Abstract
Toll/interleukin-1 (TIR)receptor-containing adapters are critical in orchestrating the different signal transduction pathways following Toll-like receptor (TLR) activation. MyD88 adapter-like (Mal), also termed TIRAP, is involved in bridging MyD88 to the receptor complex for TLR-2 and TLR4 signaling in response to bacterial infection. We have previously reported an interaction between Mal and tumor necrosis factor receptor-associated factor 6 (TRAF6) via a TRAF6-binding motif, the disruption of which inhibited TLR-mediated NF-κB-luciferase reporter activity. Given the recent report of intracellular TRAM localization promoting sequential signaling in TLR4 responses, we further characterized Mal interaction with TRAF6, the cellular localization, and the outcomes of disrupting this association on TLR inflammatory responses. We found that Mal and TRAF6 directly interact in response to TLR2 and TLR4 stimulation, although membrane localization is not necessary to facilitate interaction. Critically, reconstitution of murine Mal-deficient macrophages with MalE190A, containing a mutation within the TRAF6-binding motif, fails to reconstitute the proinflammatory response to TLR2 and TLR4 ligands compared with wild type Mal. Furthermore, Mal interaction with TRAF6 mediates Ser phosphorylation of the p65 subunit of NF-κB and thus controls transcriptional activation but not nuclear translocation of NF-κB. This study characterizes the novel role for Mal in facilitating the direct recruitment of TRAF6 to the plasma membrane, which is necessary for TLR2- and TLR4-induced transactivation of NF-κB and regulation of the subsequent pro-inflammatory response.
Toll-like receptors (TLRs)2 recognize and respond to both pathogen-associated molecular patterns and endogenous signals associated with danger (1). Upon ligand-induced dimerization the TLRs activate, via their cytosolic Toll/IL-1 receptor (TIR) domains, the homotypic recruitment of one or more proteins of a family of five cytosolic TIR-containing adapter proteins (2). All TLRs, with the exception of TLR3, recruit MyD88 to their receptor complex, as do members of the IL-1 receptor family. MyD88 recruits interleukin-1 receptor-associated kinase 1 (IRAK1), IRAK4, and then TNF receptor-associated factor 6 (TRAF6), which results in the nuclear translocation of the prototypic inflammatory transcription factor NF-κB, termed the canonical pathway (3, 4), which encodes inflammatory genes such as TNF-α and IL-6. Although TLRs induce common signaling pathways, there is specificity in recruitment of TIR-containing adapter proteins. MyD88 adapter-like (Mal)/TIRAP was the second described adapter capable of mediating NF-κB activation and was responsible for signaling selectively via TLR4 (5, 6) and TLR2 signaling (7, 8). TIR domain-containing adapter protein inducing IFNβ (TRIF, also known as TICAM1) was subsequently found to mediate the MyD88-independent pathway leading to TLR4-mediated activation of the transcription factor interferon regulatory factor 3, which regulates Type I IFN production (9). Importantly, TRIF also mediates downstream signaling from TLR3, independent of MyD88. The TRIF-related adapter molecule (TRAM, also known as TICAM2) specifically acts to bridge TLR4 with TRIF, where TRAM-deficient macrophages are ablated in their responses to TLR4 activation but not TLR3 (10, 11).
Mal and TRAM have been described as bridging adapters, responsible for specific recruitment of MyD88 and TRIF proximal to the surface localized TLR2 and TLR4 receptor complexes (10, 12, 13). Membrane localization of these bridging adapters has further demonstrated the spatial coordination required for transmission of MyD88 (12) and TRAM (14) signals, allowing specificity of downstream signaling mediators that induce cytokine production. Mal acts to bridge MyD88 to TLR2 and TLR4 specifically via its TIR domain. The recent description of a Mal functional variant associated with protection against pnuemococcal disease, bacteremia, malaria, and tuberculosis (15) has highlighted the potential importance of Mal in human disease. Furthermore, the recent depiction of the rare TIR domain variant, Mal D96N, which is unable to interact with MyD88 and impairs downstream TLR2/TLR4-mediated responses (16), has emphasized the importance of protein-protein interactions in TLR-mediated signaling and highlighted the need for a greater understanding of how Mal mediates TLR-induced inflammation.
We have previously demonstrated that Mal interacts with TRAF6 (17) via a putative TRAF6-binding motif (18) that when disrupted inhibited TLR2- and TLR4-induced NF-κB activation. However the specificity and biological consequences of this interaction were not clearly defined. Because disruption of Mal function appears to provide a protective phenotype from disease, we endeavored to investigate the specificity of Mal interaction with TRAF6, cellular localization, and what the biological outcomes of disrupting this interaction were on TLR-mediated pro-inflammatory responses.
In this study, we demonstrate the direct interaction of Mal with TRAF6 and the localization of this association to the plasma membrane. Critically, reconstitution of Mal-deficient macrophages with wild type Mal reconstituted transactivation of the p65 subunit of NF-κB, which was ablated with disruption of the TRAF6-binding motif. Importantly, NF-κB translocation to the nucleus was not affected in these cells. Significantly, whereas Mal-deficient macrophages reconstituted with wild type Mal express the expected pro-inflammatory cytokines TNF-α and IL-6 in response to TLR2 and TLR4 stimulation, cells reconstituted with a single point mutation within the TRAF6-binding motif (MalE190A) display a significantly disrupted cytokine response to TLR stimulation. Together these studies characterize the novel role for Mal in facilitating the direct recruitment of TRAF6 to the plasma membrane, which is necessary for TLR2- and TLR4-induced transactivation of NF-κB and regulation of the subsequent pro-inflammatory response.
MATERIALS AND METHODS
Cell Culture and Reagents
HEK293 cells, HEK293T cells, and murine immortalized Mal-deficient macrophages (16) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Invitrogen), 2 mm glutamine, 100 units/ml penicillin and maintained in a 37 °C humidified atmosphere. Human monocyte THP-1 cells were maintained in RPMI 1640 medium 2 mm glutamine, 100 units/ml penicillin and maintained in a 37 °C humidified atmosphere. LPS K235 (Sigma) was repurified as described previously (19), Pam3Cys was obtained from EMC Microcollections (Tuebingen, Germany), CL-75 was from InvivoGen (San Diego, CA), CpG DNA oligonucleotide was from GeneWorks (Adelaide, Australia), anti-FLAG M2-agarose beads were from Sigma, and glutathione-agarose beads were from Amersham Biosciences. GST, TRAF6, and IκBα antibodies were sourced from Santa Cruz.
Plasmids and Protein Purification
The full-length pDC304 Mal-HA, pDC304 Mal-HA N-terminal (amino acids 1–74), and pDC304 Mal-HA TIR domains (amino acids 74–235) have been described previously (5). The following Mal variants were described previously: MalE190A-HA (17), Mal-GFP, Mal 4KK-GFP (PIP2-binding domain), and Mal (1–20aa)-GFP (first 20 amino acids) (12). MalS180L-HA, MalE190A-GFP, and MalΔx3-GFP mutants were generated using the QuikChange II site-directed mutagenesis kit with Pfu Turbo (Stratagene, La Jolla, CA) using the pDC304 Mal-HA or Mal-GFP template, respectively. Lentiviral vectors encoding the various Mal-GFP gene were generated by subcloning the GFP-tagged cDNA from pEGFP-N1 into pLV-CMV. Gal4-p65(1–551) plasmid encoding the full-length p65 subunit of NF-κB fused to Gal4 DNA-binding domain was a kind gift from Lienhard Schmitz (German Cancer Research Centre, Heidelberg, Germany). The Gal-luciferase reporter gene pFA-Jun and pFA-Elk-1 fusion vectors for analysis of JNK and Erk1/2, respectively, were obtained from Stratagene.
Mal-GST and MyD88-GST purification have been described previously (20). MalE190A-GST purification was prepared the same as for Mal-GST. GST-TRAF6-FLAG protein was transformed into BL21-CodonPlusTM-RIL series Escherichia coli (Stratagene) and grown in Luria broth. A bacterial cell culture was grown to A600 = 0.5 under ampicillin selection. Protein expression was induced with 50 μm isopropyl β-d-thiogalactopyranoside and incubated overnight at 18 °C. Bacterial cells were harvested and resuspended in low salt soluble KalB buffer (50 mm Hepes, pH 7.6, 50 mm NaCl, 0.5 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, and 1 tablet of protease inhibitor mixture (Roche Applied Science). The cells were lysed by sonication and centrifuged, and cell pellet was resuspended in high salt soluble buffer (50 mm Hepes, pH 7.6, 500 mm NaCl, 0.5% Triton X-100, 0.5 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science)). Cell debris was separated by centrifugation (18,000 rpm, 4 °C). Protein purification was facilitated by affinity purification on glutathione-Sepharose and removal of GST fusion tag by PreScissionTM (Amersham Biosciences) enzyme cleavage according to the manufacturer's instructions.
Immunoprecipitation and Immunoblot Analysis
HEK293T cells (2 × 106 cells/10-cm dish) were transfected using FuGENE 6 (Roche Applied Science) with the indicated plasmids where the total amount of DNA (2.5 μg/dish) was kept constant. Twenty-four h later, the cells were lysed in KalB buffer as described (21). The indicated antibodies (2 μg) or anti-FLAG-Sepharose beads (20 μl, 50% slurry) were incubated with the cell lysates for 2 h, followed by the addition of 40 μl of 50% protein G slurry for 1 h. The immune complexes were precipitated, washed, eluted by the addition of sample buffer followed by SDS-PAGE and immunoblotted using the indicated antibodies. For GST pulldown experiments, the lysates prepared from HEK293T cells transfected with indicated vectors were used in a GST pulldown assay whereby cell lysates were incubated for 2 h at 4 °C with recombinant GST fusion protein coupled to glutathione-Sepharose. The complexes were washed three times in lysis buffer, separated by SDS-PAGE, and immunoblotted as indicated in the figure legend. For endogenous immunoprecipitation, confluent THP-1 cells (2 × 107) were resuspended in 1 ml of conditioned medium, centrifuged, and harvested as per immunoprecipitation method.
Protein samples were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the indicated antibodies. Immunocomplexes were visualized by using SuperSignal West Pico chemiluminescent substrate solution (Pierce) followed by exposure to x-ray film (Hyperfilm ECL; Amersham Biosciences) to detect chemiluminescence or visualized using the Odyssey fluorimager (LI-COR) and probed with AlexaFluor 680 (Molecular Probes) and IRDye800CW (Rockland, Gilbertsville, PA) fluorescently labeled secondary antibodies.
Confocal Imaging Analysis
HEK293 cells were cotransfected with TRAF6-FLAG and fluorophore-conjugated proteins at the concentrations indicated for 48 h at 32 °C. The cells were washed in phosphate-buffered saline, fixed in 4% paraformaldehyde, and permeabilized in 0.1% Triton X-100. The cells were immunohistostained with anti-FLAG antibody (Sigma) at 1:200 overnight at 4 °C and probed with Alexa Fluor 594 (Texas Red) or Alexa Fluor 488 (Green)-conjugated IgG secondary antibody (Invitrogen). Mounted cells on coverslips were examined with 40× oil lens objective on Leica TCS NT upright confocal microscope (Leica Microsystems GmbH, Welzlar, Germany) using Leica TCS NT LASAF software. Deconvoluation and colocalization was performed using the IMARIS software (Bit-plane AG, Zurich, Switzerland).
Luciferase Reporter Assay
HEK293 cells were seeded at 2 × 104 cells/well in a 96-well plate 24 h prior to transfection with FuGENE 6. NF-κB-dependent gene expression was determined using the 5× κB-luciferase reporter construct (Stratagene) concomitantly with indicated vectors. Using the PathDetect transient transfection kit (Stratagene), cotransfection of pFR-luciferase in combination with Gal4 fusions p65, pFA-Jun, pFA-CHOP, or pFA-Elk-1, respectively, were used to analyze activation of p65 transactivation and MAP kinase, respectively. The Renilla luciferase-thymidine kinase encoding plasmid (pRL-TK) (Promega, Madison, WI) was used to normalize for transfection efficiency, and pEF-BOS empty vector was used to maintain constant DNA. Transfected cells were lysed using passive lysis buffer (Promega) and assayed for luciferase and Renilla activity using luciferase assay reagent (Promega). Luminescence readings were corrected for Renilla activity and expressed as fold increases over nonstimulated control values.
Electrophoretic Mobility Shift Assay
Nuclear extracts (4 μg) were incubated with 10,000 cpm of a 22-bp DNA fragment oligonucleotide containing the NF-κB consensus sequence (Promega) that had previously been labeled with [γ-32P]ATP (10 mCi mmol) (PerkinElmer Life Sciences) by T4 polynucleotide kinase (Promega). Incubations were performed for 30 min at room temperature in the presence of 2 μg of poly(dI-dC) as nonspecific competitor and 10 mm Tris-HCl, pH 7.5, containing 100 mm NaCl, 1 mm EDTA, 5 mm dithiothreitol, 4% glycerol, and 100 mg/ml nuclease-free bovine serum albumin. The samples were subjected to electrophoresis on native 5% (w/v) polyacrylamide gels, which were subsequently dried and autoradiographed.
Enzyme-linked Immunosorbent Assay and Reverse Transcription-PCR
Macrophage supernatants from virally transduced cells were used for enzyme-linked immunosorbent assay to detect expression of TNF-α or IL-6 with reagents from BD PharMingen (St. Jose, CA) according to the manufacturer's instructions. Total cellular RNA was prepared using the RNeasy mini kit according to the manufacturer's instructions (Qiagen). Two μg of total RNA was treated with DNase (Promega) before cDNA synthesis using Superscript III (Invitrogen) and random primers in a volume of 40 μl. cDNA was diluted ⅓, and triplicate wells were run for each sample on the 7900HT Fast Real-Time PCR system (ABI, Foster City, CA). TaqMan® gene expression assays were conducted using premade kits from ABI, and all of the samples were multiplexed with the endogenous control 18 S ribosomal RNA. Relative gene expression was determined using the ΔΔCT cycle threshold method.
Statistical Analysis
For statistical analysis, the data obtained from independent experiments are presented as the means ± S.D. All of the statistical analysis was performed using GraphPad PRISM software (GraphPad Software, Inc., San Diego, CA). The mean differences were calculated using Student's t test. Analysis of two independent variables was calculated using a two-way analysis of variance. The levels of significance are denoted as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RESULTS
Mal Interaction with TRAF6 Is Induced by TLR2 and TLR4 Stimulation
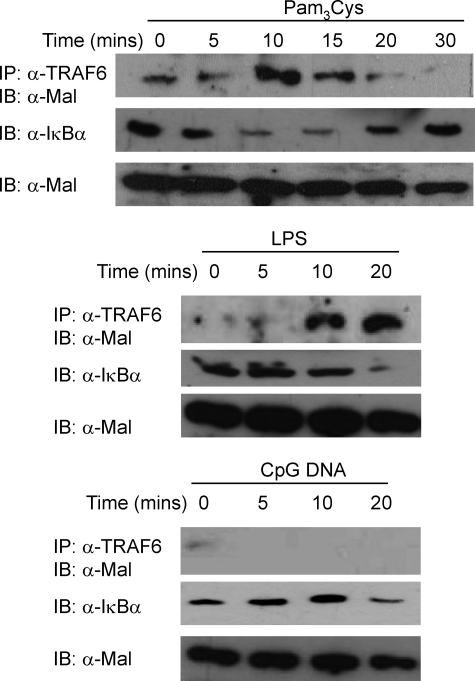
We have previously demonstrated that Mal interacted with TRAF6 in ectopic coimmunoprecipitation experiments; therefore we first wished to establish whether TLR stimulation induced association between the two proteins. As can be observed in the top set of panels in Fig. 1, TRAF6 immunoprecipitated Mal in a time-dependent manner within 10–15 min of TLR2 (Pam3Cys) stimulation of human THP-1 monocytic cells. TLR4 (LPS) activation also induced the same effect (Fig. 1, middle set of panels), although with slightly delayed kinetics, peaking at 20 min following stimulation. Importantly, TLR9 (CpG DNA) stimulation failed to induce Mal-TRAF6 interaction (Fig. 1, bottom set of panels), consistent with Mal having a specific role in TLR2 and TLR4 signaling. Degradation of IκBα was also observed in cells following stimulation, confirming that the respective ligands had activated their relevant TLR. These results demonstrate a specific interaction between Mal and TRAF6 following stimulation of TLR2 and TLR4, but not TLR9, consistent with the described role for Mal in TLR2 and TLR4 signal transduction.
FIGURE 1.
Mal immunoprecipitates with TRAF6 following TLR2 and TLR4 stimulation. Human monoctyic THP1 cells (2 × 107) were stimulated with Pam3Cys (top, 100 ng/ml), LPS (middle, 100 ng/ml), or CpG-DNA (bottom, 500 nm) for indicated times. The cells were lysed, and cellular debris was removed by centrifugation (14,000 rpm, 10 min, 4 °C). The lysates were precleared with protein A-Sepharose beads, and the lysates were probed with α-TRAF6 monoclonal antibody (1 μg) bound to α-mouse IgG beads. Detection of immunoprecipitated (IP) endogenous Mal was analyzed by immunoblotting (IB) with α-Mal antibody (Pearl-1) and IκBα detected with α-IκBα monoclonal antibody. The results are representative of three independent experiments.
Mal Interaction with TRAF6 Occurs at the Plasma Membrane
Recently, Kagan and Medzhitov (12) demonstrated the critical role of Mal localization to the plasma membrane via an N-terminal PIP2-binding domain in TLR2 and TLR4 signaling. Localization to the plasma membrane was required to facilitate MyD88 recruitment to the plasma membrane TLR receptor complex leading to initiation of the canonical signaling pathway and subsequent activation and nuclear translocation of NF-κB. Because ectopic expression of Mal is able to localize to the membrane (12) and activate NF-κB (5, 17) mimicking TLR stimulation, we wished to determine whether localization of Mal and TRAF6 occurs at the plasma membrane or the cytosol in HEK293 cells by fluorescence microscopy.
We found that GFP-tagged Mal localized to the plasma membrane by costaining with cholera toxin subunit B (supplemental Fig. S1A). We next confirmed that Mal 4KK-GFP (a kind gift from Ruslan Medzhitov, Yale University), which contains point mutations of the PIP2-binding region (K15A,K16A,K32A,K33A) and therefore disrupts Mal membrane localization, and Mal 1–20-GFP, consisting of the first 20 amino acids of Mal lacking the PIP2 membrane-binding motif and functionality, both displayed diffuse staining throughout the cell consistent with earlier reports (12) (supplemental Fig. S1, B and C, respectively). Ectopic expression of FLAG-tagged TRAF6 alone was found in discrete foci scattered throughout the cytosol and did not label the cell surface when compared with cholera toxin subunit B staining (supplemental Fig. S1D).
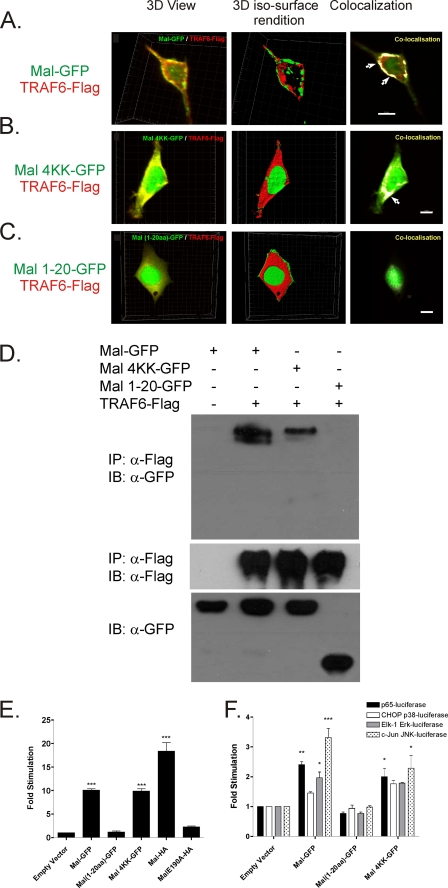
To investigate whether membrane localization was required to facilitate interaction between Mal and TRAF6, we compared wild type Mal association with TRAF6 to Mal 4KK-GFP, because this variant still contains an intact TRAF6-binding motif (Mal 188–193) but fails to localize to the membrane and Mal 1–20-GFP, which lacks the PIP2, TIR-, and TRAF6-binding domains. Initial three-dimensional view image analysis of coexpressed wild type Mal, Mal 4KK, and Mal 1–20 TRAF6 (Fig. 2, left panel) shows that all three Mal proteins are colocalizing with TRAF6 either at the plasma membrane (wild type Mal; Fig. 2A) or in the cytosol (Mal 4KK and Mal 1–20; Fig. 2, B and C). However when three-dimensional iso-surface rendition analysis is applied, where the intensities of the fluorescence of each tag are standardized to remove variance in fluorescence intensity between flurophores, it becomes apparent that although Mal and Mal 4KK colocalize in the same plane as TRAF6 and therefore appear intensely colocalized (Fig. 2, A and B, right panels), Mal 1–20 is clearly not colocalizing in the same plane and therefore is not colocalized with TRAF6 (Fig. 2C, middle and right panels). Similar colocalization of Mal and TRAF6 was observed of Mal tagged with DsRed and TRAF6-GFP (supplemental Fig. S2A) or Mal-GFP and TRAF6-DsRed (supplemental Fig. S2B), indicating that the colocalization at the plasma membrane did not result from the tag and represents a feature of Mal and TRAF6 association itself.
FIGURE 2.
TRAF6 colocalizes with Mal regardless of localization. A–C, micrographs of HEK293 cotransfected with GFP-tagged Mal (A), Mal 4KK (B), or Mal (1–20 aa) (C) and TRAF6-FLAG for 48 h at 32 °C. Immunofluorescence microscopy of Z-stack images (35–50) was taken of individual cells, and three-dimensional image reconstruction was performed using Imaris software (BitPlan). Colocalization of the Z-stack images (intense yellow) illustrates clusters of TRAF6-FLAG (Texas Red) colocalizing with concentrated Mal-GFP in plasma membrane ruffles (arrows, right panel; bar, 8 μm) following three-dimensional iso-surface rendition. Mal4KK-GFP and Mal (1–20 aa)-GFP illustrates TRAF6 localized to the cytosol. All of the images are sections taken with a 40× oil objective lens and are representative of at least three independent experiments where over 500 cells were examined per condition; >90% of the cells displayed similar staining. D, HEK293T cells were cotransfected with TRAF6-FLAG and GFP-tagged wild type Mal, Mal 4KK, or Mal (1–20 aa) where indicated. The cells were lysed, and TRAF6 was immunoprecipitated (IP) with α-M2 FLAG-agarose beads. Detection of Mal or Mal variant-GFP proteins were analyzed by immunoblotting (IB) with α-Mal antibody. Expression of TRAF6 was verified by immunoblot analysis of cellular lysates with α-FLAG M2-HRP conjugated antibody, whereas GFP-tagged protein expression was verified by α-GFP immunostaining (n = 3). E, HEK293 cells were cotransfected with 5× κB-luciferase promoter gene plasmid (100 ng), concomitantly with 10 ng of either GFP-tagged Mal, Mal 4KK, Mal (1–20 aa), or HA-tagged Mal and MalE190A constructs for stimulation. Shown is the mean relative luciferase activity ± S.E. for a representative experiment from three separate experiments. The readings are normalized for κB-luciferase expression over TK Renilla expression. F, HEK293 cells were cotransfected with components of the pRF-luciferase and either c-Jun (2 ng), CHOP (5 ng), Elk-1 (10 ng), or p651–551 (10 ng) Gal4 fusion vector, in conjunction with either GFP-tagged Mal, Mal 4KK, or Mal (1–20 aa) constructs, respectively. Shown is the mean relative luciferase activity ± S.E. for a representative experiment from three separate experiments. All of the luciferase results were comparative to constitutively expressed TK Renilla. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To further support these findings we found that both wild type Mal-GFP and Mal 4KK-GFP, but not Mal 1–20-GFP, were able to interact with TRAF6 in coexpression immunoprecipitation experiments (Fig. 2D), confirming that the GFP tag did not affect the ability of Mal and TRAF6 to interact and that association occurs independent of cellular localization.
Having established that interaction was not dependent on localization, we next assessed the effects on downstream signaling. We have previously found that Mal interaction with TRAF6 mediated the activation of the MAP kinase pathway and importantly, transactivation of the p65 subunit of NF-κB (17). We therefore tested the ability of these GFP-tagged proteins to drive NF-κB activation (Fig. 2E) compared with our previously reported results using HA-tagged Mal. Consistent with our localization and immunoprecipitation data, both wild type Mal and Mal 4KK were able to induce activation of NF-κB luciferase expression, whereas Mal 1–20 failed to induce a similar responses. Furthermore, consistent with our earlier report (17), wild type Mal and Mal 4KK were able to induce NF-κB p65 transactivation and Erk1/2 and JNK activation (Fig. 2F) via reporter assays but critically not p38. As expected, Mal 1–20 was unable to induce activation of any pathway because of its lack of PIP2 and TIR domains.
Taken together these results demonstrate that Mal and TRAF6 interact either at the plasma membrane or in the cytosol independent of localization to the plasma membrane, suggesting that the geographical constraint of Mal to the plasma membrane by PIP2 modification may be important for spatial signaling or kinetics. Alternatively, TRAF6 may have a higher preference, but not an absolute requirement for, association for the membrane-bound Mal.
Disrupting the TRAF6-binding Motif in Mal Inhibits Mal-TRAF6 Interaction
The requirement of an intact Mal TRAF6-binding motif for interaction with TRAF6 was next determined. In our previous study we found that a single amino acid change of E190A in Mal inhibited TLR2 and TLR4 signaling; however, in ectopic immunoprecipitation studies, this mutation was still able to interact with TRAF6. Therefore we wished to determine a more specific assessment of the role of the critical glutamic acid residue in the TRAF6-binding motif in mediating Mal interaction with TRAF6.
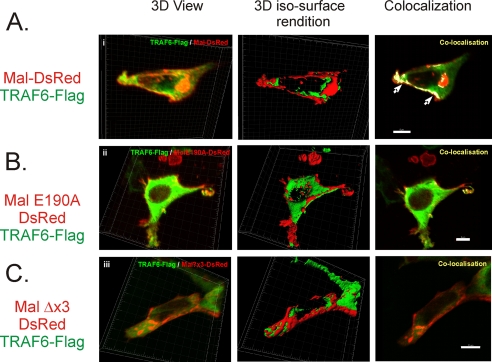
To begin with we assessed Mal and TRAF6 association by immunomicroscopy. As shown in Fig. 3, whereas it appears that both Mal (Fig. 3A) and MalE190A (Fig. 3B) are both colocalizing with TRAF6 at the plasma membrane, three-dimensional iso-surface rendition analysis shows that once the fluorescence intensities are standardized, it is apparent that Mal and TRAF6 are colocalizing in the same plane (Fig. 3A, middle panel) and as such appear intensely colocalized at the plasma membrane (right panel). Conversely, three-dimensional iso-surface rendition shows that MalE190A appears as discrete foci along the plasma membrane, whereas TRAF6 remained diffuse in the cytosol and is not in the same plane (Fig. 3B, middle panel) and as such does not colocalize (right panel). To further assess this interaction, we generated a mutation of all three key residues of the TRAF6-binding motif (PXEX(Ar/Ac)) (18) from PPELRF → QPAERA, termed MalΔx3, which interestingly, does not appear to colocalize with TRAF6, neither three-dimensional view nor iso-surface rendition, and as such does not colocalize, demonstrating a lack of association between the proteins (Fig. 3C). This protein also failed to display any discernable colocalization with TRAF6 (Fig. 3C) as rendered by three-dimensional iso-surface rendition colocalization, appearing primarily throughout the cytosol. Identical results were observed using GFP-tagged TRAF6 and DsRed-Mal constructs (data not shown), indicating that the localization and the importance of an intact TRAF6-binding motif for Mal interaction with TRAF6 were indicative of a function of Mal and not the result of the fusion tag to the proteins.
FIGURE 3.
Mal colocalization at the plasma membrane with TRAF6 requires an intact TRAF6 binding motif. HEK293 cells were cotransfected with DsRed-tagged Mal (A), MalE190A (B), or MalΔx3 (C) with TRAF6-FLAG for 48 h at 32 °C. Immunofluorescence microscopy of Z-stack (29–45) images were taken of individual cells, and three-dimensional image reconstruction was performed using Imaris software (BitPlan). Colocalization illustrates TRAF6-FLAG colocalizing with Mal-DsRed in plasma membrane ruffles (arrows, right panel; bar, 8 μm). MalE190A-DsRed and MalΔx3-DsRed mutant demonstrate plasma localization, whereas TRAF6 is diffusely distributed within the cytosol (right panel; bar, 8 μm), and minimal colocalization is observed (right panel; bar, 10 μm). All of the results are representative of three independent experiments.
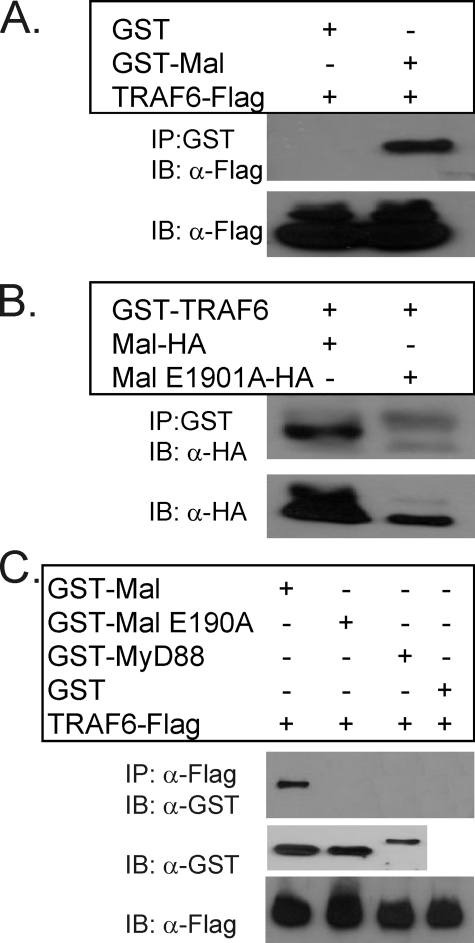
Having demonstrated an interaction between Mal and TRAF6 by microscopy, we next wished to further support the findings above and investigated the specificity of interaction and whether the association was dependent of an intact TRAF6-binding motif within Mal and independent of bridging proteins. As shown in Fig. 4A, recombinant GST-Mal was able to specifically immunoprecipitate ectopically expressed recombinant TRAF6 from cellular lysates, whereas recombinant GST alone was unable to recognize TRAF6. Conversely, recombinant GST-TRAF6 was able to precipitate ectopically expressed Mal from cellular lysates but was unable to interact with MalE190A (Fig. 4B), clearly demonstrating the requirement of an intact TRAF6-binding motif to facilitate association. To demonstrate that the Mal interaction with TRAF6 was direct, we next coincubated purified recombinant fusion proteins of TRAF6 tagged with FLAG, in conjunction with the indicated recombinant TIR-containing proteins. As can be observed in Fig. 4C, neither recombinant GST-MalE190A, GST-MyD88, nor GST alone was able to be immunoprecipitated with TRAF6, whereas GST-Mal and TRAF6 markedly formed a complex.
FIGURE 4.
Mal directly interacts with TRAF6 via an intact TRAF6-binding motif. A, HEK293T cells were transiently transfected with FLAG-tagged TRAF6 (1.25 μg) for 24 h. The cells were lysed and immunoprecipitated (IP) with either 1 μg of recombinant GST-Mal or GST protein. Mal interacting complexes were visualized by immunoblotting (IB) with α-FLAG antibody to represent TRAF6. Expression of TRAF6 in transiently transfected cells was confirmed by immunoblot of cell lysates with α-FLAG antibody. B, HEK293T cells were transfected with either HA-tagged wild type Mal or the Mal mutated TRAF6-binding motif, MalE190A (1.25 μg, respectively) where indicated. The cells were lysed and immunoprecipitated with 1 μg of recombinant TRAF6-GST. TRAF6-GST protein immunocomplexed with ectopically expressed wild type Mal but not with MalE190A when immunoblotted with α-HA antibody. Mal protein expression was confirmed by immunoblot analysis of cell lysates using α-HA antibody to detect Mal-HA. C, recombinant TRAF6-FLAG protein (0.75 μg) interacts specifically with recombinant GST-Mal (1.5 μg) but not with GST-fused MyD88 or MalE190A protein in an in vitro binding assay. TRAF6 immunocomplexes were precipitated using α-M2 FLAG beads in a high salt (400 mm) phosphate-buffered saline buffer and immunoblotted with α-GST antibody to detect interacting proteins. Recombinant proteins were confirmed by immunoblot analysis of inputs with their indicated antibodies. All of the results are representative of three independent experiments.
Taken together, these results demonstrate that Mal and TRAF6 directly interact at the plasma membrane, the interaction dependent on an intact TRAF6-binding motif within Mal. Importantly, these findings also suggest the interaction between Mal and TRAF6 are independent of post-translational modification of either Mal or TRAF6 because of formation of a specific complex of recombinant proteins.
Mal Association with TRAF6 Is Critical for TLR2- and TLR4-mediated Inflammatory Responses
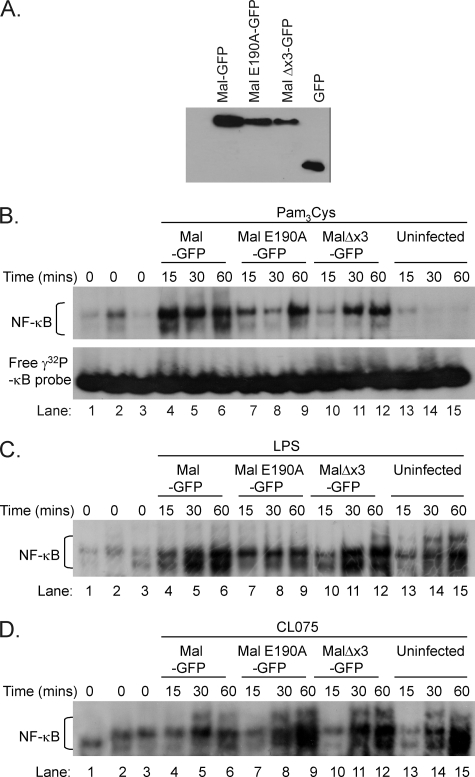
To establish the biological importance of Mal and TRAF6 interaction in regulating the immune response to TLR2 and TLR4 activation, we examined the effect of disruption of the Mal TRAF6-binding motif on TLR inflammatory responses. To perform these experiments we utilized immortalized Mal-deficient macrophages. We first established that this cell line was unresponsive to TLR2 (Pam3Cys) and TLR4 (LPS) stimulation but did respond to TLR9 (CpG DNA), TLR7 (CL75), and TLR3 (poly(I-C)) ligands (supplemental Fig. S3A) as assessed by TNF-α production. Immunoblotting also demonstrated the lack of Mal in these cells (supplemental Fig. S3B). Mal-deficient macrophages were then reconstituted with GFP-tagged Mal, MalE190A, or MalΔx3 by lentiviral transduction. GFP+ve cells were sorted to give a homogenous population of cells expressing GFP-tagged proteins (supplemental Fig. S3C). Fluorescence microscopy of cells with GFP+ve expression for each tagged protein was used to verify protein expression (data not shown) and supported by immunoblot analysis of Mal protein expression in reconstituted cells (Fig. 5A), demonstrating comparable expression levels of wild type Mal, MalE190A, MalΔx3, and GFP alone.
FIGURE 5.
Mal interaction with TRAF6 does not affect NF-κB nuclear translocation in response to TLR stimulation. A, virally transduced GFP-positive populations for each indicated gene were gated, sorted for >90% GFP positive population, and then seeded at a concentration of 3 × 104 cells/well 24 h prior to lysis in KalB buffer. Cellular debris was removed by centrifugation (13,000 rpm, 10 min, 4 °C), and equal protein was loaded onto SDS-PAGE and transferred to nitrocellulose, and the relative expression of viral reconstituted immortalized Mal-deficient macrophages assessed for GFP-expressing proteins by immunoblot of cell lysates with α-GFP antibody. Reconstituted Mal-deficient macrophages expressing the indicated genes were seeded at 1 × 104 cells/ml in 6-well plates 72 h prior to stimulation with TLR2 ligand Pam3Cys (B, 10 ng/ml), TLR4 ligand LPS (C, 10 ng/ml), or TLR7 ligand CL-75 (D, 0.5 μg/ml) for the indicated times. Nuclear extracts (4 μg) were prepared, incubated with γ-32P-labeled NF-κB DNA probe, and analyzed by a 6% native gel. Binding to the NF-κB probe was observed in all stimulated virally reconstituted nuclear extracts (lanes 1–12) but only weakly observed in nonstimulated cells (lanes 1–3). Uninfected stimulated extracts observed binding from treatment with LPS (C) and CL-75 (D) but not from Pam3Cys (B) (lanes 13–15). Unbound free NF-κB probe was shown in lower panel of B. The results are representative of two independent experiments.
Initially we wished to determine whether an intact TRAF6-binding domain was required for NF-κB nuclear translocation to the nucleus following TLR stimulation. As can be observed in Fig. 5, macrophages reconstituted with either wild type Mal (lanes 1 and 4–6), MalE190A (lanes 2 and 7–9), or MalΔx3 (lanes 3 and 10–12), respectively, display comparable NF-κB translocation to the nucleus in response to TLR2 (Fig. 5A), TLR4 (Fig. 5B), and TLR7 (Fig. 5C) stimulation, which suggests that interaction between Mal and TRAF6 is not required for efficient NF-κB translocation via the MyD88 canonical pathway.
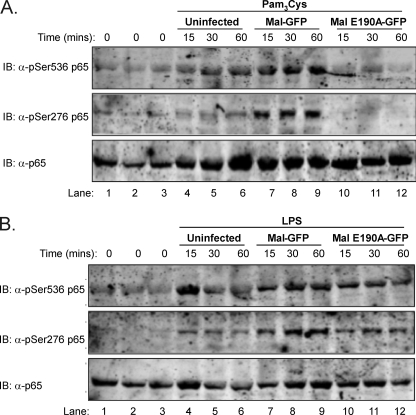
We have previously reported that Mal interaction with TRAF6 is required for transactivation of NF-κB (17) and that Mal is crucial for Ser536 phosphorylation of the p65 subunit of NF-κB (21). Because transactivation of NF-κB is a critical process in the transcriptional regulation of the inflammatory response (22), we next investigated whether Mal interaction with TRAF6 was critical to this response. As can be observed in Fig. 6A, only cells reconstituted with Mal were able to demonstrate robust Ser(P)536 p65 in response to TLR2 stimulation with Pam3Cys (lanes 2 and 7–9) compared with uninfected cells (lanes 1 and 4–6) in a time-dependent manner. Critically, reconstitution of Mal-deficient macrophages with MalE190A (lanes 3 and 10–12) failed to recapitulate TLR2-induced phosphorylation of the p65 subunit, thereby ablating transactivation of NF-κB. Notably, we observed that cells reconstituted with Mal demonstrate TLR-induced Ser276 phosphorylation of p65, which is absent in MalE190A reconstituted cells (compare lanes 7–9 with lanes 10–12). Mal has not been implicated previously in this transactivation event. Interestingly, in response to TLR4 stimulation, MalE190A reconstituted cells display partially restored p65 Ser536 or Ser276 phosphorylation (Fig. 6B), perhaps because of compensation by the MyD88-independent pathway.
FIGURE 6.
Mal interaction with TRAF6 is required for TLR2- and TLR4-mediated transactivation of the p65 subunit of NF-κB. Immortalized Mal-deficient macrophages were virally reconstituted with either GFP-tagged Mal (lanes 2 and 7–9) or MalE190A (lanes 3 and 10–12) as described previously in Fig. 5. GFP-positive populations were challenged with either 10 ng/ml of Pam3Cys (A) or LPS (B) for indicated time points and lysed, and protein was separated by SDS-PAGE followed by immunoblot (IB) for p65, p65 Ser276, or p65 Ser536 phosphorylation where indicated. The results represent three independent experiments.
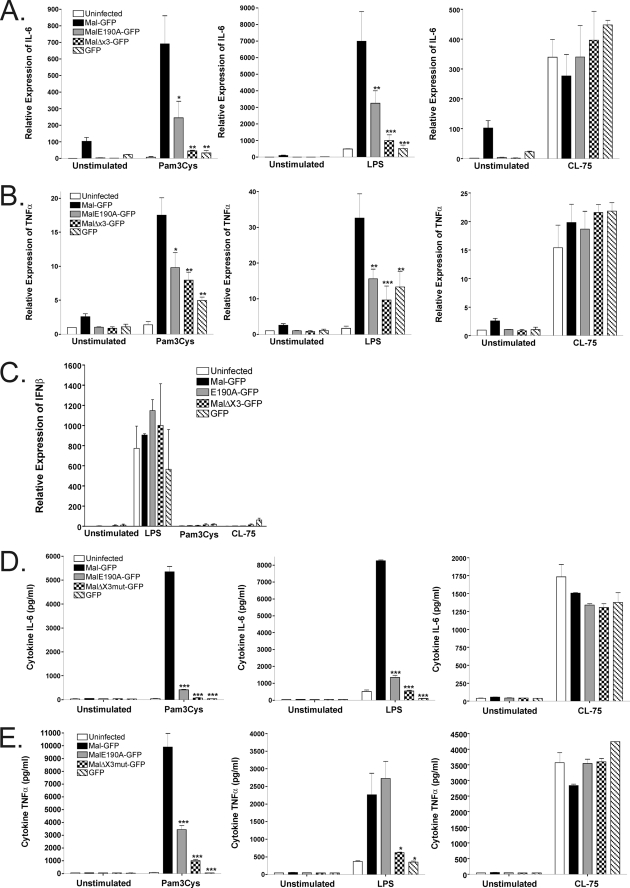
To determine whether Mal-TRAF6-mediated NF-κB transactivation plays a role in the inflammatory cytokine response to TLR stimulation, we next investigated cytokine expression in reconstituted Mal-deficient macrophages. Mal-deficient macrophages were reconstituted with wild type Mal, MalE190A, or MalΔx3 and stimulated with LPS, Pam3Cys, or the TLR7/8 agonist CL-75 (23) to induce IL-6 or TNF-α mRNA and protein gene expression. Although reconstitution with Mal was able to mediate potent IL-6 (Fig. 7, A and D) and TNF-α (Fig. 7, B and E) expression compared with the GFP reconstituted or noninfected cells alone, both MalE190A and MalΔx3 displayed significantly inhibited TLR2 and TLR4 proinflammatory responses. Importantly, as expected, TLR7/8 stimulation induced a robust response with all constructs and was independent of the absence or presence of Mal. Significantly, TLR4 induction of IFNβ mRNA was also not affected either in the presence or the absence of Mal (Fig. 7C), consistent with Mal-independent TLR4-induced IFNβ expression (8).
FIGURE 7.
A disrupted TRAF6-binding domain in Mal inhibits TLR2- and TLR4-mediated NF-κB proinflammatory responses. Mal-deficient macrophages were reconstituted with either no virus, GFP, GFP-tagged Mal, MalE190A, or MalΔx3 as indicated were seeded at 3 × 104 cells/well 72 h prior to stimulation with 10 ng/ml of Pam3Cys, LPS, or CL-75 (0.5 μg/ml) ligand, respectively. RNA was extracted from macrophages after 3 h of challenge, and mRNA induction levels of IL-6 (A), TNF-α (B), and IFNβ (C) were measured using quantitative real time PCR where the relative abundance was compared with 18 S levels. The supernatants were collected from macrophages 24 h after stimulation and assessed for IL-6 (D) or TNF-α (E) expression by enzyme-linked immunosorbent assay. Mal variants lacking an intact TRAF6-binding motif demonstrate defective for TLR2 and TLR4-induced pro-inflammatory cytokine expression. The results are the means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Taken together, these results clearly demonstrate the critical role Mal interaction with TRAF6 plays in generating the pro-inflammatory response as a consequence of TLR2 and TLR4 activation and that the mechanism is via a pathway of NF-κB transactivation, independent of the canonical MyD88-dependent NF-κB nuclear translocation.
DISCUSSION
In this study we have extended and characterized our earlier finding that the novel interaction of Mal with TRAF6 has a critical role in TLR2- and TLR4-induced inflammatory responses. Localization studies demonstrate that the geographical constraint of Mal to the plasma membrane may be important in the spatial control of Mal-mediated signaling. Significantly, the data presented characterize the direct interaction between Mal and TRAF6, which regulates transactivation of the p65 subunit of NF-κB but does not affect NF-κB nuclear translocation. The association of Mal and TRAF6 therefore appears to control the transcriptional expression of NF-κB-dependent pro-inflammatory cytokines. Reconstitution of Mal-deficient macrophages with MalE190A containing a point mutation within the TRAF6-binding motif significantly inhibits both TLR2- and TLR4-mediated inflammatory responses compared with wild type Mal. These results describe a specific role for Mal in TLR signaling and identify a direct interaction between Mal and the key signaling mediator TRAF6 that is separate from that of acting simply as a bridging adapter for MyD88 signaling.
The importance of Mal in TLR-mediated signal transduction in human disease was suggested by the recent description of the S180L functional variant and its association with protection against pneumoccal disease, tuberculosis, malaria, bacteremia, and systemic lupus erythematosus (15, 24). We found no effect of this variant on the ability of Mal to recruit TRAF6 (data not shown); however, our studies with MalE190A and those by Hill and co-workers (15) combined with the inability of the rare Mal D96N SNP to interact with MyD88 (16) highlight the fact that structural modifications to Mal can have important implications in the formation of the plasma membrane proximal signaling complex. Interestingly, whereas Mal has been reported to undergo Tyr phosphorylation (25, 26), caspase-1-mediated cleavage (27), and now the physiological impact of the S180L variant, none of these events altered the ability of Mal to interact with TRAF6 in immunoprecipitation experiments because ectopically expressed single amino substitution variants were still recognized by recombinant TRAF6 (data not shown), which combined with the direct interaction of the recombinant Mal and TRAF6 proteins strongly suggests that post-translational modification is not required for the interaction to occur. This suggests therefore that there is still more to be understood as to the signaling capabilities of Mal. Critically, this study illustrates the importance of a single amino acid substitution within the critical TRAF6-binding motif of Mal and demonstrates how the immune response to TLR2 and TLR4 is profoundly affected. Consequently, Mal does not act solely as a bridging adapter, facilitating MyD88 recruitment to the membrane proximal receptor complex, but directly recruits TRAF6 to the plasma membrane to regulate NF-κB transactivation and ultimately transcriptional activation of the inflammatory response, independent of NF-κB nuclear translocation.
Interestingly, the TRAF6-binding motif in Mal is situated within its TIR domain raising the possibility of MyD88 and TRAF6 competing for binding sites. A recent paper by Ohnishi et al. (28) demonstrated through complex structural modeling that two Mal proteins bound MyD88 based on residues responsible for TIR-TIR interaction. Their model suggested that MyD88-TIR binding would not interfere with interactions between Mal-TIR and TLR4, TLR2, or TRAF-6 because the Glu190 in Mal was distal to the Mal-MyD88 interaction surface. This model is therefore suggestive of the formation of a signaling complex comprising TLR4, Mal, TRAF6, and MyD88 to initiate the signaling cascade.
It has previously been demonstrated that the myristoylation sequence in the N terminus of TRAM is critical for its plasma localization and the ability to mediate TLR4 signaling (29). Kagan et al. (14) demonstrated that TRAM also requires further phosophoinositide modification that facilitates recruitment of TRIF and subsequently TRAF3 to the complex to promote other signaling pathways. This modification or geographical constraint appears to provide the TRAM·TRIF complex with the opportunity to specifically recruit TRAF3, an adapter required for induction of IFN. This description of “spatial” signaling control of TRAM (14, 30) and its geographical restriction to the endosomal membrane as a means of organizing and coordinating the two TLR4 adapter systems may have similarities to what we have observed in this study for Mal. Consequently, selective recruitment of TRAF family members to TIR adapter proteins because of localization may provide spatial control over signaling, which integrates signals and allows coordination of NF-κB, MAP kinase, and interferon regulatory factor signaling pathways. Indeed our study found that the Mal 4KK variant that was unable to localize to the plasma membrane could still activate MAP kinases and NF-κB transactivation commensurate with wild type Mal, suggesting that membrane localization was not critical for mediating this aspect of Mal function. Analogous to that observed with TRAM, Mal localization to the membrane via PIP2 activation may provide an optimal platform for Mal and TRAF6 to associate, which would be distinct from the MyD88·IRAK·TRAF6 complex utilized by other TIR signaling pathways. The experiments described herein suggest that Mal interaction with TRAF6 does not affect MyD88-dependent pathway signal transduction leading to NF-κB nuclear translocation. However, the nuclear translocated NF-κB would not appear to undergo transactivation without the Mal-TRAF6 interaction. It would appear that Mal interaction with TRAF6 is implicit in transactivation of the p65 subunit of NF-κB, seemingly via the MAP kinase pathway (17). Thus the plasma membrane localization of Mal and TRAM appears to allow the spatial recruitment of specific signaling components and, as such, coordinating downstream signaling events (30) analogous to that described with TRAM and TRAF3 to regulate IFN induction.
Using complementation experiments in Mal-deficient macrophages, we were able to clearly demonstrate the importance of interaction between Mal and TRAF6 for a robust inflammatory response to TLR2 and TLR4 ligands. Disruption of the TRAF6-binding motif in Mal inhibited both NF-κB transactivation (phospho-p65 Ser276 and Ser536) (22) and significantly ablated cytokine gene expression and production in response to TLR2 and TLR4 stimulation. Interestingly, whereas the TRAF6-binding motif is important for some of the functionality of Mal, all three critical amino acids of the motif required mutation to significantly inhibit LPS-mediated TNF-α production. This result suggests that other pathways may play a role in TLR4-induced TNF-α secretion. This disparity in effect may be a combination of the complexity of TLR4 signaling (i.e. MyD88-dependent and MyD88-independent signaling) combined with the intricacies of TNF secretion requiring expression, trafficking, and transmembrane cleavage, which may explain the discrepancy between these results. Interestingly, Baltimore and co-workers (31) have suggested that TNF-α is expressed by the MyD88-independent pathway via interferon regulatory factor 3, which was time-dependent. Additionally, p38 has been shown to play a role in regulating both TNF-α message stability and protein expression (32, 33). We have described previously that Mal interaction with TRAF6 mediates Erk1/2 and JNK activation (17), but not p38, which may explain the discrepancy that the MalE190A mutant is still able to induce TNF-α expression. Moreover, we also demonstrated that reconstitution of Mal-deficient cells with MalE190A did not affect the ability of TLR4 to drive the MyD88-independent pathway (as witnessed by a lack of an effect on IFNβ induction). Therefore, it may be possible that inhibition of TNF-α mRNA expression by MalE190A represents “early” activation, but the “late” MyD88-independent response has compensated for this inhibition.
Overall, this study clearly demonstrates a critical role for Mal in TLR2- and TLR4-mediated inflammatory responses that are novel to its role as a bridging adapter for MyD88. Mal regulation of the transcriptional control of NF-κB via TRAF6 interaction and the subsequent expression of inflammatory cytokines is a key regulatory step in TLR2 and TLR4 inflammatory responses. This interaction allows activation of both the MyD88-dependent canonical and Mal-TRAF6-dependent transactivation pathways, culminating in the coordinated translocation and subsequent transactivation of NF-κB, both of which are critical to production of pro-inflammatory cytokines.
Significantly, a single point mutation within the TRAF6-binding motif of Mal is able to potently inhibit the pro-inflammatory response to TLR2 and TLR4 ligands, the primary response mechanism for Gram-positive and Gram-negative bacteria. This study identifies Mal as a key regulator of TLR2 and TLR4 signal transduction via a novel interaction with TRAF6, the possible disruption of which may provide a putative specific therapeutic intervention for TLR2- and TLR4-mediated inflammatory diseases without impacting on MyD88-dependent or TRIF/TRAM-dependent signaling pathways.
Supplementary Material
Acknowledgments
We thank Prof. Ruslan Medzhitov (Yale University) for the Mal 4KK and Mal 1–20 plasmid constructs and Prof. Luke O'Neill (School of Biochemistry and Immunology, Trinity College, Dublin) for the generous gift of the anti-Mal antibody.
This work was supported by Australian National Health and Medical Research Council Grants 334023 and 465116 (to A. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TLR
- Toll-like receptor
- TIR
- Toll/IL-1 receptor
- GST
- glutathione S-transferase
- IFN
- interferon
- IL
- interleukin
- IRAK
- interleukin-1 receptor-associated kinase
- LPS
- lipopolysaccharide
- Mal
- MyD88 adapter-like
- NF-κB
- nuclear factor κB
- PIP2
- phosphatidylinositol 4,5-biphosphate
- TNF
- tumor necrosis factor
- TRAF
- TNF receptor-associated factor
- TRIF
- TIR domain-containing adapter protein inducing IFNβ
- TRAM
- TRIF-related adapter molecule
- HEK
- human embryonic kidney
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- MAP
- mitogen-activated protein
- Erk
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase.
REFERENCES
- 1.Kawai T., Akira S. (2007) Semin. Immunol. 19, 24–32 [DOI] [PubMed] [Google Scholar]
- 2.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 3.O'Neill L. A. (2008) Immunol. Rev. 226, 10–18 [DOI] [PubMed] [Google Scholar]
- 4.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 6.Horng T., Barton G. M., Medzhitov R. (2001) Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 7.Horng T., Barton G. M., Flavell R. A., Medzhitov R. (2002) Nature 420, 329–333 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., Takeda K., Akira S. (2002) Nature 420, 324–329 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., Akira S. (2003) Nat. Immunol. 4, 1144–1150 [DOI] [PubMed] [Google Scholar]
- 12.Kagan J. C., Medzhitov R. (2006) Cell 125, 943–955 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 14.Kagan J. C., Su T., Horng T., Chow A., Akira S., Medzhitov R. (2008) Nat. Immunol. 9, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khor C. C., Chapman S. J., Vannberg F. O., Dunne A., Murphy C., Ling E. Y., Frodsham A. J., Walley A. J., Kyrieleis O., Khan A., Aucan C., Segal S., Moore C. E., Knox K., Campbell S. J., Lienhardt C., Scott A., Aaby P., Sow O. Y., Grignani R. T., Sillah J., Sirugo G., Peshu N., Williams T. N., Maitland K., Davies R. J., Kwiatkowski D. P., Day N. P., Yala D., Crook D. W., Marsh K., Berkley J. A., O'Neill L. A., Hill A. V. (2007) Nat. Genet. 39, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagpal K., Plantinga T. S., Wong J., Monks B. G., Gay N. J., Netea M. G., Fitzgerald K. A., Golenbock D. T. (July10, 2009) J. Biol. Chem. 10.1074/jbc.M109.014886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansell A., Brint E., Gould J. A., O'Neill L. A., Hertzog P. J. (2004) J. Biol. Chem. 279, 37227–37230 [DOI] [PubMed] [Google Scholar]
- 18.Ye H., Arron J. R., Lamothe B., Cirilli M., Kobayashi T., Shevde N. K., Segal D., Dzivenu O. K., Vologodskaia M., Yim M., Du K., Singh S., Pike J. W., Darnay B. G., Choi Y., Wu H. (2002) Nature 418, 443–447 [DOI] [PubMed] [Google Scholar]
- 19.Hirschfeld M., Kirschning C. J., Schwandner R., Wesche H., Weis J. H., Wooten R. M., Weis J. J. (1999) J. Immunol. 163, 2382–2386 [PubMed] [Google Scholar]
- 20.Dunne A., Ejdeback M., Ludidi P. L., O'Neill L. A., Gay N. J. (2003) J. Biol. Chem. 278, 41443–41451 [DOI] [PubMed] [Google Scholar]
- 21.Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 22.Vermeulen L., De Wilde G., Notebaert S., Vanden Berghe W., Haegeman G. (2002) Biochem. Pharmacol. 64, 963–970 [DOI] [PubMed] [Google Scholar]
- 23.Gorden K. B., Gorski K. S., Gibson S. J., Kedl R. M., Kieper W. C., Qiu X., Tomai M. A., Alkan S. S., Vasilakos J. P. (2005) J. Immunol. 174, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 24.Castiblanco J., Varela D. C., Castaño-Rodríguez N., Rojas-Villarraga A., Hincapié M. E., Anaya J. M. (2008) Infect. Genet. Evol. 8, 541–544 [DOI] [PubMed] [Google Scholar]
- 25.Gray P., Dunne A., Brikos C., Jefferies C. A., Doyle S. L., O'Neill L. A. (2006) J. Biol. Chem. 281, 10489–10495 [DOI] [PubMed] [Google Scholar]
- 26.Piao W., Song C., Chen H., Wahl L. M., Fitzgerald K. A., O'Neill L. A., Medvedev A. E. (2008) J. Biol. Chem. 283, 3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miggin S. M., Pålsson-McDermott E., Dunne A., Jefferies C., Pinteaux E., Banahan K., Murphy C., Moynagh P., Yamamoto M., Akira S., Rothwell N., Golenbock D., Fitzgerald K. A., O'Neill L. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3372–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi H., Tochio H., Kato Z., Orii K. E., Li A., Kimura T., Hiroaki H., Kondo N., Shirakawa M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowe D. C., McGettrick A. F., Latz E., Monks B. G., Gay N. J., Yamamoto M., Akira S., O'Neill L. A., Fitzgerald K. A., Golenbock D. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6299–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts C. (2008) Nat. Immunol. 9, 343–345 [DOI] [PubMed] [Google Scholar]
- 31.Covert M. W., Leung T. H., Gaston J. E., Baltimore D. (2005) Science 309, 1854–1857 [DOI] [PubMed] [Google Scholar]
- 32.Clark A. R., Dean J. L., Saklatvala J. (2003) FEBS Lett. 546, 37–44 [DOI] [PubMed] [Google Scholar]
- 33.Saklatvala J., Dean J., Clark A. (2003) Biochem. Soc. Symp. 70, 95–106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.