities (Leu256, Arg382, Leu384, and eight residues in the Cα3 FG loop) between the IgA receptor-binding site on IgA and the equivalent amino acids (by sequence alignment) in IgY (Fig. 6). In the lower hinge region of IgY there is some conservation of the residues involved in IgG and IgE receptor binding also, but a binding site here is difficult to reconcile with the 2:1 stoichiometry. An x-ray structure for the IgY-smCHIR-AB1 complex is clearly needed. Our previous finding that the Cυ2 domains play little or no role in the kinetics of IgY binding to receptor (6), unlike the homologous Cϵ2 domains in IgE (30), is also consistent with an IgA-like mode of binding. An ancient IgA-like interaction would indicate that the mode of Fc receptor binding of IgY orthologues IgG and IgE is a relatively recent evolutionary change.
FIGURE 6.
Conservation of Fc-region residues between human IgA and chicken IgY. Shown is amino acid sequence alignment of the Cα2 and Cα3 domains of human IgA1 and IgA2 (accession numbers P01876-1 and P01877-1) and the Cυ3 and Cυ4 domains of chicken IgY (accession number X07174), performed using ClustalW (36). Residues in IgA1 that contact FcαRI in the crystal structure of the complex (Protein Data Bank code 1OW0) and their conserved equivalents in IgA2 and IgY sequences are shown in green; semi-conserved residues (i.e. residues with similar properties) are shown in blue.
It is not clear what effect the 2:1 stoichiometry of CHIR-AB1:IgY might have on signaling. It has been suggested that 2:1 stoichiometry of receptor to IgA exists before activation (29). Dephosphorylation of FcαRI by protein phosphatase 2A has now been identified as the activation switch for IgA bound to its receptor on neutrophils (31). A mutant IgG, binding only one molecule of FcRn, the fetal and IgG homeostasis receptor (32), has been used to show that 1:1 receptor:antibody stoichiometry does decrease the efficiency of transcytosis and recycling and also does increase the proportion of antibody targeted for degradation (33). Thus, antibody-receptor stoichiometry has been shown to act as a modulator, but not a switch of function.
Receptors with genes in the LRC have several features not seen in those from the classical Fc Receptor Cluster. Unlike receptors for IgG and IgE, both inhibitory and activatory signals can be due to a single IgA receptor immunoreceptor tyrosine-based activatory motif (34); the siting of the CHIR-AB1 gene in the LRC and its use of the FcϵRγ adaptor (5) suggest that the immunoreceptor tyrosine-based activatory motif on chicken FcϵRγ may also transmit inhibitory signals. CHIR-AB1 and one human receptor, KIR2DL4, in the LRC are unique in possessing both an immunoreceptor tyrosine-based activatory motif and an immunoreceptor tyrosine-based inhibitory motif, although inhibitory signaling has not yet been observed for either. In addition, KIR2DL4 is the only receptor for which two separate activatory signals have been detected (35). If the structural similarities between CHIR-AB1 and both FcαRI and KIR2DL4 are predictive of function, this chicken IgY receptor has the potential for more activatory and inhibitory signaling mechanisms than any other Fc receptor to date. Exploring these mechanisms with CHIR-AB1 is certain to offer a fresh perspective on antibody-Fc receptor interactions.
Footnotes
- CHIR-AB1
- chicken leukocyte receptor AB1
- Fcα/γ/ϵ/υ
- Fc fragment of IgA/IgG/IgE/IgY
- LRC
- leukocyte receptor cluster
- FcαRI
- the leukocyte receptor for IgA (CD89)
- FcγRIII
- a low affinity IgG receptor (CD16)
- FcϵRI
- the high affinity IgE receptor
- smCHIR-AB1
- soluble monomeric chicken leukocyte receptor
- sfpCHIR-AB1
- soluble fusion protein of CHIR-AB1 and IgG-Fc
- Fcυ2–4
- IgY-Fc fragment containing heavy chain constant domains 2, 3, and 4
- TBSa
- Tris-buffered saline with sodium azide
- RU
- resonance units
- FACS
- flow cytometry
- MQ-NCSU
- a chicken monocyte cell line
- Cα3
- IgA heavy chain constant domain 3
- Cυ2
- IgY heavy chain constant domain 2
- Cϵ2
- IgE heavy chain constant domain 2
- HPLC
- high pressure liquid chromatography
- SPR
- surface plasmon resonance
- FITC
- fluorescein isothiocyanate.
REFERENCES
- 1.Warr G. W., Magor K. E., Higgins D. A. (1995) Immunol. Today 16, 392–398 [DOI] [PubMed] [Google Scholar]
- 2.Davison T. F. (2003) Br. Poult. Sci. 44, 6–21 [DOI] [PubMed] [Google Scholar]
- 3.Faith R. E., Clem L. W. (1973) Immunology 25, 151–164 [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi M. A., Miller L., Lillehoj H. S., Ficken M. D. (1990) Vet. Immunol. Immunopathol. 26, 237–250 [DOI] [PubMed] [Google Scholar]
- 5.Viertlboeck B. C., Schweinsberg S., Hanczaruk M. A., Schmitt R., Du Pasquier L., Herberg F. W., Göbel T. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11718–11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor A. I., Gould H. J., Sutton B. J., Calvert R. A. (2008) J. Biol. Chem. 283, 16384–16390 [DOI] [PubMed] [Google Scholar]
- 7.Taylor A. I., Gould H. J., Sutton B. J., Calvert R. A. (2007) Immunogenetics 59, 323–328 [DOI] [PubMed] [Google Scholar]
- 8.Laun K., Coggill P., Palmer S., Sims S., Ning Z., Ragoussis J., Volpi E., Wilson N., Beck S., Ziegler A., Volz A. (2006) PLoS Genet. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayngerts S. A., Najakshin A. M., Taranin A. V. (2007) Immunogenetics 59, 493–506 [DOI] [PubMed] [Google Scholar]
- 10.Barrow A. D., Trowsdale J. (2008) Immunol. Rev. 224, 98–123 [DOI] [PubMed] [Google Scholar]
- 11.Viertlboeck B. C., Schmitt R., Hanczaruk M. A., Crooijmans R. P., Groenen M. A., Göbel T. W. (2009) J. Immunol. 182, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 12.Woof J. M., Burton D. R. (2004) Nat. Rev. Immunol. 4, 89–99 [DOI] [PubMed] [Google Scholar]
- 13.Herr A. B., Ballister E. R., Bjorkman P. J. (2003) Nature 423, 614–620 [DOI] [PubMed] [Google Scholar]
- 14.Sánchez L. M., Penny D. M., Bjorkman P. J. (1999) Biochemistry 38, 9471–9476 [DOI] [PubMed] [Google Scholar]
- 15.Taylor A. I., Fabiane S. M., Sutton B. J., Calvert R. A. (2009) Biochemistry 48, 558–562 [DOI] [PubMed] [Google Scholar]
- 16.Arnon T. I., Kaiser J. T., West A. P., Jr., Olson R., Diskin R., Viertlboeck B. C., Göbel T. W., Bjorkman P. J. (2008) J. Mol. Biol. 381, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng J., Carstens R. P. (2006) Protein Expr. Purif. 48, 142–150 [DOI] [PubMed] [Google Scholar]
- 18.Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M. ed) pp.571–607, Humana Press, Totowa, NJ [Google Scholar]
- 20.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 21.Shi J., Ghirlando R., Beavil R. L., Beavil A. J., Keown M. B., Young R. J., Owens R. J., Sutton B. J., Gould H. J. (1997) Biochemistry 36, 2112–2122 [DOI] [PubMed] [Google Scholar]
- 22.Ghirlando R., Keown M. B., Mackay G. A., Lewis M. S., Unkeless J. C., Gould H. J. (1995) Biochemistry 34, 13320–13327 [DOI] [PubMed] [Google Scholar]
- 23.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science ( Harding S. E., Rowe A. J., Horton J. C. eds) pp.90–125, Royal Society of Chemistry, Cambridge [Google Scholar]
- 24.Hummel J. P., Dreyer W. J. (1962) Biochim. Biophys. Acta 63, 530–532 [DOI] [PubMed] [Google Scholar]
- 25.Nice E. C., Catimel B. (1999) Bioessays 21, 339–352 [DOI] [PubMed] [Google Scholar]
- 26.Suzuki N., Lee Y. C. (2004) Glycobiology 14, 275–292 [DOI] [PubMed] [Google Scholar]
- 27.Young R. J., Owens R. J., Mackay G. A., Chan C. M., Shi J., Hide M., Francis D. M., Henry A. J., Sutton B. J., Gould H. J. (1995) Protein Eng. 8, 193–199 [DOI] [PubMed] [Google Scholar]
- 28.Raychaudhuri G., McCool D., Painter R. H. (1985) Mol. Immunol. 22, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 29.Herr A. B., White C. L., Milburn C., Wu C., Bjorkman P. J. (2003) J. Mol. Biol. 327, 645–657 [DOI] [PubMed] [Google Scholar]
- 30.McDonnell J. M., Calvert R., Beavil R. L., Beavil A. J., Henry A. J., Sutton B. J., Gould H. J., Cowburn D. (2001) Nat. Struct. Biol. 8, 437–441 [DOI] [PubMed] [Google Scholar]
- 31.Bakema J. E., Bakker A., de Haij S., Honing H., Bracke M., Koenderman L., Vidarsson G., van de Winkel J. G., Leusen J. H. (2008) J. Immunol. 181, 4080–4088 [DOI] [PubMed] [Google Scholar]
- 32.Martin W. L., Bjorkman P. J. (1999) Biochemistry 38, 12639–12647 [DOI] [PubMed] [Google Scholar]
- 33.Tesar D. B., Tiangco N. E., Bjorkman P. J. (2006) Traffic 7, 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasquier B., Launay P., Kanamaru Y., Moura I. C., Pfirsch S., Ruffié C., Hénin D., Benhamou M., Pretolani M., Blank U., Monteiro R. C. (2005) Immunity 22, 31–42 [DOI] [PubMed] [Google Scholar]
- 35.Miah S. M., Hughes T. L., Campbell K. S. (2008) J. Immunol. 180, 2922–2932 [DOI] [PubMed] [Google Scholar]
- 36.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]