Abstract
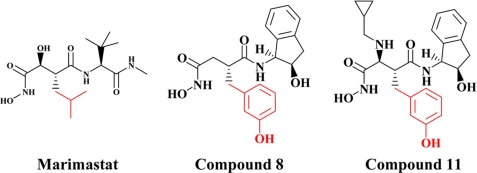
Several inhibitors of a series of cis-1(S)2(R)-amino-2-indanol-based compounds were reported to be selective for the aggrecanases, ADAMTS-4 and -5 over other metalloproteases. To understand the nature of this selectivity for aggrecanases, the inhibitors, along with the broad spectrum metalloprotease inhibitor marimastat, were independently bound to the catalytic domain of ADAMTS-5, and the corresponding crystal structures were determined. By comparing the structures, it was determined that the specificity of the relative inhibitors for ADAMTS-5 was not driven by a specific interaction, such as zinc chelation, hydrogen bonding, or charge interactions, but rather by subtle and indirect factors, such as water bridging, ring rigidity, pocket size, and shape, as well as protein conformation flexibility.
Osteoarthritis (OA)3 pathology includes degradation of articular cartilage, along with subchondral bone sclerosis and osteophyte formation, all contributing to impaired joint function. Pain, restricted movement, and joint instability accompany these structural changes and often result in the need for total joint replacement. Current therapies alleviate the mild to moderate pain and inflammation associated with OA, but do not protect the cartilage from further damage and have not demonstrated an effect on disease progression (1). Therefore, therapeutics that prevent or slow the alteration of joint structure and function will address a major unmet medical need.
Loss of aggrecan, a macromolecular proteoglycan providing cartilage with its properties of compressibility and resilience, is a major phenotype associated with OA and is believed to be a critical event in driving disease progression (2, 3). Both ex vivo and in vivo proof of concept studies support ADAMTS-4 and ADAMTS-5, commonly referred to as aggrecanase-1 and -2, respectively, as the two major enzymes responsible for the proteolytic breakdown of cartilage aggrecan (reviewed in Ref. 4). Blocking their activity may be an attractive strategy to stop or slow down the progression of the disease, as suggested by studies in knock-out mice (5). Given the chronic nature of the disease, long term treatment will be likely, demanding very safe therapeutic interventions only achievable with ADAMTS-4- and ADAMTS-5-specific inhibitors lacking off-target side effects. Designing selectivity has been very challenging and a major source of difficulty is that at least 57 metalloproteases (MP) divided in three major families, 1) matrix metalloproteases (MMP); 2) a disintegrin and metalloproteinase (ADAM); and 3) a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), are present in humans. ADAMTS-4 and -5 belong to the ADAMTS family and share common catalytic and structural features with the other MP members. These features include the highly conserved amino acid sequence, HEXXHXXGXXH, harboring a catalytic zinc cation, required for activation of the peptide bond toward hydrolysis. In addition, many MPs share significant structural topology in the active site, such as a flexible S1′ loop. To complicate matters further, only a handful of MP structures, mostly in the MMP family, have been determined, and the functions of most MPs still remain unknown, a fact that has earned them an orphan status denomination. Lack of structural information has hindered the design for inhibitor specificity and without known substrates for many MPs, assays for screening inhibitors are often not available, making determinations of selectivity difficult (6, 7).
Yao et al. (8, 9) reported the discovery of a series of (2R)-N4-hydroxy-2-(3-hydroxybenzyl)-N1-[(1S,2R)-2-hydroxy-,3-dihydro-1H-inden-1-yl]butanediamide derivatives as potent and selective inhibitors of aggrecanase activity. Using a homology model of aggrecanase based on the active site of atrolysin C and adamalysin II and docking compound 8, shown in Fig. 1, the authors concluded that the 3-hydroxyl group of inhibitor 8 achieved selectivity through a specific hydrogen-bonding interaction with Thr440 (numerical numbering is based on the human sequence of ADAMTS-5) in the S1′ pocket of aggrecanase.
FIGURE 1.
Aggrecanase inhibitors. Structures of the inhibitors evaluated in these studies. The P1 moiety of each inhibitor is marked in red.
Whereas both ADAMTS-4 and -5 have a threonine at this position, MMP-1, -2, -3, -7, -8, -9, -10, -13, -14, -16, and ADAM-17 have a valine, lending credence to the proposed hypothesis around selectivity. Recently, our group has established a protocol for crystallizing the catalytic domain of ADAMTS-5 and determined its three-dimensional structure (10). Thus, experimental validation or invalidation of the hypothesis that inhibitor 8 and related molecules form a specific hydrogen bond with the hydroxyl group of threonine in the S1′ pocket of aggrecanases could now be determined by protease-inhibitor crystallographic analysis.
In the current study, we wanted to confirm and extend the selectivity profile of compound 8 and 11 against a wide array of MPs. Moreover, we wanted to elucidate the key molecular interactions responsible for the enhanced selectivity profile of this series of compounds. For this purpose we generated co-crystals and solved the structures of marimastat, compound 8, and compound 11 in complex with the catalytic domain of recombinant human ADAMTS-5.
EXPERIMENTAL PROCEDURES
Bovine Articular Cartilage (BAC) Assay
Cartilage was dissected from the metatarsophalangeal joints of young cows obtained from the slaughterhouse. Cartilage was allowed to equilibrate for 3 days in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 units/ml), and streptomycin (100 μg/ml), all purchased from Invitrogen (Carlsbad, CA). Subsequently, cartilage was cut into 3 × 3-mm explants, weighing ∼10–20 mg each, and incubated in 96-well plates for 48 h with either control medium (serum-free DMEM), medium containing interleukin-1β (IL-1β produced at Pfizer, as described, Ref. 11) (100 ng/ml) or medium containing IL-1β (100 ng/ml) plus marimastat, compound 8, or compound 11 at 30 to 10,000 nm. At the end of the culture period, supernatants were collected and frozen at −20 °C until further assay. Glycosaminoglycan content of the supernatants was determined using the dimethylmethylene blue assay, as described by Farndale et al. (12). Shark chondroitin sulfate was used as a standard, and results were expressed as μg of GAG per mg of wet weight cartilage.
Aggrecanase Inhibition Assay
Inhibition of ADAMTS-4 or ADAMTS-5 cleavage at the Glu373/Ala374 site of aggrecan was measured as follows. Full-length recombinant ADAMTS-4 and -5 were expressed in SF9 cells and purified as previously described (13, 14). Reaction mixtures of 100-μl final volume in 96-well polypropylene plates contained purified bovine aggrecan, 0.2 nm ADAMTS-4 or ADAMTS-5, and inhibitor or 1.0% dimethyl sulfoxide vehicle control in 50 mm Tris buffer at pH 7.5, containing 100 mm NaCl and 5 mm CaCl2. The concentration of aggrecan was 125 nm for IC50 determinations. After a 6-h incubation at 37 °C, the reaction was stopped with the addition of 10 μl of 0.5 m EDTA, and 75 μl were transferred to a 96-well polyvinylidene difluoride (PVDF) membrane plate (Millipore, Billerica, MA) containing 75 μl of 20 mm carbonate-bicarbonate buffer at pH 9.6 (Sigma). Prior to the addition of the carbonate-bicarbonate buffer, the PVDF plates had been conditioned for 5 min with 100 μl 70% ethanol in water and washed two times with 200 μl of water. Plate washing was facilitated by using a Tecan Columbus Plus plate washer (Research Triangle Park, NC) modified with the addition of a vacuum manifold by Flush Tec (Cathedral City, CA). The samples were allowed to bind to the PVDF plates overnight at room temperature, washed twice with 200 μl of Tris-buffered saline (TBS) (Bio-Rad product 170-6435), and incubated for 3 h at 37 °C with 0.1 units/ml chondroitinase ABC, 0.1 units/ml keratanase, and 0.01 units/ml keratanase II (Associates of Cape Cod, East Falmouth, MA), in 100 μl of 50 mm Tris, 100 mm sodium acetate buffer at pH 6.5, containing 1% bovine serum albumin (BSA). The plates were washed twice with 200 μl of TBS and incubated for 1 h at room temperature with 2 μg/ml BC-3 antibody and 1 μg/ml goat anti-mouse IgG conjugated to alkaline phosphatase (Promega, Madison, WI) in 100 μl TBS containing 1% BSA buffer. The monoclonal neoepitope antibody BC-3 recognizes the new N terminus 374ARGS on aggrecan fragments produced by cleavage at the Glu373/Ala374 bond (15) and was licensed from Dr. B. Caterson (University of Wales, Cardiff, UK). The plate was then washed three times with 200 μl of TBS, and alkaline phosphatase was detected by the addition of 100 μl of p-nitrophenyl phosphate substrate (Sigma-Aldrich Co., product P7998). After 20 min of incubation at room temperature, the reaction was stopped with the addition of 100 μl of 0.5 m EDTA, and 150 μl were transferred to a flat-bottomed 96-well transparent plate (Corning) for reading absorbance at 405 nm.
Metalloproteinase Inhibition Assays
Full-length MMP-1, MMP-2, MMP-3, MMP-9, MMP-13, and catalytic domain MMP-7, MMP-8, and MT1-MMP were expressed, purified, and activated as referenced previously (16). Recombinant catalytic domain of human MMP-10 and MMP-26 were expressed and purified in similar manner (16, 17). The catalytic domain of MMP12 was purchased from R and D Systems (Minneapolis, MN). MT2-MMP (MMP-15) (catalytic domain), MT3-MMP (MMP-16) (catalytic domain), and MT5-MMP (MMP-24) were purchased from Chemicon International (Temecula, CA); ADAM-17 was purchased from Oncogene Science. Marimastat was synthesized as described by Pratt et al. (18). The fluorogenic, methoxycoumarin-containing polypeptide substrate, Mca-Arg-Pro-Leu-Gly↓Leu-Dpa-Ala-Arg-Glu-Arg-NH2 was used for all MMP enzymes except MMP3, for which Mca-Arg-Pro-Lys-Pro-Val-Glu↓Nva-Trp-Arg-Lys(Dnp)-NH2 (Bachem M-2110) was used. The substrate Mca-Pro-Leu-Ala-Gln-Ala↓Val-Dpa-Arg-Ser-Ser-Ser-Arg-NH2 (Bachem M-2255) was used for ADAM-17, Here, “Mca” is 7-methoxycoumarin-4-yl acetyl, “Dpa” is N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl, and “Dnp” is 2,4-dinitrophenyl. The site of cleavage by the enzyme in indicated by ↓. Cleavage separates the highly fluorescent methoxycoumarin moiety from the 2,4-dinitrophenyl quencher, resulting in increase of fluorescent intensity. The assay was performed at room temperature using the buffer and general procedure of Knight et al. (19) except that the Brij-35 concentration was 0.005%. After a 60-min preincubation of enzyme with inhibitor, substrate was added to 4 μm final concentration and the initial rate of fluorescence increase determined. Apparent inhibition constants were determined by non-linear regression of reaction velocity as a function of inhibitor and enzyme concentration using the two-stage approach of fitting the Morrison equation described by Kuzmic et al. (20). Inhibition constants (Ki values) were calculated by correcting the apparent constants for the effect of substrate competition using the relationship of Cheng and Prusoff for competitive inhibitors (21) .
Co-crystallization of Compounds with ADAMTS-5
A previously reported procedure from our group was adopted for ADAMTS-5 catalytic domain refolding from Escherichia coli inclusion bodies, and for its purification to homogeneity (10). Crystallization and crystal growth were performed as previously described (10). Specifically, drops of equal ratio were set up by the sitting drop method with 10 mg/ml ADAMTS-5 in 25 mm Hepes, pH 7.1, 5 mm CaCl2, and 6 mm compound against a reservoir containing 30% PEG 3350, 0.1 m Tris, pH 8.5. After 2–3 weeks, crystals reached full size and were stabilized in a drop containing 33% PEG 3350, 0.1 m Tris pH 8.5 for at least 2 h. Crystals were then transferred to soaks containing 2 mm marimastat, compound 8, or compound 11 in stabilization solution. The new compounds displaced compound 1 (10) within 2 days to 1 week. Cryo-protection was performed by adding 20% ethylene glycol directly to the drop prior to freezing in liquid nitrogen.
Structural Analysis
The crystal information, x-ray data, refinement, and final structure statistics of these five complex structures are listed in Table 1. Data were collected using synchrotron radiation in the IMCA-CAT beamlines 17ID and 17BM of the Advanced Photon Source at Argonne National Lab. The computer programs used for data process, refinement, ligand docking, and electron fitting are HKL2000, REFMAC, AFITT, and COOT, respectively.
TABLE 1.
Data collection and refinement statistics
| TS5/Marimastat | TS5/Compound 8 | TS5/Compound 11 | MMP13/Marimastat | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21212 |
| Cell dimensions | 52.61, 44.46, 76.35 (Å) | 52.82, 44.49, 76.66 (Å) | 52.69, 44.44, 76.46 (Å) | 119.83, 93.00, 37.00 (Å) |
| β = 89.81° | β = 90.26° | β = 89.81° | ||
| Z | 4 | 4 | 4 | 8 |
| Resolution (Å) | 50.0-1.70 (1.76-1.70)a | 30.0-1.40 (1.45-1.40)a | 50.0-2.02 (2.09-2.02)a | 30.0-2.40 (2.49-2.40)a |
| Total unique reflections | 39599 | 70352 | 23516 | 16839 |
| Rmerge | 0.059 (0.350) | 0.060 (0.259) | 0.044 (0.098) | 0.107 (0.417) |
| I/σI | 19.4 (2.3) | 24.1 (3.3) | 29.1 (7.8) | 10.4 (2.1) |
| Completeness (%) | 96.9 (82.3) | 99.5 (97.3) | 99.8 (98.2) | 95.2 (03/5) |
| Redundancy | 3.4 (2.1) | 3.6 (3.3) | 3.6 (2.1) | 3.8 (3.5) |
| Refinement | ||||
| Resolution (Å) | 26.31-1.70 | 25.55-1.40 | 26.31-1.70 | 28.51-2.40 |
| No. reflections | 39569 | 70344 | 23500 | 16834 |
| Rwork/Rfree | 0.173/0.224 | 0.181/0.210 | 0.159/0.238 | 0.196/0.286 |
| No. non-hydrogen atoms (molecule A/molecule B) | ||||
| Protein | 1668/1668 | 1668/1668 | 1668/1668 | 1304/1304 |
| Ligand | 23/23 | 27/27 | 32/32 | 23/23 |
| Ion | 4/4 | 4/4 | 4/4 | 6/6 |
| HEPES | ||||
| Sulfate | ||||
| Water | 284/253 | 293/294 | 238/249 | 189/151 |
| B-factors (average) (molecule A/molecule B) | ||||
| Protein (backbone atoms only) | 18.29/17.35 | 13.88/10.70 | 19.45/15.54 | 34.21/35.88 |
| Ligand | 20.84/20.76 | 14.71/12.82 | 28.16/26.37 | 20.84/21.79 |
| Ion | 14.29/13.56 | 9.88/7.80 | 14.88/13.57 | 43.41/49.04 |
| HEPES | ||||
| Sulfate | ||||
| Water | 35.71/34.18 | 33.79/28.31 | 36.23/31.08 | 40.47/42.22 |
| R.m.s. deviations | ||||
| Bond length (Å) | 0.009 | 0.009 | 0.010 | 0.009 |
| Bond angles (°) | 1.2 | 1.4 | 1.4 | 1.2 |
a Values in parentheses are for highest resolution shell. The data were obtained using one crystal.
RESULTS AND DISCUSSION
Potency and Selectivity Profile of Compounds against a Panel of Metalloproteases
Yao et al. (8, 9) showed 2 orders of magnitude increased selectivity of compounds 8 and 11 for endogenous aggrecanase activity, using a partially purified preparation from IL-1β stimulated bovine cartilage, over MMP-1, -2, -8, and -9. In the current study, both compounds were screened against recombinant human ADAMTS-4 and -5, the proteases responsible for the endogenous aggrecanase activity observed in IL-1β stimulated bovine cartilage, as well as an extensive range of other MP, including the soluble MMPs (MMP-1, -2, -3, -7, -8, -9, -10, -12, -13, -26), the membrane associated MMPs (MT1-, MT2-, MT3-, and MT5-MMP) and ADAM-17. The potency and selectivity profile of compounds 8 and 11 were compared with marimastat, a known non-selective, broad spectrum metalloprotease inhibitor (Fig. 1 and Table 2).
TABLE 2.
Potency and selectivity profile of marimastat, compound 8, and compound 11 against a panel of metalloproteases
Ki values are reported for each MP with the exception of ADAMTS-4 and -5 and BAC where IC50 values are reported.
| Metalloproteinase | Km | Calculated Ki values |
||
|---|---|---|---|---|
| Marimastat | Compound 8 | Compound 11 | ||
| μm | nm | |||
| ADAMTS-4 (IC50) | 79 | 65 | 3 | |
| ADAMTS-5 (IC50) | 106 | 93 | 17 | |
| BAC (IC50) | 5,290 | 1,700 | 624 | |
| Assayed with substrate 1a | ||||
| MMP-1 | 16 | 0.2 | >8,000 | >8,000 |
| MMP-2 | 17 | 0.2 | >8,095 | 4,995 |
| MMP-7 | 26 | 1.3 | >8,667 | 3,519 |
| MMP-8 | 20 | 0.2 | 3,800 | 321 |
| MMP-9 | 5 | 0.4 | 4144 | 1,261 |
| MMP-10 | >100 | 18 | >10,000 | NDb |
| MMP-12 | 22 | 0.2 | 553 | ND |
| MMP-13 | 4 | 0.3 | >5,000 | 2,970 |
| MMP-26 | >100 | 0.4 | 3,980 | ND |
| MT1-MMP | 8 | 1.1 | 5,300 | >6,667 |
| MT2-MMP | 17 | 1.1 | 6,387 | ND |
| MT3-MMP | 7 | 0.4 | 6,077 | ND |
| MT5-MMP | 12 | 0.8 | 5,228 | ND |
| Assayed with substrate 2c | ||||
| MMP-3 | 13 | 15 | >7,647 | 4,680 |
| Assayed with substrate 3d | ||||
| ADAM-17 | 66 | 11 | 5,572 | ND |
a Substrate 1: Mca-Arg-Pro-Leu-Gly↓Leu-Dpa-Ala-Arg-Glu-Arg-NH2.
b ND, not determined.
c Substrate 2: Mca-Arg-Pro-Lys-Pro-Val-Glu↓Nva-Trp-Arg-Lys(Dnp)-NH2.
d Substrate 3: Mca-Pro-Leu-Ala-Gln-Ala↓Val-Dpa-Arg-Ser-Ser-Ser-Arg-NH2.
Compound 8 inhibited both ADAMTS-4 and -5 with IC50 values of 65 and 93 nm, respectively, potencies comparable to marimastat. However, unlike marimastat, compound 8 showed a significant degree of selectivity over every other MP tested in the panel, ranging from 10- to >100-fold, whereas marimastat inhibited every MP in the panel with similar affinity. Compound 11 demonstrated increased potency over compound 8 against both ADAMTS-4 and -5, with IC50 values of 3 and 17 nm, and like compound 8 showed approximately one to two orders of magnitude selectivity against many other MPs tested. Collectively, the screening data confirm that the series of molecules previously described by Yao (8, 9) are indeed selective for aggrecanases over other MPs.
To confirm the potency of these compounds against endogenous aggrecanase activity, each molecule was tested for efficacy in blocking aggrecan breakdown/release in live BAC explants following stimulation with IL-1β. Previously, we and others have shown that the majority of aggrecanase activity produced in bovine cartilage following IL-1β treatment is a combination of both ADAMTS-4 and -5, with ADAMTS-4 accounting for ∼70% and ADAMTS-5 for ∼30% of the total activity (22). The most efficacious molecule in blocking aggrecan catabolism was compound 11, followed by compound 8 and then marimastat, with IC50 values of 0.6, 1.7, and 5.3 μm, respectively (Table 2). Not surprisingly, the rank order of efficacy in the BNC assay was consistent with the rank order of potency against both ADAMTS-4 and -5 enzyme activity (Table 2), which is in step with the hypothesis that ADAMTS-4 and -5 are the major aggrecanases responsible for aggrecan breakdown in stimulated bovine cartilage. The large difference between the potencies of marimastat, compound 8, and compound 11, for inhibition of ADAMTS-4 and -5 and for inhibition in the cartilage degradation assay has also been observed for other inhibitors. Although assumed to be due to competition by high levels of endogenous substrate, a recent report suggests that inhibitors that are competitive with small peptide substrates may in contrast appear noncompetitive with the native aggrecan substrate (23). This suggests that other factors, such as nonspecific protein binding or a hindered ability of the inhibitor to diffuse into the cartilage matrix, may be important for the observed potency differences.
Structural Analysis of Marimastat, Compound 8, and Compound 11 in Complex with the Catalytic Domain of ADAMTS-5
Homology model studies by Yao et al. (8, 9) based on atrolysin C and adamalysin II predicted that the 2-OH groups from compound 8 or 11 interacted with Thr440. To determine whether compounds 8 and 11 achieve their increased selectivity for ADAMTS-4 and -5 over other MP via hydrogen bonding to Thr440 as hypothesized, the structures of marimastat, compound 8 and compound 11 in complex with ADAMTS-5 were generated. From the high resolution co-crystal structures, a detailed analysis of the molecular interactions of each molecule with the active site of ADAMTS-5 was determined as well as insights gained on the molecular selection and recognition of compounds 8 and 11.
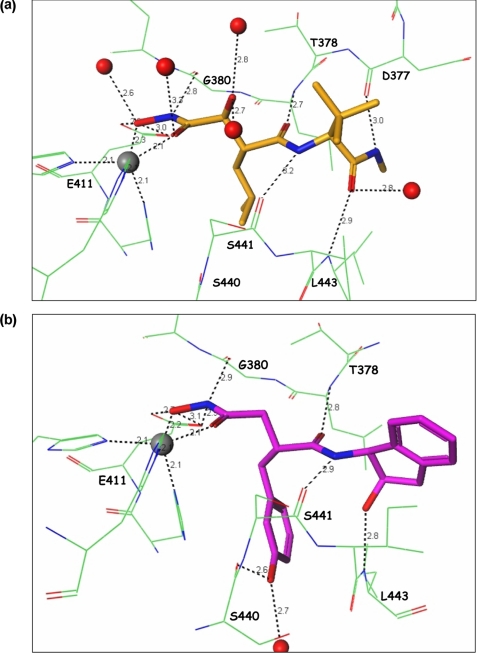
As expected and described previously, the hydroxamate group of marimastat, compound 8, and compound 11 coordinated with the catalytic zinc and hydrogen bonded to Glu411 in the active site of ADAMTS-5 (Fig. 2, a–c). However, unlike that predicted by the homology model based on atrolysin C and adamalysin II, neither 2-OH groups from compound 8 or 11 interacted with Thr440, but instead formed several hydrogen-bonding interactions with the protein backbone of ADAMTS-5. Based on sequence homology, similar interactions with marimastat and compounds 8 and 11 toward ADAMTS-4 are expected.
FIGURE 2.
Crystal structure of ADAMTS-5 with inhibitors. Illustration of the interactions between (a) marimastat, (b) molecule 8, and (c) molecule 11 with the active site of the catalytic domain of ADAMTS-5.
Significance of the Cyclopropyl-N-methymethanamine Chain
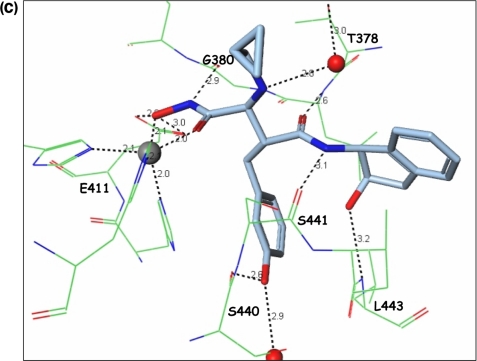
The only difference between compounds 8 and 11 is the cyclopropyl-N-methymethanamine chain at the Cα position from the hydroxamate group. In the structure, the cyclopropyl ring does not seem to engage any role, but the amino group of compound 11 forms a hydrogen bond with an adjacent water molecule, which links to Thr378 through another hydrogen bond (Fig. 3). This indirect interaction linking the inhibitor molecule to the protein probably explains the additional affinity of molecule 11. In addition, the Thr378 residue is quite unique to ADAMTS members and probably confers some selectivity to the molecule. In most MMPs, the corresponding residue is a nonpolar amino acid.
FIGURE 3.
Interactions of the cyclopropyl chain. Structure depicting the imino group of molecule 11 forming a hydrogen bond with an adjacent water molecule and linking to Thr378 by way of another hydrogen bond.
Potency and Selectivity from the 2-Indanol Ring
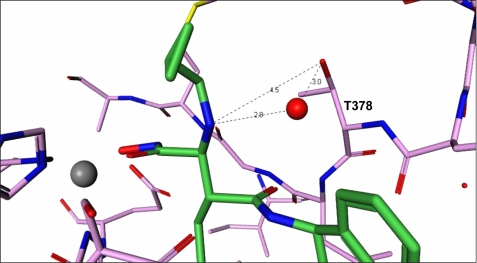
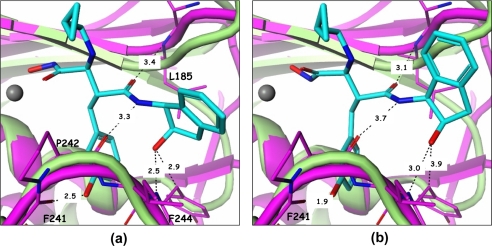
From the 2-indanol and on, inhibitor molecules 8 and 11 form a netted hydrogen bond network (Fig. 2, b and c). The hydroxyl group of the indanol ring forms a hydrogen bond with the backbone -NH- of Leu443. The amide -NH- in the inhibitor connects with the backbone carbonyl group of Ser441, the amide carbonyl group hydrogen bonds with the backbone -NH- of Leu379, and the meta-hydroxyl of the phenyl ring makes additional hydrogen bonds with Ser440 and water. This hydrogen-bonding network rigidifies the conformation of this part of the molecule, making it difficult for compounds 8 and 11 to fit into different pockets with different shapes and is likely a major reason why these compounds exhibit selectivity toward other MPs (Table 1). In fact, the low affinity of these two compounds toward MMP-13 explains the lack of success in attempts to crystallize them with this enzyme. The superposition of the MMP-13 structure (PDB code 830c) with that of ADAMTS-5 in complex with compound 11 (Fig. 4a) clearly shows that the hydroxyl group from the indanol ring is in close contact with the Cα atom of Phe244 in MMP-13. In addition, the hydrogen bonds generated from the middle amides of compound 11 are weak. Modeling efforts based on other similar structures4 were able to place compound 11 with a slightly different conformation and in a slightly different location in the MMP-13 pocket as shown in Fig. 4b. In this configuration, the contact with Phe244 is totally eliminated, but the meta-hydroxyl group in the phenyl ring creates a serious clash with the carbonyl group of the Phe241 backbone. The hydrogen bonds of the middle amides are still very weak in this configuration, thus the conclusion from these studies is that the indanol hydroxyl and the phenyl meta-hydroxyl groups of compound 11 cannot simultaneously fit into the MMP-13 pocket properly. This conclusion can also be reached for MMP-1 (PDB code 966c) and likely extended to other MPs. Thus, structural studies conducted herein do not support selectivity arising from the binding of the meta-hydroxyl group with the ADAMTS-specific Thr440 as proposed earlier (8, 9).
FIGURE 4.
Compound 11 harbored in the pockets of ADAMTS-5 and MMP-13. a, direct superposition of ADAMTS-5 (green) and MMP-13 (PDB code 830c) reveals that compound 11 will clash with Phe244 in MMP-13 (purple). b, modeling based on other similar in-house structures shows that compound 11 can release from the close contact with Phe244, but generates a more serious clash with the carbonyl backbone of Phe241.
Recognition of the P1′ Moiety
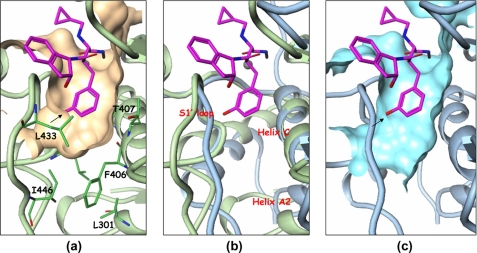
The S1′ pocket, also referred to as the specificity pocket, is the recognition site for the different substrates for metalloproteinases. Marimastat, shown in Fig. 1, has a very short P1′ moiety, hence it can be recognized by most of the metalloproteinases. The S1′ pocket in ADAMTS-5 is enclosed by a flexible loop including residues 426–436, the helix C defined by residues 402 to 416, and part of helix A2. Several key residues, Leu438, Leu443, and Ile446 in the flexible loop and Phe406, Thr407, His410, and Leu301 in the helices help shape/form the pocket, as shown in Fig. 5a. The back side of the pocket in Fig. 5a is walled off essentially by the zinc-chelating His410 residue. The most significant feature of the ADAMTS-5 pocket is its shallowness due to the presence of Leu438, Ile446, Leu301, and Phe406. Another feature is the restriction on the left side of the pocket due to the S1′ loop backbone. The S1′ loop of ADAMTS-5 is smaller compared with the loop found in MMP-13 since it is two residues shorter, making penetration through the loop difficult. The bigger S1′ loop found in MMP-13 adopts a different conformation and its helix A2 tilts away from the pocket as shown in Fig. 5b, in such a manner as to open up the channel. The channel pocket in MMP-13 actually extends through the loop and reaches out into the solvent region. However, the extension point does not start from the meta-position of the terminal phenyl ring, but rather from its para-position as seen in Fig. 5c. The para-position exhibits more space, and the extension from this position has more room to maneuver to find the best fit with the protein in other parts of the molecule. It will not be tethered to a limit or prohibited area as in the meta-position. Many para-extended compounds in this series of compounds do show strong affinity toward MMP-13.4
FIGURE 5.
The S1′ pocket of ADAMTS-5 and MMP-13. a, compound 11 in the S1′ pocket of ADAMTS-5. b, comparison of the S1′ pockets of ADAMTS-5 with MMP-13 (PDB code 830c). ADAMTS-5 is shown in pale green and MMP-13 in pale blue. Compound 11 is kept as a reference. c, S1′ pocket of MMP-13 with compound 11 positioned as a reference.
Conclusion
These studies pave the way for the design and development for potent and selective inhibitors of ADAMTS-4 and -5, which will be required for chronic OA therapy.
Acknowledgments
Use of the IMCA-CAT beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago.
The atomic coordinates and structure factors (codes 3HY7, 3HYG, and 3HY9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
H. S. Shieh, unpublished findings.
- OA
- osteoarthritis
- ADAMTS
- a disintegrin and metalloproteinase with thrombospondin motifs
- ADAM
- a disintegrin and metalloproteinase
- MMP
- matrix metalloproteinases
- BNC
- bovine nasal cartilage
- IL-1β
- interleukin-1β
- PEG
- polyethylene glycol
- PDB
- Protein Data Bank
- R.m.s.
- root mean square
- BAC
- bovine articular cartilage.
REFERENCES
- 1.Felson D. T., Kim Y. J. (2007) Arthritis Rheum. 56, 1378–1383 [DOI] [PubMed] [Google Scholar]
- 2.Mankin H. J., Lippiello L. (1970) J. Bone Joint Surg. 52, 424–434 [PubMed] [Google Scholar]
- 3.Pratta M. A., Yao W., Decicco C., Tortorella M. D., Liu R. Q., Copeland R. A., Magolda R., Newton R. C., Trzaskos J. M., Arner E. C. (2003) J. Biol. Chem. 278, 45539–45545 [DOI] [PubMed] [Google Scholar]
- 4.Tortorella M. D., Malfait A. M. (2008) Curr. Pharm Biotechnol. 9, 16–23 [DOI] [PubMed] [Google Scholar]
- 5.Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. (2005) Nature 434, 644–648 [DOI] [PubMed] [Google Scholar]
- 6.Hooper N. M. L., Uwe (ds). ( 2005) The ADAM Family of Proteases, Vol 4,Birkhaűser Verlag [Google Scholar]
- 7.Lagente V., Boichot E. e. (2008) Matrix Metalloproteinases in Tissue Remodelling and Inflammation, Vol. XI, Birkhaűser [Google Scholar]
- 8.Yao W., Chao M., Wasserman Z. R., Liu R. Q., Covington M. B., Newton R., Christ D., Wexler R. R., Decicco C. P. (2002) Bioorg Med. Chem. Lett 12, 101–104 [DOI] [PubMed] [Google Scholar]
- 9.Yao W., Wasserman Z. R., Chao M., Reddy G., Shi E., Liu R. Q., Covington M. B., Arner E. C., Pratta M. A., Tortorella M., Magolda R. L., Newton R., Qian M., Ribadeneira M. D., Christ D., Wexler R. R., DeCicco C. P. (2001) J. Med. Chem. 44, 3347–3350 [DOI] [PubMed] [Google Scholar]
- 10.Shieh H. S., Mathis K. J., Williams J. M., Hills R. L., Wiese J. F., Benson T. E., Kiefer J. R., Marino M. H., Carroll J. N., Leone J. W., Malfait A. M., Arner E. C., Tortorella M. D., Tomasselli A. (2008) J. Biol. Chem. 283, 1501–1507 [DOI] [PubMed] [Google Scholar]
- 11.Yem A. W., Curry K. A., Tomich C. S., Deibel M. R., Jr. (1988) Immunol. Invest. 17, 551–559 [DOI] [PubMed] [Google Scholar]
- 12.Farndale R. W., Buttle D. J., Barrett A. J. (1986) Biochim. Biophys Acta 883, 173–177 [DOI] [PubMed] [Google Scholar]
- 13.Abbaszade I., Liu R. Q., Yang F., Rosenfeld S. A., Ross O. H., Link J. R., Ellis D. M., Tortorella M. D., Pratta M. A., Hollis J. M., Wynn R., Duke J. L., George H. J., Hillman M. C., Jr., Murphy K., Wiswall B. H., Copeland R. A., Decicco C. P., Bruckner R., Nagase H., Itoh Y., Newton R. C., Magolda R. L., Trzaskos J. M., Burn T. C. (1999) J. Biol. Chem. 274, 23443–23450 [DOI] [PubMed] [Google Scholar]
- 14.Tortorella M. D., Burn T. C., Pratta M. A., Abbaszade I., Hollis J. M., Liu R., Rosenfeld S. A., Copeland R. A., Decicco C. P., Wynn R., Rockwell A., Yang F., Duke J. L., Solomon K., George H., Bruckner R., Nagase H., Itoh Y., Ellis D. M., Ross H., Wiswall B. H., Murphy K., Hillman M. C., Jr., Hollis G. F., Newton R. C., Magolda R. L., Trzaskos J. M., Arner E. C. (1999) Science 284, 1664–1666 [DOI] [PubMed] [Google Scholar]
- 15.Hughes C. E., Caterson B., Fosang A. J., Roughley P. J., Mort J. S. (1995) Biochem. J. 305, 799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tortorella M. D., Arner E. C., Hills R., Gormley J., Fok K., Pegg L., Munie G., Malfait A. M. (2005) Arch. Biochem. Biophys. 444, 34–44 [DOI] [PubMed] [Google Scholar]
- 17.Rosenfeld S. A., Ross O. H., Corman J. I., Pratta M. A., Blessington D. L., Feeser W. S., Freimark B. D. (1994) Gene 139, 281–286 [DOI] [PubMed] [Google Scholar]
- 18.Pratt L. M., Beckett R. P., Bellamy C. L., Corkill D. J., Cossins J., Courtney P. F., Davies S. J., Davidson A. H., Drummond A. H., Helfrich K., Lewis C. N., Mangan M., Martin F. M., Miller K., Nayee P., Ricketts M. L., Thomas W., Todd R. S., Whittaker M. (1998) Bioorg Med Chem Lett 8, 1359–1364 [DOI] [PubMed] [Google Scholar]
- 19.Knight C. G., Willenbrock F., Murphy G. (1992) FEBS Lett. 296, 263–266 [DOI] [PubMed] [Google Scholar]
- 20.Kuzmic P., Elrod K. C., Cregar L. M., Sideris S., Rai R., Janc J. W. (2000) Anal. Biochem. 286, 45–50 [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 22.Tortorella M. D., Malfait A. M., Deccico C., Arner E. (2001) Osteoarthritis and Cartilage 9, 539–552 [DOI] [PubMed] [Google Scholar]
- 23.Wittwer A. J., Hills R. L., Keith R. H., Munie G. E., Arner E. C., Anglin C. P., Malfait A. M., Tortorella M. D. (2007) Biochemistry 46, 6393–6401 [DOI] [PubMed] [Google Scholar]