Abstract
Propofol is the most widely used injectable general anesthetic. Its targets include ligand-gated ion channels such as the GABAA receptor, but such receptor-channel complexes remain challenging to study at atomic resolution. Until structural biology methods advance to the point of being able to deal with systems such as the GABAA receptor, it will be necessary to use more tractable surrogates to probe the molecular details of anesthetic recognition. We have previously shown that recognition of inhalational general anesthetics by the model protein apoferritin closely mirrors recognition by more complex and clinically relevant protein targets; here we show that apoferritin also binds propofol and related GABAergic anesthetics, and that the same binding site mediates recognition of both inhalational and injectable anesthetics. Apoferritin binding affinities for a series of propofol analogs were found to be strongly correlated with the ability to potentiate GABA responses at GABAA receptors, validating this model system for injectable anesthetics. High resolution x-ray crystal structures reveal that, despite the presence of hydrogen bond donors and acceptors, anesthetic recognition is mediated largely by van der Waals forces and the hydrophobic effect. Molecular dynamics simulations indicate that the ligands undergo considerable fluctuations about their equilibrium positions. Finally, apoferritin displays both structural and dynamic responses to anesthetic binding, which may mimic changes elicited by anesthetics in physiologic targets like ion channels.
Most general anesthetics alter the activity of ligand-gated ion channels, and electrophysiology, photolabeling, and transgenic animal experiments imply that this effect contributes to the mechanism of anesthesia (1–9). Although the molecular mechanism for this effect is not yet clear, photolabeling studies indicate that anesthetics bind within the transmembrane regions of Cys-loop ligand-gated ion channels such as the nicotinic acetylcholine and the γ-aminobutyric acid (GABA)2 type A receptors (2, 9–11). Practical difficulties associated with overexpression, purification, and crystallization of ion channels have thus far stymied investigation of the structural and energetic bases underlying anesthetic recognition. However, general anesthetics also bind specifically to sites in soluble proteins, including firefly luciferase, human serum albumin (HSA), and horse spleen apoferritin (HSAF) (12–14), and x-ray crystal structures have been determined for complexes of these proteins with several general anesthetics (14–16). In particular, HSAF is an attractive model for studying anesthetic-protein interactions because it has the highest affinity for anesthetics of any protein studied to date, has a unique anesthetic binding site, and is a multimer of 4-helix bundles, much like the putative anesthetic binding regions in ligand-gated channels. In addition, apoferritin is commercially available and crystallizes readily. Most importantly, however, the affinity of HSAF for a broad range of general anesthetics is highly correlated with anesthetic potency, confirming the utility and relevance of this model system (17).
Ferritin is a 24-mer iron-binding protein. It sequesters free iron ions, thereby helping to maintain non-toxic levels of iron in the cell and functioning as a cellular iron reservoir (18, 19). Each subunit has a molecular mass of ∼20 kDa and adopts a 4-helix bundle fold. The 24-mer forms a hollow, roughly spherical particle with 432 symmetry. Two ferritin isoforms are found in mammals, heavy (H) and light (L), and 24-mers can contain all H chains, all L chains, or mixtures of varying stoichiometry; the biological significance of the H/L ratio is not yet clear (20).
In addition to the large central cavity, the apoferritin 24-mer contains additional, smaller cavities at the dimer interfaces; these smaller cavities are of an appropriate size to accommodate anesthetics. X-ray crystallography has confirmed that this interfacial cavity is the binding site for the inhalational anesthetics halothane and isoflurane, and isothermal titration calorimetry (ITC) measurements have shown that this interfacial site has a relatively high affinity for these anesthetics (Ka values ∼105 m−1) (14).
General anesthetics fall into at least two broad classes, inhalational and injectable. Whereas both classes of drugs can induce the amnesia, immobility, and hypnosis associated with anesthesia, molecules in the two classes differ substantially in their chemical and physical properties. Prior to this work, only one crystal structure has been available for an injectable general anesthetic complexed with a protein-propofol, bound to HSA (16). This structure revealed that the propofol binding sites on this protein do not, by and large, overlap with the binding sites for inhalational anesthetics. This raises the question of whether the two types of drug invariably bind to separate sets of targets, or whether they could possibly transduce their effects by binding to a single protein site. To address this question we assessed whether propofol binds to the apoferritin site that had been previously identified as the binding site for inhalational anesthetics. Using x-ray crystallography, calorimetry, and molecular modeling, we show that the two types of anesthetics do indeed share a common binding site. We also investigated structure-binding relationships for a homologous series of propofol-like compounds and found that, remarkably, the energetics of binding to apoferritin precisely match the compound's abilities to potentiate GABA effects at GABAA receptors, suggesting that similar structural and physicochemical factors mediate anesthetic recognition by both apoferritin and ligand-gated ion channels. This argues for the possibility that anesthetic binding might trigger structural and dynamic alterations in GABAA receptors similar to those observed in apoferritin, and that these changes underlie anesthetic effects.
EXPERIMENTAL PROCEDURES
Materials
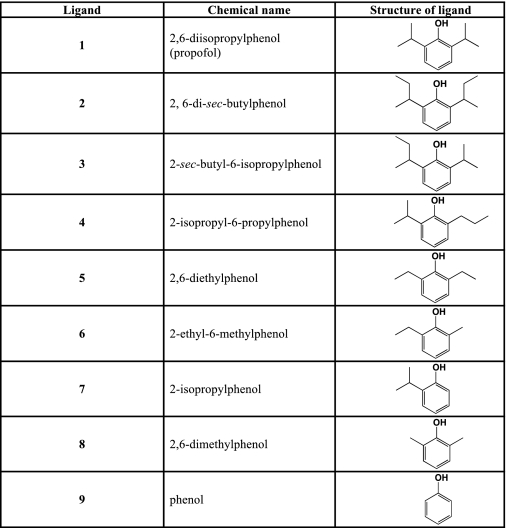
HSAF was purchased from Sigma-Aldrich. Propofol and analogs were purchased from Sigma-Aldrich or Acros (Geel, Belgium); compounds 2, 3, and 5 were synthesized as previously described (21, 22). Ligand structures are given in Table 1.
TABLE 1.
Structures of the compounds studied
Isothermal Titration Calorimetry (ITC)
ITC measurements were carried out essentially as previously described (14) using a MicroCal VP-ITC instrument (Northampton, MA); details are given under supplemental data.
Protein Crystallization and Structure Determination
Apoferritin was purified by gel filtration chromatography (23) and co-crystallized with ligands using hanging drop vapor diffusion at 18 °C. Briefly, reservoir solutions containing 0.2–1.2 m (NH4)2SO4 and 0.1–0.225 m CdSO4 plus and minus 1–5 mm ligand were mixed with 12 mg/ml HSAF in 0.2 m sodium acetate pH 5.0 and equilibrated over 1 ml of reservoir solution. Diffraction data were collected at beamlines X6A and X25 of the National Synchrotron Light Source and 17-ID of the Advanced Photon Source. Duplicate structures were determined independently for two of the complexes, to provide some sense of the precision associated with the structure determination. Data collection and refinement statistics are shown in Table 2. Additional details of crystallization and structure determination are given under supplemental data. Coordinates and structure factors for all models have been deposited with the Protein Data Bank.
TABLE 2.
Data collection and refinement statistics
| Ligand | Unliganded | 1 | 5 (#1) | 5 (#2) | 7 | 8 (#1) | 8 (#2) | 9 |
|---|---|---|---|---|---|---|---|---|
| PDB ID | 3F32 | 3F33 | 3F35 | 3F34 | 3F36 | 3F38 | 3F37 | 3F39 |
| Resolution range (Å)a | 20.97–1.70(1.76–1.70) | 37.25–1.70(1.76–1.70) | 28.92–1.92(1.99–1.92) | 32.08–1.68(1.74–1.68) | 25.54–1.70(1.76–1.70) | 20.88–1.75(1.81–1.75) | 37.0–1.54(1.60–1.54) | 20.97–1.70(1.76–1.70) |
| Wavelength (Å) | 0.9795 | 1.1 | 1.0 | 0.9537 | 1.0 | 0.9795 | 0.9537 | 0.9795 |
| Cell constantsb (a = b = c) | 181.60 | 182.49 | 182.93 | 181.46 | 182.37 | 181.99 | 181.25 | 181.53 |
| Completeness | 99.5 (95.5) | 99.8 (99.9) | 99.9 (100.0) | 97.9 (85.7) | 99.8 (100.0) | 97.9 (87.6) | 99.5 (96.1) | 99.4 (96.4) |
| Rmerge | 0.102 (0.615) | 0.070 (0.319) | 0.079 (0.433) | 0.099 (0.527) | 0.057 (0.317) | 0.064 (0.397) | 0.053 (0.460) | 0.072 (0.460) |
| <I/σ(I)> | 15.5 (3.6) | 23.5 (4.9) | 18.4 (5.7) | 13.0 (2.0) | 24.9 (5.3) | 23.1 (5.5) | 30.0 (2.5) | 17.1 (3.1) |
| No. of non-hydrogen atoms in ASU | ||||||||
| Protein | 1354 | 1354 | 1354 | 1354 | 1354 | 1354 | 1354 | 1354 |
| Water | 201 | 210 | 135 | 189 | 153 | 179 | 203 | 149 |
| Metal ions | 4 | 6 | 7 | 5 | 8 | 6 | 7 | 8 |
| Ligand | 0 | 13 | 11 | 11 | 10 | 9 | 9 | 7 |
| Other | 10 | 5 | 15 | 19 | 10 | 10 | 19 | 10 |
| No. of reflections used in refinement (work/free) | 27172/1445 | 27620/1480 | 19281/1038 | 27587/1490 | 27347/1458 | 24679/1320 | 36122/1900 | 21259/1147 |
| R/Rfree | 0.178/0.209 | 0.189/0.22 | 0.190/0.236 | 0.198/0.247 | 0.185/0.21 | 0.172/0.222 | 0.174/0.198 | 0.184/0.216 |
| Deviations from ideality | ||||||||
| Bond lengths (Å) | 0.013 | 0.013 | 0.020 | 0.015 | 0.013 | 0.014 | 0.010 | 0.018 |
| Bond angles (degrees) | 1.236 | 1.230 | 1.751 | 1.392 | 1.367 | 1.311 | 1.167 | 1.396 |
| Mean B values (Å2) | 22.9 | 22.2 | 33.5 | 30.9 | 31.6 | 24.3 | 24.7 | 27.0 |
| Protein | 20.7 | 19.9 | 32.1 | 29.3 | 30.0 | 24.8 | 22.0 | 25.4 |
| Water/ions | 37.0 | 35.7 | 42.3 | 40.3 | 42.7 | 36.6 | 40.0 | 38.8 |
| Ligand | – | 40.9 | 71.8 | 45.9 | 58.1 | 59.8 | 55.1 | 52.3 |
| Ramachandran plot statisticsc % residues in: | ||||||||
| Most favored regions | 95.4 | 94.7 | 96.1 | 95.4 | 94.7 | 95.4 | 95.4 | 94.7 |
| Additionally allowed regions | 4.6 | 5.3 | 3.9 | 4.6 | 5.3 | 4.6 | 4.6 | 5.3 |
a Values in parentheses refer to the highest resolution shell.
b All crystals belong to space group F432.
c Calculated using PROCHECK (38).
Volume Calculations
Cavity and ligand volumes were calculated using the programs VOIDOO and FLOOD (24); details are given under supplemental data.
Molecular Dynamics
Three distinct systems were studied using molecular dynamics simulations performed in VMD (25). Each system consisted of an apoferritin dimer set in a water box, with the protein containing either no ligand (control), phenol, or propofol bound in the orientation indicated by the crystal structure. The propofol system was simulated under two distinct equilibration protocols (A and B); in the A protocol, protein Ca atoms were restrained to their initial positions for the first 500 ps, followed by unrestrained simulation; in the B protocol, both the ligand and protein were restrained initially, and the restraints were relaxed more slowly. Details are given under supplemental data.
RESULTS
Isothermal Titration Calorimetry
Calorimetric measurements demonstrated that all of the compounds tested bound specifically to apoferritin (Table 3), with dissociation constants ranging from ∼2–600 micromolar. The concentration of propofol in the serum is generally 1–2 μm during normal anesthetic applications (26), and hence apoferritin represents a good model for a high affinity anesthetic binding site that is significantly occupied during anesthesia. The calorimetric data were well fit using a single class of sites model with variable stoichiometry. The number of binding sites per apoferritin 24-mer was consistently estimated as ∼1–4, even though 12 sites are predicted from the crystal structure. The origin of this discrepancy is not yet clear, but it is likely that a single class, non-interacting site model may be an oversimplification, especially given that the HSAF 24-mer contains both L (85%) and H (15%) subunits; however, the relatively low affinity of the ligands renders more complex binding models difficult to justify or prove.
TABLE 3.
Apoferritin-ligand complexes
| Ligand | Vol. of liganda | Vol. of cavityb | Packing density | R.m.s.d. vs. unliganded apoferritinc | Solvent accessible surface area | Ka |
|---|---|---|---|---|---|---|
| Å3 | Å3 | Å3 | Å3 | ×10−410m−1m | ||
| None | – | 363 ± 12 | – | – | – | – |
| 1 | 186.3 | 417 ± 8c | 0.45 | 0.134 (0.74) | 370.8 | 20.80 ± 1.1 |
| 2d | 218 | – | – | – | 426.4 | 17.75 ± 3.3 |
| 3d | 201.6 | – | – | – | 401.2 | 50.33 ± 8.2 |
| 4d | 186.8 | – | – | – | 380.6 | 21.15 ± 1.3 |
| 5e | 156 | 397 ± 10 | 0.39 | 0.104 (0.41) | 332.8 | 8.14 ± 0.4 |
| 390 ± 9 | 0.40 | 0.134 (0.73) | ||||
| 6d | 142 | – | – | – | 311 | 3.31 ± 0.1 |
| 7 | 139.8 | 377 ± 10 | 0.37 | 0.077 (0.39) | 306.3 | 6.04 ± 0.3 |
| 8e | 125.1 | 363 ± 9 | 0.34 | 0.060 (0.63) | 286.2 | 0.94 ± 0.02 |
| 352 ± 9 | 0.36 | 0.086 (0.57) | ||||
| 9 | 94.7 | 395 ± 14 | 0.24 | 0.116 (0.63) | 239.6 | 0.18 ± 0.006 |
a Volume of ligand determined using VOIDOO (24).
b Probe-occupied volume of cavity calculated using VOIDOO (24).
c Calculated for Cα atoms of the entire protein; values in parentheses calculated for all atoms of cavity-lining residues.
d X-ray crystal structures were not determined for these complexes.
e Structures were determined in duplicate (see Table II).
General Features of the Apoferritin Anesthetic Binding Cavity
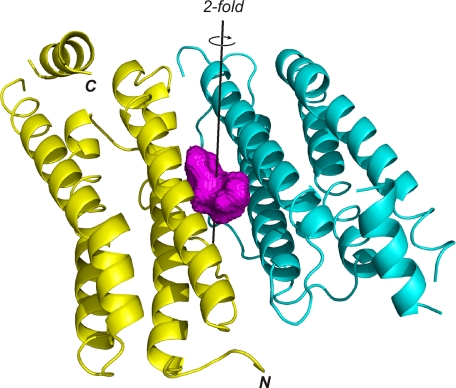
The asymmetric unit of our apoferritin crystals contains a single protomer, and application of the 432 crystallographic symmetry generates the 24-mer. The apoferritin structures reported here are essentially identical to the previously reported structures of HSAF and recombinant horse L apoferritin (27–30). The protomer forms a bundle of four long helices; a fifth short helix at the molecular C terminus caps the bundle at one end. A long extended crossover strand bridges the second and third helices (Fig. 1).
FIGURE 1.
The apoferritin dimer. The two subunits are shown in yellow and cyan, while the surface of the anesthetic binding cavity is shown in magenta. The position of the 2-fold symmetry axis relating the two subunits is shown as a black line, running through the center of the binding cavity. Figs. 1 and 2 were made using PyMOL (39).
The anesthetic binding site lies at the interface between two apoferritin subunits and is centered about a crystallographic 2-fold symmetry axis that relates the two subunits. The sides of the cavity are formed by the first and second helices of both subunits, while the base of the cavity is formed by the strands that connect helices 2 and 3 of each subunit. The top of the cavity is capped by the side chains of two symmetry-related copies of Arg-59. Small openings on either side of these residues connect the cavity with the surface of the protein; these openings face the interior of the hollow sphere that is formed by the 24-mer. The cavity is lined largely by apolar and mildly polar residues. These include Leu-81 at the base of the cavity and Ser-27, Tyr-28, Leu-24, Leu-31, and Ala-55 around the sides of the cavity. Several charged residues are found at the mouth of the cavity, including the Arg-59 residues mentioned above, as well as Glu-56 and Glu-63 (Fig. 2). Two copies of each of these residues (one from each symmetry-related apoferritin protomer) contribute to forming the cavity. The 2-fold symmetric nature of the binding pocket means that two positions are seen for each ligand, related to one another by a 180° rotation. Only one of the two symmetry-related positions can be occupied in any given cavity, due to steric overlap, and so the maximum possible occupancy for each ligand position is 0.5.
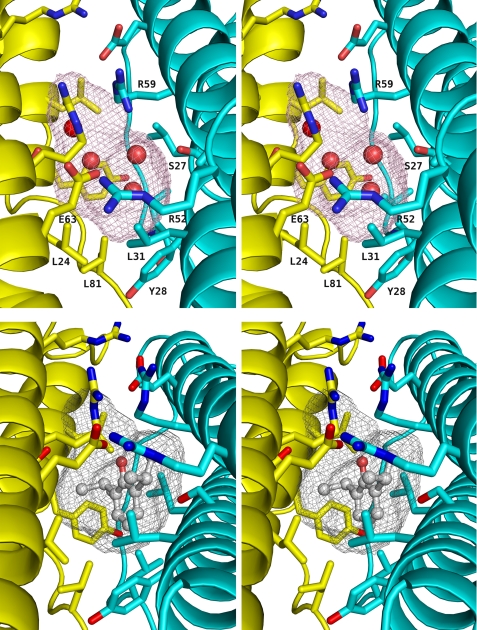
FIGURE 2.
Stereoviews of the apoferritin anesthetic binding site. Upper panel, unliganded apoferritin. The two protein protomers making up the dimer are shown in the same color scheme as Fig. 1, with the side chains of the cavity-lining residues shown in a stick representation. The surface of the cavity is shown as a mesh. The four water molecules observed in the cavity are shown as red spheres. Lower panel, apoferritin-propofol complex. The viewpoint is similar to that of the upper panel; again, the cavity surface is shown in a mesh representation. The propofol molecule is shown in ball-and-stick representation. In both panels, oxygen atoms are shown in red and nitrogen atoms in blue.
In the absence of ligand, electron density was observed in the anesthetic binding pocket and modeled as two water molecules per asymmetric unit (Fig. 2). These waters do not overlap their symmetry mates, and so each cavity contains four ordered waters. One of these waters is well positioned to make hydrogen bonds with Arg-59 (oxygen-nitrogen distance 2.9 Å) and Ser-27 (oxygen-oxygen distance 2.8 Å). The second is within hydrogen bonding distance of the first (3.2 Å), but is not within hydrogen-bonding distance of any potential donor or acceptor on the protein.
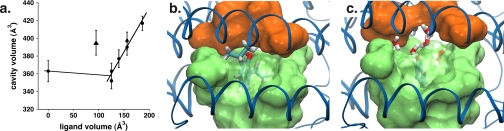
Addition of ligands to the cavity does not alter the global structure of apoferritin appreciably. The r.m.s. difference in Cα positions between any of the liganded structures and unliganded apoferritin is ∼0.1 Å, comparable to the expected coordinate error (31), and r.m.s. differences for all atoms of the residues lining the cavity are still quite small, 0.4–0.7 Å (Table 3). The volume of the cavity in the different structures ranges from 352 to 417 Å3; it appears independent of ligand volume for small ligands, but increases in direct proportion to ligand volume for larger ligands (Fig. 3). Phenol departs from this trend, with the apoferritin-phenol complex showing an unexpectedly large cavity volume; interestingly, this effect is predicted by the molecular dynamics simulations (vide infra).
FIGURE 3.
a, dependence of cavity volume upon ligand volume in the x-ray crystal structures of different apoferritin-ligand complexes. Circles correspond to unliganded apoferritin and the complexes with compounds 1, 5, 7, and 8; the triangle corresponds to the complex with compound 9, phenol. b and c, structures of the apoferritin complexes with propofol and phenol, respectively, obtained from molecular dynamics simulations. For panels b and c, the viewpoint is such that the top of the figure corresponds to the surface of the protein; each panel represents a single snapshot from the simulation. In both panels the surface defined by the side chains lining the sides of the cavity is shown as a transparent green surface, with the ligand visible within. The Arg-59 side chains are seen at the top of both panels, colored orange. Note how in the propofol structure (b), the Arg-59 side chains pack against the top of the ligand, whereas in the phenol-bound structure (c), the two arginine side chains arch upward, expanding the cavity and allowing additional water molecules to enter (shown in light gray and red).
Details of Specific Ligand Complexes
Propofol (1) sits in the anesthetic binding cavity with the plane of its aromatic ring almost parallel to the crystallographic 2-fold symmetry axis (Fig. 2). The carbon-hydroxyl bond is tilted ∼30° away from the symmetry axis. The hydroxyl group points toward the top of the cavity, and toward Arg-59; however, the hydroxyl does not participate in a hydrogen bond or other polar interaction with any part of the protein. One of the isopropyl groups packs against the hydrophobic portions of the two Arg-59 side chains, the side chain of Ser-27, the side chain of Glu-63 and the backbone carbonyl oxygen of Ala-55; the other isopropyl group points into the interior of the cavity, and most closely approaches the side chain of the symmetry-related Ser-27 and the carbonyl oxygen of Leu-24. The aromatic ring is not positioned so as to allow the π electrons to form any specific interactions with the protein; the closest protein moieties to the ring are the backbone and side chain of Tyr-28 and the side chains of Leu-81, which approach to within 4 Å of the aromatic ring. No water molecules are seen in the cavity together with propofol, which presumably displaces all four of the waters found in the unliganded apoferritin cavity. As was the case with all of the ligands, the positions of the propofol molecule and its symmetry mate overlap extensively, and so only one ligand may occupy any given cavity.
Other 2,6-Substituted Phenols
The two di-substituted analogs 5 and 8 adopt orientations within the cavity that are roughly similarly to that of propofol. For both of the ligands, one alkyl substituent packs against the Ser-27 side chain and the alkyl portions of the symmetry-related Arg-59 residues, while the other points inward into the cavity, lying between the two symmetry-related copies of Leu-81. The aromatic rings of both 5 and 8 lie close to both backbone and side chain atoms of Tyr-28 and to the side chain of Leu-81, again similar to the propofol complex. Compounds 5 and 8 also displace all waters observed in unliganded apoferritin. Neither of these compounds uses its hydroxyl group to form a hydrogen bond with the protein; the closest hydrogen bond partners are the guanidino nitrogens of Arg-59 and the Ser-27 hydroxyl, all of which are at least 4 Å away from the phenolic hydroxyl oxygen.
Smaller Ligands
The monosubstituted ligand 7 is oriented like the disubstituted ligands, with the isopropyl group occupying the same position as one of the isopropyl substituents of propofol. However, the absence of a substituent at the 6-position of the ring allows two of the four waters seen in the unliganded structure to remain in the cavity. These waters appear to bridge the ligand to the protein, forming hydrogen bonds with the ligand hydroxyl group and the side chains of Ser-27 and Arg-59.
Phenol (9) binds in the same cavity as the other ligands, but is oriented differently, with its hydroxyl group pointing toward the side of the cavity, not toward Arg-59 and the top of the cavity as is seen with the other ligands. This orientation allows phenol to form a hydrogen bond with the Ser-27 hydroxyl, making it the only ligand studied that forms a direct polar interaction with apoferritin. Like 7, phenol displaces half of the waters seen in the unliganded structure. Unlike the case with 7, the remaining water molecules do not form hydrogen bonds with phenol, as modeled in our structure. However, the small size of phenol may allow it to rotate relatively freely within the cavity; indeed, the molecular dynamics studies described below suggest that phenol can alternate between hydrogen bonding to Ser-27 and to waters within the cavity. Like all of the other ligands studied, there was no evidence for polar interactions between the protein molecule and the π-electrons of the phenol aromatic ring.
Molecular Dynamics
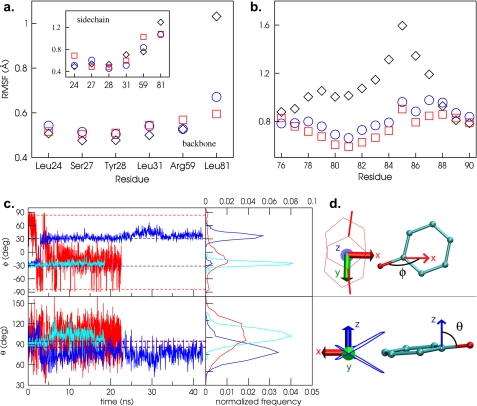
Molecular dynamics simulations were conducted to investigate the dynamics of the ligand within the apoferritin binding site and to judge consequences of binding on protein structure and dynamics. Once equilibrated, the structure of the apoferritin dimer remains stable over the 20–40-ns time scale of our simulations, and the overall shape and size of the anesthetic binding cavity do not change throughout the simulations. Ligand binding affects the dynamic behavior of most of the cavity-forming residues, causing some to become less mobile while others show increased dynamic behavior in the presence of ligands (Fig. 4).
FIGURE 4.
Protein and ligand dynamics from molecular dynamics simulations. a, RMSF for backbone (main panel) and side chain (inset) atoms of cavity-forming residues over the course of the simulations. Black diamonds, no ligand; red squares, phenol; blue circles, propofol. b, RMSF values for backbone atoms in strand connecting helices 2 and 3. c, orientation of the ligands versus time, as characterized by the polar angle ϕ and the azimuthal angle θ (defined in panel d). Red, phenol simulation; blue and cyan, propofol A and B simulation, respectively. The orientations seen in the crystal structures are represented by the symmetric dashed lines. Probability distributions extracted from the simulation data are shown in the right hand panel. d, definition of ϕ and θ. ϕ is the angle between the ligand C-O bond and the 2-fold symmetry axis that relates the two protomers that form the binding site. θ is the angle between the ligand C-O bond and the z axis, where z is defined as the normal to a plane that lies exactly halfway between the planes of the aromatic groups of the two symmetry-related ligand positions seen in the crystal structure.
Arg-59 shows markedly higher r.m.s. fluctuations in the presence of phenol than with propofol or no ligand. In the propofol and control simulations, the Arg-59 side chains form a tight cap to the cavity. However, when phenol is bound, one of the Arg-59 side chains tilts away from the cavity for an extended period, increasing the cavity volume. This movement appears to reflect the polarity of the phenol ligand: when Arg-59 tilts away from the cavity several waters (typically between 4 and 8) enter the cavity and hydrate the phenol hydroxyl. Cavity swelling caused by ligand hydration likely explains the anomalously large cavity volume seen in the crystal structure of the phenol complex (Fig. 3). Large numbers of waters are not seen sharing the cavity with phenol in the crystal structure, but this is not unexpected because the simulations suggest the water is not ordered.
Leu-24, which forms two sides of the cavity, is another residue that shows increased mobility in the phenol simulation, as compared with the control or propofol simulations. Phenol induces a rotation of the side chain χ2 dihedral that packs the leucine side chain against the ligand aromatic ring, shielding it from water in the cavity. Since the concentration of water in the cavity is higher when phenol is bound than when propofol is present, this shielding is expected to be more advantageous for phenol than in the presence of propofol (lower water concentration, hence less need for shielding) or in the control (no ligand to shield).
Leu-81 is a residue for which ligand binding reduces motion. This residue is located in the crossover strand that connects the second and third helices. Ligand binding significantly reduces the mobility of both the backbone and side chain of Leu-81; in fact, backbone fluctuations are dampened for the entire strand (Fig. 4). The ligands insert their aromatic rings tightly between the two symmetry-related copies of the Leu-81 side chain, and this tight packing appears to be the cause of the reduction in protein mobility. The Leu-81 rotameric probability distributions are very similar for propofol and phenol, indicating that Leu-81 is not particularly sensitive to the substituents on the phenol ring and that the dominant driving force is attraction between the ligand ring and leucine methyl groups.
During the propofol simulations, the ligand showed substantial fluctuations about its equilibrium position, but the angle between the C-O bond and the symmetry axis (Φ) remained close to ±45 °, in reasonable agreement with the crystal structure (Fig. 4). Only one transition between the two symmetry-related sites was ever observed. Taken together with the narrow distribution of Φ around the values observed in the crystal structure, this suggests that there is a barrier to rotation of propofol within the cavity. However, the azimuthal or tilt angle, θ (defined in Fig. 4), shows a much broader distribution of values than Φ, indicating that while switching from one binding position to the other may be disfavored, tilting and other localized movements of propofol within the cavity are allowed.
The phenol simulation shows a much wider distribution in both the polar and tilt angles than propofol (Fig. 4), suggesting the ligand is essentially fluid within the cavity. The Φ distribution does show a small peak at the orientation specified by the crystal structure (± 82°), but there is a much larger peak around Φ −20°, corresponding to an orientation roughly analogous to that of propofol. Furthermore, there are very fast transitions between the preferred orientation and one occurring at Φ = 0 in which the phenol hydroxyl is pointed toward the cavity exit, allowing hydration of the hydroxyl, and in which the central phenol ring is wedged between the two Leu-81 side chains at the bottom of the cavity.
Throughout the course of the simulations, neither ligand showed a strong tendency to form hydrogen bonds with the protein residues lining the cavity. Propofol participated in hydrogen bonds less than 0.1% of the simulation time, while the hydroxyl of phenol participated in a hydrogen bond with the Ser-27 10% of the simulation time; no other hydrogen bonds were observed. Both ligands formed hydrogen bonds with water more frequently than to the cavity, forming at least one hydrogen bond with water 3% of the time for propofol and 73% of the time for phenol.
DISCUSSION
Propofol is now the most widely used of the injectable general anesthetics. It provides rapid induction of unconsciousness, swift emergence and few of the problems associated with other drugs, such as delirium and nausea (32). Its desired effects are thought to arise from potentiation of GABAergic transmission. Structure-activity relationships have been probed for various propofol analogs (33), but the atomic details of the structure-binding relationship, necessary to guide further drug design, have not been reported. To elucidate such details for the anesthetic-binding protein apoferritin, we examined a homologous series of compounds that all share the same phenolic skeleton, but differ in the size of the aliphatic substituents at the 2- and 6-positions.
The apoferritin system provides a clear example in which polar interactions are not required for specific, high-affinity recognition of propofol and related molecules. Apart from phenol, none of the compounds studied participate in direct polar interactions with the protein, even though multiple polar residues are available within the apoferritin cavity, and despite the fact that all of the compounds are capable of participating in hydrogen bonds and polar interactions involving their pi electrons. The molecular dynamics simulations show that the ligands prefer to form hydrogen bonds to water molecules in the cavity, rather than to their protein target. It is surprising that hydrogen bonding or other polar interactions do not play key roles in propofol recognition, given that the hydroxyl group has been considered an essential component of the propofol pharmacophore (34, 35). However, our results strongly suggest that the hydroxyl may serve primarily to enhance water solubility, rather than to mediate protein recognition.
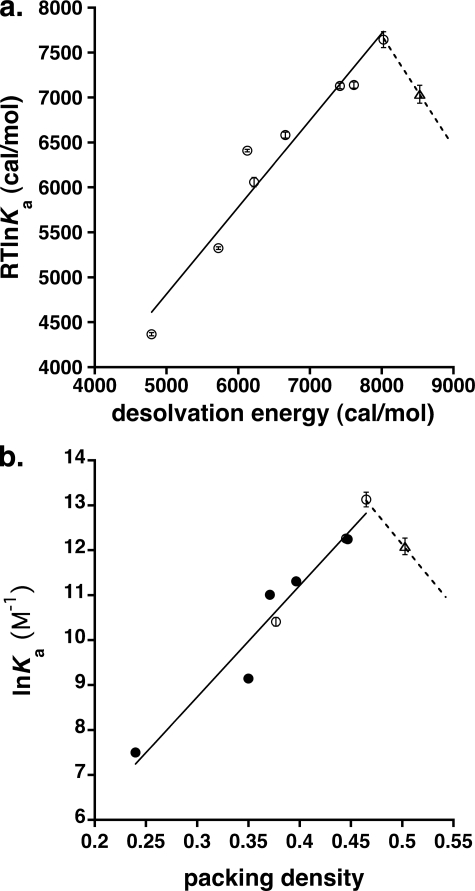
In the absence of strong polar interactions such as hydrogen bonds, affinity should be governed by enthalpic contributions from relatively weak van der Waals forces and entropic contributions from dehydration of the ligand and cavity upon binding. We estimated the free energy contribution from the latter effect using a value of 20 cal/mol per Å2 of hydrophobic surface area (36) and found excellent agreement with experimental values, indicating that the hydrophobic effect is the principal force driving anesthetic recognition by apoferritin (Fig. 5).
FIGURE 5.
a, calculated versus measured affinities of propofol analogs for apoferritin. The x axis shows the calculated binding energy for desolvation of a hydrophobic molecule, assuming a value of 20 cal/mol per Å2 of hydrophobic surface area (36), and the y axis shows the observed binding energy derived from the ITC data. The solid line represents a least-squares fit to all data points except the point corresponding to compound 2, which is represented by a triangle. b, natural logarithm of the binding affinity shown as a function of the packing density. The solid line represents a least squares fit to all data points except compound 2 (open triangle). Solid circles represent experimental data. Open circles and triangle were extrapolated or interpolated from experimental data. The dotted line is meant as an aid to the eye, suggesting that optimal binding occurs at a packing density of ∼0.48, and that affinity decreases for higher values of the packing density.
Of the ligands studied, only the largest compound, 2, departs from this trend, with a measured binding affinity that is markedly lower than predicted by the hydrophobic effect. This discrepancy probably reflects the entropic cost of immobilizing a large ligand in the absence of strong offsetting enthalpic contributions, and is entirely consistent with the results from our molecular dynamics simulations, which suggest that “optimal” ligands still exhibit a considerable degree of mobility within the cavity.
A similar enthalpy-entropy calculus governs the recognition of guest molecules by encapsulating hosts, which is also controlled by weak forces (37), and suggests that optimal affinity corresponds to a packing density of guests within the host of about 0.55, similar to the packing density of most organic liquids. Looser packing fails to fully exploit favorable enthalpic contacts between host and guest, while too-tight packing restricts guest motion and exacts too great an entropic cost. Packing densities for our apoferritin-ligand complexes range from 0.35 to ∼0.52, with binding affinity peaking at a packing density of 0.48 and decreasing for higher values. Considering that apoferritin cavity volume calculations include nooks inaccessible to ligand, these values likely underestimate the true packing density and are therefore in remarkable agreement with the predictions of the host-guest model (37) (Fig. 5). Unfortunately, larger ligands with higher predicted packing densities are only sparingly soluble in water, complicating experimental confirmation of this relationship.
To test generality of this relationship in another anesthetic-binding protein, we examined the packing density and binding affinity of propofol for HSA (13, 16). Propofol binds two distinct sites in HSA; an average Ka of 15, 300 m−1 has been estimated for the two sites (13), which is more than 10-fold weaker than the apoferritin affinity for propofol. Only one of the two HSA sites is a buried cavity and therefore permits volume determination (16); packing densities fall in the range 0.50–0.55. This is higher than the “optimal” value found for the apoferritin-ligand complexes, suggesting that the HSA lower affinity for propofol may stem in part from a too-small binding cavity. However, examination of other HSA-ligand complexes reveals that this cavity is capable of enlargement; the fact that it adopts the volume that it does in the HSA-propofol complex may signal that the free energy gained from a hydrogen bond formed between propofol and HSA partially compensates for the entropic cost of immobilizing the ligand.
Careful analysis of both the structure and the dynamics of the apoferritin model target reveal a complex interplay between anesthetic and protein. The crystal structures clearly show that the protein cavity is able to expand, at least to a limited degree, to accommodate larger ligands (induced fit), implying that models wherein anesthetics simply bind to preformed cavities may be overly simplistic. The molecular dynamics simulations complement this structural analysis, providing an explanation for the unexpectedly large cavity volume observed for the phenol-apoferritin complex. Given that the cavity occurs at a subunit interface, it is not difficult to hypothesize that expansions of similar cavities in the transmembrane regions of multisubunit ion channels could influence the energetics of conductance or gating.
The utility of complementing structural analyses with dynamics simulations is further demonstrated by the pronounced effects of anesthetic binding on residue and strand dynamics, observed even for the lowest affinity ligand. While crystallography does not identify significant global differences (other than cavity size) between unliganded and liganded apoferritin, molecular dynamics simulations reveal that the local effects of anesthetic binding translate into substantial global effects on protein dynamics, even at loci distant from the binding site (e.g. the crossover strand connecting helices 2 and 3). It is easy to envision how similar allosteric effects, triggered by anesthetic binding to LGIC targets, might modulate agonist affinity or channel function.
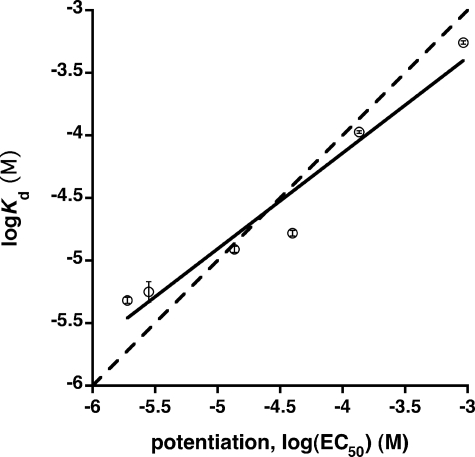
Because apoferritin is itself unlikely to transduce anesthesia, its utility as a model system lies in its ability to predict pharmacologically relevant anesthetic effects. Structure-function data for propofol and related compounds reveal a good correlation between anesthetic potency (as measured by loss of righting reflex in Xenopus laevis tadpoles) and the ability to potentiate GABA responses at GABAA receptors (33), suggesting a GABAergic mechanism for propofol-like molecules. It is therefore interesting to note that in the propofol series, binding affinity for apoferritin is strongly correlated with potentiation of GABA responses (Fig. 6). Further, this correlation approximates the line of identity, implying that occupancy of an apoferritin-like site in the GABAA receptor complex is sufficient for potentiation. A significant correlation is also observed between apoferritin binding affinity and anesthetic potency in tadpoles, although not as strong as with GABA potentiation. This is not surprising, given that systems level responses such as the loss of righting reflex almost certainly reflect contributions from multiple targets.
FIGURE 6.
Correlation between apoferritin binding affinity and the EC50 for potentiation of GABA responses at GABAA receptors (units are mol/liter). Binding affinity was measured by ITC and is expressed as the dissociation constant Kd; GABA potentiation data are taken from Ref. (33). The points shown correspond to compounds 1, 2, 5, 7, 8, and 9. The solid line represents a least squares fit to the data; a slope of unity (dashed line) lies within the 95% confidence interval for the slope of the regression line.
Even aside from these functional correlations, the notion that apoferritin can act as a model for the GABAA receptor is consistent with our current knowledge of LGIC architecture. Mutagenesis and photolabeling experiments (1–9), coupled with homology modeling (9), have established that the anesthetic binding site of the GABAA receptor lies within its transmembrane region, at or near the interface between subunit 4-helix bundles. Similarly, the apoferritin anesthetic binding site is found in an interfacial location, sandwiched between two four-helix bundles. Obviously, the structural homology cannot extend to the atomic level, because there is no primary sequence homology between apoferritin and the GABAA receptor, nor is the 2-fold symmetry axis found in apoferritin consistent with what we know of the architecture of the transmembrane domains of ligand-gated ion channels. Nonetheless, apoferritin displays a striking level of mimicry at the secondary, tertiary, and quaternary structural levels.
In conclusion, apoferritin binding affinity recapitulates anesthetic potency in a propofol-based homologous series. Recognition of these molecules, based almost entirely on van der Waals forces and the hydrophobic effect, occurs at the same binding site previously shown to bind inhalational general anesthetics such as halothane and isoflurane. When taken together with the structural mimicry described above, these results argue strongly that the apoferritin anesthetic binding site bears a high degree of physicochemical and architectural similarity to sites that exist in the GABAA receptor and other clinically relevant targets. Our results will allow the development of specific structural and dynamical hypotheses to explain anesthetic mechanisms within pharmacologically relevant ion channel targets.
Supplementary Material
Acknowledgments
We thank Vivian Stojanoff, Anand Saxena, and Lisa Keefe for access to data collection facilities. X-ray diffraction data for this study were measured at beamlines X6A and X25 of the National Synchrotron Light Source (NSLS) and the IMCA-CAT beamline 17-ID of the Advanced Photon Source at Argonne National Laboratory. Financial support for the NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Dept. of Energy, and from the National Center for Research Resources of the National Institutes of Health. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source is supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago.
This work was supported, in whole or in part, by National Institutes of Health Grants GM51595 and GM55876 (to R. G. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. S1 and S2.
- GABA
- γ-aminobutyric acid
- HSA
- human serum albumin
- HSAF
- horse spleen apoferritin
- ITC
- isothermal calorimetry
- LGIC
- ligand-gated ion channel
- r.m.s.
- root mean square
- RMSF
- root mean squared fluctuations.
REFERENCES
- 1.Mascia M. P., Trudell J. R., Harris R. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9305–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenkins A., Greenblatt E. P., Faulkner H. J., Bertaccini E., Light A., Lin A., Andreasen A., Viner A., Trudell J. R., Harrison N. L. (2001) J. Neurosci. 21, RC136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krasowski M. D., Nishikawa K., Nikolaeva N., Lin A., Harrison N. L. (2001) Neuropharmacology 41, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins A., Andreasen A., Trudell J. R., Harrison N. L. (2002) Neuropharmacology 43, 669–678 [DOI] [PubMed] [Google Scholar]
- 5.Siegwart R., Jurd R., Rudolph U. (2002) J. Neurochem. 80, 140–148 [DOI] [PubMed] [Google Scholar]
- 6.Chang C. S., Olcese R., Olsen R. W. (2003) J. Biol. Chem. 278, 42821–42828 [DOI] [PubMed] [Google Scholar]
- 7.Jurd R., Arras M., Lambert S., Drexler B., Siegwart R., Crestani F., Zaugg M., Vogt K. E., Ledermann B., Antkowiak B., Rudolph U. (2003) FASEB J. 17, 250–252 [DOI] [PubMed] [Google Scholar]
- 8.Schofield C. M., Harrison N. L. (2005) Brain Res. 1032, 30–35 [DOI] [PubMed] [Google Scholar]
- 9.Li G. D., Chiara D. C., Sawyer G. W., Husain S. S., Olsen R. W., Cohen J. B. (2006) J. Neurosci. 26, 11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiara D., Dangott L. J., Eckenhoff R. G., Cohen J. B. (2003) Biochemistry 42, 13457–13467 [DOI] [PubMed] [Google Scholar]
- 11.Miyazawa A., Fujiyoshi Y., Unwin N. (2003) Nature 423, 949–955 [DOI] [PubMed] [Google Scholar]
- 12.Franks N. P., Lieb W. R. (1984) Nature 310, 599–601 [DOI] [PubMed] [Google Scholar]
- 13.Liu R., Meng Q., Xi J., Yang J., Ha C. E., Bhagavan N. V., Eckenhoff R. G. (2004) Biochem. J. 380, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R., Loll P. J., Eckenhoff R. G. (2005) FASEB J. 19, 567–576 [DOI] [PubMed] [Google Scholar]
- 15.Franks N. P., Jenkins A., Conti E., Lieb W. R., Brick P. (1998) Biophys. J. 75, 2205–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya A. A., Curry S., Franks N. P. (2000) J. Biol. Chem. 275, 38731–38738 [DOI] [PubMed] [Google Scholar]
- 17.Butts C. A., Xi J., Brannigan G., Saad A. A., Venkatachalan S. P., Pearce R. A., Klein M. L., Eckenhoff R. G., Dmochowski I. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6501–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theil E. C. (2001) in Handbook of Metalloproteins ( Messerschmidt A., Huber R., Poulos T., Wieghardt K. eds),John Wiley & Sons, Ltd., Chichester [Google Scholar]
- 19.Takagi H., Shi D., Ha Y., Allewell N. M., Theil E. C. (1998) J. Biol. Chem. 273, 18685–18688 [DOI] [PubMed] [Google Scholar]
- 20.Arosio P., Levi S. (2002) Ferritins: Structural and Functional Aspects, Marcel Dekker, Inc., New York [Google Scholar]
- 21.James R., Glen J. B. (1980) J. Med. Chem. 23, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 22.Breslow R., Groves K., Mayer M. U. (2002) J. Am. Chem. Soc. 124, 3622–3635 [DOI] [PubMed] [Google Scholar]
- 23.Thomas B. R., Carter D., Rosenberger F. (1998) J. Crystal Growth 187, 499–510 [Google Scholar]
- 24.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 25.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 26.Ko S. H., Yu C. W., Lee S. K., Choe H., Chung M. J., Kwak Y. G., Chae S. W., Song H. S. (1997) Anesth. Analg. 85, 719–724 [DOI] [PubMed] [Google Scholar]
- 27.Granier T., Gallois B., Dautant A., Langlois , d'Estaintot B., Précigoux G. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 580–587 [DOI] [PubMed] [Google Scholar]
- 28.Hempstead P. D., Yewdall S. J., Fernie A. R., Lawson D. M., Artymiuk P. J., Rice D. W., Ford G. C., Harrison P. M. (1997) J. Mol. Biol. 268, 424–448 [DOI] [PubMed] [Google Scholar]
- 29.Gallois B., d'Estaintot B. L., Michaux M. A., Dautant A., Granier T., Precigoux G., Soruco J. A., Roland F., ChavasAlba O., Herbas A., Crichton R. R. (1997) J. Biol. Inorg. Chem. 2, 360–367 [Google Scholar]
- 30.Yoshizawa K., Mishima Y., Park S. Y., Heddle J. G., Tame J. R., Iwahori K., Kobayashi M., Yamashita I. (2007) J. Biochem. 142, 707–713 [DOI] [PubMed] [Google Scholar]
- 31.Cruickshank D. W. (1999) Acta Crystallogr D Biol Crystallogr. 55, 583–601 [DOI] [PubMed] [Google Scholar]
- 32.Kotani Y., Shimazawa M., Yoshimura S., Iwama T., Hara H. (2008) CNS Neurosci. Ther. 14, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krasowski M. D., Jenkins A., Flood P., Kung A. Y., Hopfinger A. J., Harrison N. L. (2001) J. Pharmacol. Exp. Ther. 297, 338–351 [PubMed] [Google Scholar]
- 34.Sewell J. C., Sear J. W. (2004) Br. J. Anaesth. 92, 45–53 [DOI] [PubMed] [Google Scholar]
- 35.Krasowski M. D., Hong X., Hopfinger A. J., Harrison N. L. (2002) J. Med. Chem. 45, 3210–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds J. A., Gilbert D. B., Tanford C. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 2925–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mecozzi S., Rebek J., Jr. (1998) Chem. Eur. J. 4, 1016–1022 [Google Scholar]
- 38.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 39.DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.