Abstract
Adaptation to tumor hypoxia is mediated in large part by changes in protein expression. These are driven by multiple pathways, including activation of the hypoxia inducible factor-1 (HIF-1) transcription factor and the PKR-like endoplasmic reticulum kinase PERK, a component of the unfolded protein response. Through gene expression profiling we discovered that induction of the HIF-1 target gene CA9 was defective in mouse embryo fibroblasts derived from mice harboring an eIF2α S51A knock-in mutation. This finding was confirmed in two isogenic human cell lines with an engineered defect in eIF2α phosphorylation. We show that impaired CA9 expression was not due to changes in HIF activity or CA9 mRNA stability. Using chromatin immunoprecipitation we show that the eIF2α-dependent translationally regulated gene ATF4 binds directly to the CA9 promoter and is associated with loss of the transcriptional repressive histone 3 lysine 27 tri-methylation mark. Loss or overexpression of ATF4 confirmed its role in CA9 induction during hypoxia. Our data indicate that expression of CA9 is regulated through both the HIF-1 and unfolded protein response hypoxia response pathways in vitro and in vivo.
Mammalian cells have evolved a number of sophisticated mechanisms to allow adaptation to changes in the supply of essential metabolic factors. These include mechanisms for sensing the presence of nutrients, energy, and oxygen. Many of these mechanisms are exploited by tumor cells to facilitate survival in the unique conditions found within the microenvironment of solid human tumors (1). In particular, adaptation to hypoxia is considered to be an important factor that contributes to the poor prognosis of patients with high levels of tumor hypoxia (2). The hypoxia-inducible factor (HIF)2 plays an essential role in this adaptation by transcriptionally regulating the expression of a large number of genes involved in angiogenesis, glycolysis, invasion, and metastasis (3).
HIF-1 is a heterodimeric transcription factor consisting of α and β subunits that are both ubiquitously expressed. Under aerobic conditions HIF-1α is ubiquitinated by the von Hippel-Lindau tumor suppressor protein causing its rapid degradation within the 26 S proteasome (4). This process relies on hydroxylation of proline residues 402 and 564 by a family of oxygen-sensitive prolylhydroxylases 1–3. Under hypoxic conditions hydroxylation cannot occur, resulting in HIF-1α stabilization and formation of a functional transcription factor with HIF-1β. This complex drives transcription of HIF-responsive genes by binding to hypoxia-responsive elements (HREs) in their promoters.
One of the best characterized targets of HIF-1 is carbonic anhydrase 9 (CA9). The CA9 promoter contains a single HRE element that is essential for its transcriptional increase during hypoxia (5). CA9 is frequently overexpressed in human tumors and is associated with poor prognosis (6, 7). It belongs to a family of zinc metalloenzymes involved in regulating pH by catalyzing the reversible conversion of carbon dioxide to bicarbonate and H+. CA9 is found in the plasma membrane with its active site on the extracellular domain. Consequently, it is has been shown to contribute to the acidification of the extracellular tumor environment during hypoxia (8). Low extracellular pH in tumors is associated with increased invasion and metastasis (9, 10) and can also negatively affect cancer treatment by modulating uptake of anticancer drugs (11). On the other hand, the bicarbonate produced by this reaction can be used by bicarbonate transporters for import to cytoplasm and neutralization of intracellular pH. Recently it has been elegantly shown that CA9 coordinates internal pH by stimulating CO2 diffusion in a three-dimensional spheroid model (12, 13). The potential importance of CA9 as a marker of hypoxia and regulator of tumor pH has made it an attractive diagnostic and therapeutic target (12–16).
In addition to regulation by HIF through the HRE element, five potentially important regions within the CA9 promoter have been identified by DNase I footprinting (PR1–PR5) (17). PR1 plays an important role in CA9 induction during culture at high cell density via binding of SP1/3 transcription factors (18). In vitro cell culture at high cell density leads to a small decrease in the pericellular oxygen concentration due to high rates of cellular oxygen consumption. Under these conditions, transcriptional activation of CA9 requires cooperation of SP1 with minimal HIF-1 activity and operates through the PI3K pathway (18). The MAPK/ERK pathway has also been implicated in regulation of CA9 expression in response to both hypoxia and culture at high cell density (19, 20). Simultaneous inhibition of PI3K and MAPK pathways is sufficient to prevent CA9 expression during both conditions (19).
Changes in protein expression during hypoxia also arise through HIF-independent mechanisms (21). Several recent reports indicate that hypoxia activates an adaptive program called the unfolded protein response (UPR) (22–24). Activation of the UPR during hypoxia results in an immediate but reversible reduction in overall protein synthesis (25–27). This is mediated through phosphorylation of the eukaryotic translation initiation factor eIF2α by the endoplasmic reticulum resident kinase PERK. Mouse embryonic fibroblasts (MEFs) deficient in PERK or expressing an allele of eIF2α mutated at the PERK phosphorylation site (S51A) are unable to inhibit translation during hypoxia and demonstrate increased hypoxia sensitivity and profoundly decreased tumor growth compared with their WT controls (23). Although activation of the UPR causes a global decrease in mRNA translation, the degree of inhibition is highly gene-specific (25, 28, 29). The sensitivity of any particular mRNA to inhibition of translation is influenced by sequences in its 5′- and 3′-untranslated regions. Paradoxically, the translation of some mRNAs, including the transcription factor ATF4, are stimulated under conditions where eIF2α is phosphorylated (25, 28).
In this study we investigated the effect of UPR-dependent signaling during hypoxia on the regulation of CA9. We found that CA9 expression is regulated through a PERK/eIF2α/ATF4-dependent pathway. We show that ATF4 directly binds to the CA9 promoter and is required for transcriptional induction of CA9 during hypoxia. Our data indicate that regulation of CA9 is achieved through independent activation of both the HIF-1 and UPR hypoxia response.
EXPERIMENTAL PROCEDURES
Cell Culture
Exponentially growing U373 (human glioblastoma-astrocytoma) and HCT116 (human colon carcinoma) cells were grown in minimal essential medium α and Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal calf serum (Sigma-Aldrich). MEFs wild type or with a homozygous knock-in mutation for eIF2α (S51A) (30) were grown in Dulbecco's modified Eagle's medium plus 10% fetal calf serum and supplemented with 55 μm β-mercaptoethanol and non-essential amino acids. For hypoxic exposure cells were transferred into a MACS VA500 microaerophilic workstation (Don Whitley Scientific, Shipley, UK). The atmosphere in the chamber consisted of 5% H2, 5% CO2, 0% or 0.2% O2, and residual N2.
RNA Extraction and Quantitative Reverse Transcription- PCR
RNA was isolated using SV Total RNA Isolation System (Promega, Madison, WI) according manufacturers' instructions. RNA samples were reverse transcribed using iScript kit as described by the manufacturer (Bio-Rad). Real-time PCR was performed in ABI 7500 (Applied Biosystems). The abundance of human and mouse transcripts was determined using TaqMan Gene Expression Arrays (Applied Biosystems) or with primers in combination with SYBR Green I (Applied Biosystems) as described in supplemental Material and Methods. Relative gene expression was normalized by 18 S rRNA. For determination of CA9 mRNA half-life cells were treated with 5 μg/ml actinomycin D (Sigma) for the indicated times.
Western Blot Analysis
Cells were washed twice with cold PBS and scraped in 50 mm Tris HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS supplemented with protease inhibitors (Roche Applied Science). After centrifugation at 10,000 × g supernatants were boiled in Laemmli buffer for 10 min, and proteins were resolved by SDS-PAGE. Proteins were subsequently transferred onto nitrocellulose membranes and blocked overnight at 4 °C in PBS containing 0.1% Tween 20 (PBS-T) supplemented with 5% skim milk powder. Membranes were probed for 1 h with antibodies directed against CA9 (M75 (31)), HIF-1α (BD Transduction Laboratories), or β-actin (Sigma). Bound antibodies were visualized using horseradish peroxidase-linked secondary antibodies (anti-rabbit (Cell Signaling Technologies) and anti-mouse (Sigma)) and ECL luminescence (Pierce).
Transfection of Reporter Constructs and siRNA
U373 and MEF cells were transiently cotransfected with CA9 promoter constructs and pcDNALacZ using Lipofectamine (Invitrogen) as described by the manufacturer. Transfected cells were subcultured 16 h post transfection, exposed to hypoxia, and finally harvested 48 h after transfection. Luciferase and β-galactosidase activity was measured using commercial luciferase kit (Promega) and galacto light system kit (Applied Biosystems) in a LB9507 luminometer (Berthold). For transient knockdown U373 cells were transfected with 100 nm siRNA duplexes using Oligofectamine according to the manufacturer's instructions. siRNA duplex targeting ATF4 122372, CHOP 146321, ATF6 115889, and HIF1α 42840 was obtained from Ambion, and scramble control siRNA was from Dharmacon. Transfected cells were subcultured 24 h post transfection and harvested 72 h after transfection. The ATF4 open reading frame was amplified from pCMV_XL5 (Origen) using following primers ATF4_Fw5′-gtatgcggaggatccccgcaacatgaccgaaatgagc-3′ and ATF4_Rv5′-tccgcatacgaattcctcctgactatcctcaactagg-3′ and cloned into pcDNA5 (Invitrogen) using BamHI and EcoRI restriction sites. Transfection of U373 cells with pcDNA5-ATF4 or empty vector control was done as for the reporter constructs described above.
Chromatin Immunoprecipitation Assays
ChIPs were performed and analyzed essentially as described previously (32), see supplemental Materials and Methods.
Tumor Xenograft Growth
Animal experiments were performed using adult NMRI (nu/nu) female mice (28–32 g) at the animal facility of the Catholic University of Leuven in Belgium. The experiments were performed in accordance with and after approval of the local institutional Animal Care and Ethics Committee. In short, 1.5 million U373 pcDNA3, U373 HIF-1α, and U373 S51A cells were resuspended in 50 μl of Matrigel (BD Biosciences) and injected subcutaneously in the bilateral flanks of recipient mice. Tumors were excised after they reached ∼500 mm3.
Immunohistochemical Analysis of CA9
Frozen, acetone-fixed sections were stained using anti-CA9, anti-pimonidazole polyclonal rabbit antibody (E. Oosterwijk, Dept. of Urology, Radboud University Nijmegen Medical Centre) and 9F1 (rat monoclonal antibody to mouse endothelium, Dept. of Pathology, University Medical Center St. Radboud, The Netherlands) as described previously (33). For quantitative analysis, the slides were scanned by a computerized digital image processing system using a high resolution intensified solid-state camera on a fluorescence microscope (Zeiss Axioskop) with a computer-controlled motorized stepping stage. The scanning method has been described previously (34).
Statistical Analysis
Unpaired Student's t test or one-way analysis of variance with Bonferroni multiple comparison test was used to test significance between populations. p < 0.05 was considered significant. Points and error bars plotted in the graphs of all figures represent the mean ± S.D.
RESULTS
Hypoxic Induction of CA9 Requires eIF2α Phosphorylation
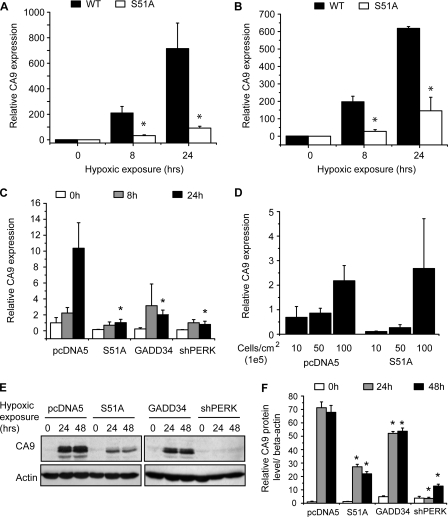
In an ongoing screen to characterize changes in gene expression during hypoxia mediated by the UPR we compared transcriptional profiles of wild-type and eIF2α S51A mutant MEFs. We identified CA9 as one of the top differentially regulated genes between the WT and S51A cells, which are unable to phosphorylate eIF2α (30) (data not shown). Quantitative reverse transcription-PCR analysis confirmed that CA9 mRNA levels during hypoxia were ∼5- to 8-fold lower in the S51A MEFs compared with their WT controls (Fig. 1, A and B). The reduction in CA9 expression in the S51A mutants is observed during culture under both severe (<0.02% O2) (Fig. 1A) and more moderate (0.2% O2) (Fig. 1B) hypoxia.
FIGURE 1.
CA9 expression is impaired in cells defective in eIF2α phosphorylation during hypoxic conditions. WT and S51A MEFs were exposed to severe (0.0% O2) (n = 3, Student's t test; *, p < 0.05, WT versus S51A) (A) or moderate (0.2% O2) (n = 3, Student's t test; *, p < 0.05, WT versus S51A) (B) hypoxia and CA9 mRNA levels were assessed using Q-PCR after 0, 8 and 24 h. C, CA9 levels measured in isogenic U373 cells expressing dominant negative eIF2α (S51A), GADD34, or short hairpin RNA against PERK after 0, 8, and 24 h of severe hypoxia (n = 3–6, one-way analysis of variance; *, p < 0.05, WT versus UPR impaired cells). D, U373 pcDNA5 and S51A were seeded at different densities and analyzed with Q-PCR for CA9 mRNA expression (n = 3). E, CA9 protein expression was assesses by Western blotting in isogenic U373 cell lines after exposure to hypoxia. F, quantitation of CA9 levels normalized to actin (n = 4, one-way analysis of variance; *, p < 0.001, WT versus UPR-impaired cells).
We also assessed hypoxia-induced CA9 expression in a series of human cell lines engineered to have defects in eIF2α phosphorylation. We created isogenic stable cell lines expressing different constructs at a single genomic site that effectively prevent eIF2α phosphorylation (supplemental Fig. S1).3 These included expression of a dominant negative eIF2α (S51A) (35) that is insensitive to PERK signaling, a C-terminal fragment of GADD34 that promotes dephosphorylation of eIF2α, and a short hairpin RNA against PERK. Fig. 1C shows that expression of each of these three constructs in U373 astrocytoma-glioblastoma cells effectively prevents induction of CA9 during hypoxia. Similar results were observed in HCT116 colon carcinoma cells carrying the same constructs (supplemental Fig. S2). The requirement for eIF2α phosphorylation seems specific to hypoxia, because up-regulation of CA9 during culture at high cell density under aerobic conditions is similar in wild-type and S51A-expressing cells (Fig. 1D). Importantly, the reduction in CA9 mRNA levels in the eIF2α phosphorylation-deficient cells also results in reduced CA9 protein expression during hypoxia (Fig. 1E).
Dependence upon eIF2α Phosphorylation on CA9 Is Independent of HIF
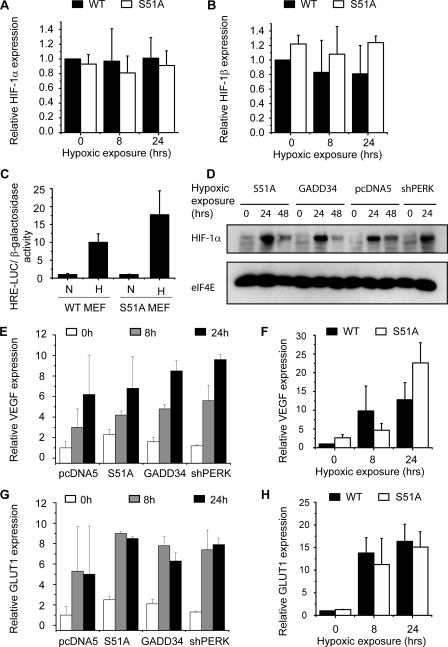
Because the transcription of CA9 is strongly regulated by HIF-1, we investigated the possibility that HIF-1 signaling was aberrant in cells with defective eIF2α phosphorylation. However, no differences in the levels of HIF-1α (Fig. 2A) or HIF-1β (Fig. 2B) mRNA were observed between S51A MEF and their WT controls. HIF activity as assessed using an HRE luciferase reporter plasmid was not significantly different in the two cell lines (p = 0.17) (Fig. 2C). We additionally assessed HIF-1α protein levels in the human isogenic U373 cell lines after exposure to hypoxia and found comparable levels in the three eIF2α defective cell lines as in the empty vector control cells (Fig. 2D). Finally, we assessed the transcriptional induction of two additional endogenous HIF-dependent genes, GLUT-1 and VEGF. In contrast to CA9, the expression of both VEGF (Fig. 2, E and F) and GLUT-1 (Fig. 2, G and H) was unaffected in the cell lines with defective eIF2α phosphorylation. Together these data suggest that HIF-1 activity is not affected by eIF2α phosphorylation.
FIGURE 2.
HIF-1 expression and activity does not require UPR signaling. Analysis of HIF-1α (A) and HIF-1β (B) mRNA levels in WT and S51A MEF cells (n = 2). C, HIF activity was measured in WT and S51A MEF cells using HRE-luciferase constructs normalized to co-transfected β-galactosidase (n = 4). D, Western blot analysis of HIF-1α levels in isogenic U373 cell lines. Induction of HIF-1α target genes VEGF (E) and GLUT-1 (G) in U373 cells after 0, 8, and 24 h of hypoxia and VEGF (F) and GLUT-1 (H) in WT and S51A MEF cells measured by Q-PCR (n = 3).
UPR Activation Is Required but Not Sufficient for CA9 Induction
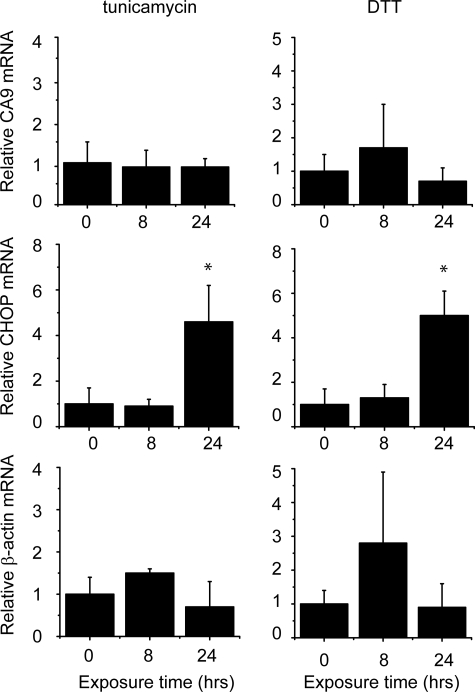
These data suggested that activation of PERK and phosphorylation of eIF2α contributes to CA9 transcription during hypoxia. We therefore investigated whether other activators of PERK and the UPR might also regulate CA9. To this end we induced endoplasmic reticulum stress in U373 cells by treatment with either dithiothreitol or tunicamycin. Although both drugs clearly caused activation of the UPR, as demonstrated by induction of the UPR responsive gene CHOP (Fig. 3), neither substantially affected CA9 mRNA levels.
FIGURE 3.
Endoplasmic reticulum stress does not induce CA9 expression during normoxia. U373 cells were stimulated with 1 μg/ml tunicamycin or 1 mm dithiothreitol for 8 or 24 h. Expression levels for CA9, CHOP, and actin were determined using Q-PCR (n = 6–8, Student's t test; *, p < 0.05, control versus dithiothreitol or tunicamycin).
CA9 Is Regulated at the Transcriptional Level by ATF4
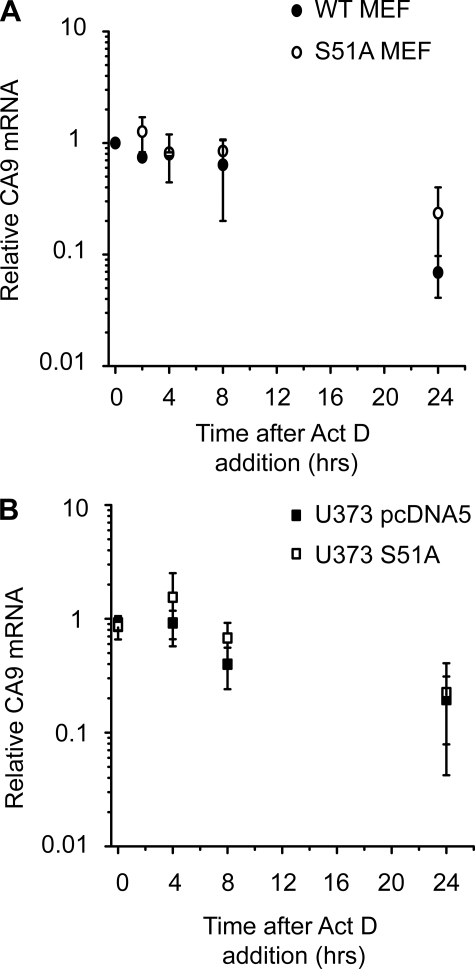
To explain the reduced CA9 mRNA levels in UPR impaired cells we first examined whether mRNA stability was dependent upon eIF2α phosphorylation. To this end we determined CA9 mRNA half-life after blocking transcription with actinomycin D for various lengths of time. However, the results presented in Fig. 4, A (MEFs) and B (U373), indicate that CA9 mRNA half-life was not altered in the eIF2α phosphorylation-deficient cells.
FIGURE 4.
Stability of CA9 mRNA is not dependent on eIF2α phosphorylation. A, CA9 mRNA levels were assessed using Q-PCR in WT and S51A MEF cells after induction with 100 μm cobalt chloride for 16 h, CA9 mRNA levels were plotted on a log scale at different time points after addition of actinomycin D (n = 2). B, CA9 mRNA stability in U373 pcDNA5 and U373 S51A cells was determined after exposure to 16 h of hypoxia (n = 5).
These data suggest that differences in CA9 expression are likely due to changes in transcription. Therefore we investigated CA9 promoter activity using luciferase reporter constructs. Promoter region −174 to +37 (pGL3-PR5) contains the 5 previously identified regulatory promoter elements (PR1–PR5) and an HRE that influence transcriptional activation of CA9 (5, 17). The minimal promoter region necessary for hypoxia-induced expression of CA9 is −50 to +37 (pGL3-PR1). Essential for this are the SP1 transcription factor in PR1 and the HRE immediately downstream of PR1. In our experiments both promoter sequences (−174 to +37) and (−50 to +37) showed basal levels of activity in UPR-impaired cells and their controls under normoxic conditions (Fig. 5A). However, exposure to hypoxia resulted in a significant induction of promoter activity in all cell lines examined. As expected, mutations in either the SP1 binding site or the HRE element completely abolished hypoxia inducibility. The fact that we did not observe any differences between UPR-impaired cells and control suggests that either elements outside the (−174 to +37) promoter region are responsible for the UPR-dependent effect on CA9 expression or that the UPR does not influence transcription of these non-chromatin containing plasmids.
FIGURE 5.
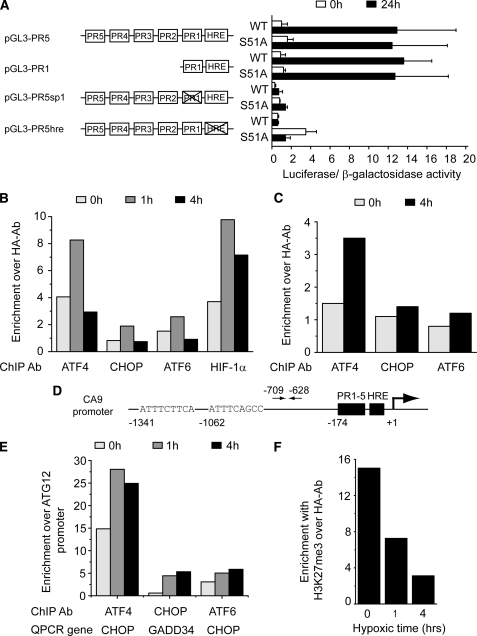
ATF4 binds to the CA9 promoter region. A, WT CA9 promoter construct or mutant versions with PR2–5 deleted, sp1 binding site or HRE inactivated were transiently cotransfected with pcDNA3-LacZ into WT MEF or S51A MEF cells. Luciferase activity was measured relative to β-galactosidase expression after 24 h of hypoxia after which -fold induction was calculated for each construct. Bars represent the average of four independent experiments plus standard deviation. ChIP analysis was performed on U373 (B) and HCT116 (C) cells after exposure to normoxia or hypoxia using indicated antibodies. Q-PCR was used to determine -fold enrichment of the CA9 promoter region over that obtained with negative control antibody HA. D, localization of putative ATF4 binding sites in CA9 promoter region, arrows indicate location of primers. E, positive controls for ChIP analysis. The -fold enrichment of CHOP and GADD34 promoter regions for ATF4, ATF6, and CHOP, respectively, over unrelated promoter region of ATG12 gene. F, ChIP analysis using U373 cells with antibodies against H3K27me during hypoxia, -fold enrichment over negative control anti-HA antibody.
ATF4 Binds the CA9 Promoter
Because our reporter constructs did not cover the complete promoter, we attempted to identify transcription factors that are able to bind the CA9 promoter using ChIP. Using this approach we tested three transcription factors that are known to be activated by the UPR: ATF4, CHOP, and ATF6. ATF4 is induced at the translational level in response to PERK-dependent phosphorylation of eIF2α during hypoxia (23, 28, 36). CHOP is a transcriptional target of both ATF4 and ATF6 and is also preferentially translated during hypoxia (25, 36). Interestingly, CHOP has previously been implicated in the regulation of CA6 (37). ATF6 on the other hand is also induced by the UPR, but has no known dependence upon PERK or eIF2α phosphorylation. U373 and HCT116 cells were exposed to aerobic or hypoxic conditions (4 h) after which ChIPs were performed using antibodies against ATF4, CHOP, ATF6, and the HA epitope tag. We observed enrichment of the CA9 promoter after performing the ChIP using ATF4 antibody in U373 (Fig. 5B) and HCT116 (Fig. 5C). In contrast, there was no enrichment using CHOP or ATF6. ATF4 binding could be explained by the presence of two putative amino acid response elements in the CA9 promoter region that closely resemble the ATF4 DNA binding sequence in the CHOP promoter (ATTGCATCA) (38) (Fig. 5D). Furthermore we show that we could efficiently immunoprecipitate the CA9 promoter using an antibody directed against HIF-1α (Fig. 5B). As additional controls for the ChIP procedures we determined association of the precipitated transcription factors with previously identified interacting promoter regions. The ATF4 and ATF6 ChIPs effectively pulled down the CHOP promoter (up to 28-fold and 6-fold enrichment, respectively), and CHOP pulled down the GADD34 promoter (∼5-fold enriched) (Fig. 5E).
We also performed ChIP analysis using an antibody directed against the histone H3 methylation mark on lysine 27 (H3K27me). The presence of H3K27me correlates with repression of gene expression (39). With U373 cells cultured under aerobic conditions we obtained significant enrichment of the CA9 promoter using an antibody against the H3K27me mark compared with an unrelated control antibody (Fig. 5F). Strikingly this H3K27me mark at the CA9 promoter disappears rapidly upon exposure to hypoxia. Similar results were obtained using HCT116 cells (data not shown). These data suggest that ATF4 mediates endogenous transcriptional activation of CA9 by direct binding to its promoter and that activation occurs following a change in histone methylation.
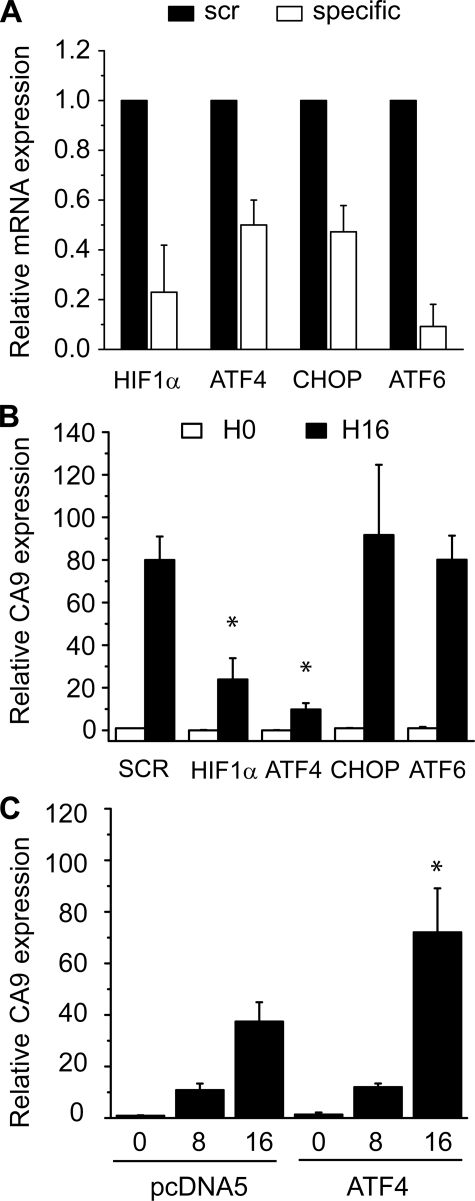
To investigate the functional importance of ATF4 in regulating CA9 expression during hypoxia we knocked down ATF4 using siRNA. In addition we knocked down the ATF4-regulated transcription factor CHOP as well as HIF-1α and ATF6 as positive and negative controls, respectively. We achieved 50–90% knockdown of our target genes compared with scrambled siRNA control in U373 (Fig. 6A) and HCT116 cells (supplemental Fig. S3A). U373 cells depleted of HIF-1α or ATF4 exhibited a strong reduction in CA9 mRNA compared with the scrambled controls after 16 h of hypoxia (Fig. 6B). Depletion of the other UPR-related transcription factors CHOP and ATF6 did not affect CA9 induction during hypoxia.
FIGURE 6.
ATF4 is required for CA9 mRNA expression during hypoxia. U373 cells were transiently transfected with siRNA duplexes directed against HIF1α, ATF4, CHOP, and ATF6. 24 h after transfection cells were exposed to 16 h of hypoxia. A, the knockdown efficiency was determined for gene-specific siRNAs using Q-PCR. Expression was normalized to mRNA levels in cells transfected with scrambled control (n = 3). B, relative expression of CA9 mRNA post transfection with siRNA after culturing under normoxia or 16 h of hypoxia (n = 3, Student's t test; *, p < 0.05, scrambled versus knock down). C, exogenous ATF4 was overexpressed in U373 cells and exposed to 0, 8, and 16 h of hypoxia. Relative levels of CA9 mRNA were determined using Q-PCR compared with empty vector control under aerobic conditions (n = 5, Student's t test; *, p < 0.05, pcDNA versus ATF4).
To determine if cellular levels of ATF4 were directly influencing endogenous CA9 expression we also overexpressed ATF4 in U373 cells and exposed them to hypoxic conditions. Again basal mRNA levels of CA9 were unaffected under aerobic conditions however after 16 h of hypoxia CA9 mRNA levels were ∼2-fold higher compared with cells bearing an empty vector control (Fig. 6C). Taken together, our data support a model in which hypoxic induction of CA9 is dependent on the PERK controlled arm of the UPR involving direct binding of ATF4 to the CA9 promoter.
The UPR Mediates Expression of CA9 in Vivo
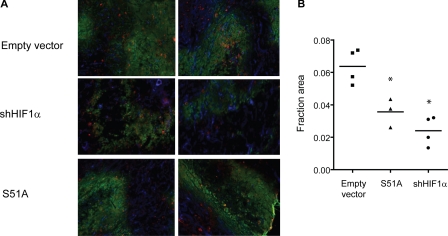
Finally to investigate the role of the UPR on induction of CA9 expression in vivo, xenograft tumors were grown from U373 cells bearing empty vector, eIF2α S51A, or short hairpin RNA against HIF-1α as a positive control. As expected immunohistochemical stainings using an antibody against CA9 revealed an overall decreased staining intensity in the HIF-1α knockdown tumor sections compared with the empty vector controls (Fig. 7, A and B). Similarly, tumors derived from the U373 cells with defective eIF2α phosphorylation also showed a reduction in CA9 levels that was comparable to that observed in the HIF-1α knockdown tumors (Fig. 7, A and B). Thus, in both in vitro and in vivo experiments expression of CA9 appears to be as dependent on UPR activation as it is on HIF1.
FIGURE 7.
Overall CA9 expression is reduced in sections of U373 S51A xenograft tumors. A, immunohistochemical analysis of CA9 (red), hypoxia (green), and vessels (blue) in xenograft tumors derived from U373 cells bearing empty vector (n = 4), eIF2α S51A (n = 3) or short hairpin RNA against HIF-1α (n = 4). B, quantitation of CA9 expression in multiple xenograft tumors (one-way analysis of variance; *, p < 0.05, empty vector versus S51A or shHIF1α).
DISCUSSION
It is well established that hypoxic conditions induce CA9 transcription through the transcription factor HIF, which binds to the HRE element in the CA9 promoter (5). Here we showed that in addition to HIF, PERK signaling to eIF2α is necessary for CA9 induction during hypoxia. This was demonstrated using several different genetic models in both human and mouse cells. The requirement for UPR signaling during hypoxia does not extend to CA9 induced by high cell density under aerobic conditions (40), which has been shown to involve the PI3K pathway (18) and MAPK signaling (19). These results support the existence of two distinct mechanisms for CA9 transcriptional regulation, an HIF-dependent mechanism activated under low oxygen levels and a PI3K/MAPK-dependent pathway at high cell density. Here we provide evidence that the former mechanism relies on simultaneous PERK/eIF2α signaling.
The dependence of CA9 expression on eIF2α phosphorylation is directly mediated by ATF4. Knockdown or overexpression of ATF4 decreased or increased CA9 expression during hypoxia, respectively. Furthermore, we showed that ATF4 directly binds to the CA9 promoter. Interestingly published microarray data from several studies has shown that CA6 is also responsive to UPR activation (37, 41, 42). In contrast to CA9 this effect is dependent on the ATF4-responsive transcription factor CHOP and not directly through ATF4 (37, 42). Nonetheless these data suggest an important and broader role for PERK/eIF2α/ATF4 in regulating pH levels through up-regulation of carbonic anhydrases.
The function of ATF4 on the CA9 promoter and its potential co-operation with HIF1 is not fully understood. Induction of the UPR under non-hypoxic conditions using dithiothreitol or tunicamycin was unable to induce CA9. Interestingly we found that the CA9 promoter contains tri-methylation marks on lysine residue 27 of histone H3 under aerobic conditions. These marks are generally correlated to a transcriptional repressive state (39). We found that this repressive mark was lost after exposure to hypoxia for periods as short as 1 h. These data suggest that hypoxia influences CA9 expression in part through an epigenetic mechanism. This result is interesting in relation to a recent report suggesting that hypoxia can affect global changes in histone methylation (43). In part this might be mediated by the jumonji family of histone demethylases that have recently been shown to be up-regulated during hypoxia via HIF (44, 45). It will be interesting to determine whether ATF4 also co-regulates the expression of these genes together with HIF as it does for CA9.
ATF4 may also influence CA9 through its known interaction with the transcriptional co-activator p300/CBP (46) or p300/CBP-associated factor (47). Both p300/CBP and p300/CBP-associated factor contain histone acetyltransferase activity and function as co-activators in the transcriptional regulation of many genes. Our data indicate that recruitment of HIF during hypoxia is essential but not sufficient for complete transcriptional activation of CA9. Because p300/CBP is required for HIF-mediated transcription, it is attractive to suggest a model in which ATF4 and HIF cooperate through their ability to recruit p300/CBP and/or p300/CBP-associated factor. Such cooperation would be meaningful in a certain hypoxic range, which simultaneously activates both pathways.
Adaptation to hypoxic conditions through activation of the UPR is an important survival mechanism for tumor cells. The Ire1/Xbp1 and PERK/eIF2α arms of the UPR have been shown to play a critical role in determining hypoxia sensitivity (21, 23, 48). PERK knock-out MEFs have decreased hypoxia tolerance and tumor growth in xenograft studies due to their inability to activate eIF2α phosphorylation (23). It is tempting to speculate that this growth defect could be due in part to decreased CA9 expression. It has previously been shown that knockdown of CA9 in MDA468 and MDA231 breast carcinoma cell lines using siRNA resulted in a significant reduction in clonogenic survival after hypoxic exposure compared with control cells (16). However, whether decreased CA9 expression is responsible for the decrease growth rate observed with the eIF2α-deficient tumors needs further investigation. PERK/eIF2α phosphorylation and up-regulation of ATF4 are also involved in other important processes, including angiogenesis (41), and maintenance of energy homeostasis (49). Thus, as is the case for HIF, the contribution of the UPR to hypoxia tolerance is likely to be multifactorial.
Our data also contribute to a growing overlap in the contribution of UPR- and HIF-dependent responses to the hypoxic tumor phenotype. pH regulation, angiogenesis, and energy homeostasis are also all strongly regulated by HIF (3, 21). In this regard it is important to note that the oxygen dependences and activation kinetics of HIF and the UPR are not the same. Whereas HIF is activated at relatively moderate hypoxic conditions (1–2%), full activation of the UPR requires more severe hypoxia (22, 50). Consequently, the UPR and HIF pathways may allow cells to adjust the strength of these responses as a function of oxygen concentration.
CA9 has become widely adopted as a surrogate marker of tumor hypoxia used to classify patients that likely respond poorly to therapy (51, 52). Although CA9 expression generally correlates well with the exogenous marker of hypoxia pimonidazole, its correlation with HIF-1α is much weaker (53, 54). CA9 expression is often detected in the perinecrotic regions of solid tumors where HIF1α levels are reduced (54). Several possible explanations have been suggested for this poor co-localization (54), including the large difference in protein half-life between HIF-1α and CA9. Due to its long half-life CA9 expression may be present in areas that have been reperfused with oxygen and no longer express HIF-1α. Our results provide an additional mechanism to explain high CA9 expression in perinecrotic tumor areas. Tumor cells in these regions are likely to exhibit activation of PERK due to the extremely hypoxic nature of these regions. Therefore, simultaneous activation of the UPR, together with more moderate HIF expression, might drive high CA9 expression in these areas. The requirement of UPR signaling may also explain why some HIF-1-positive tumors fail to express CA9.
Supplementary Material
Acknowledgments
We thank Natasja Lieuwes and Hans Peters for technical assistance.
This work was supported by the Dutch Science Organization (ZonMW-NWO Top Grant 912-03-047 to B. W. and Grants 908-02-040 and 016.046.362 to J. W. V.), the Dutch Cancer Society (KWF Grant UM 2003-2821 to B. W.), the European Union 6th framework program (Euroxy program to B. W.), and ZonMW VENI Grants 016.056.015 (to M. K.) and 916.76.158 (to K. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
K. M. A. Rouschop and B. G. Wouters, in preparation.
- HIF
- hypoxia-inducible factor
- HRE
- hypoxia-responsive element
- PERK
- PKR-like endoplasmic reticulum kinase
- CA9
- carbonic anhydrase 9
- PI3K
- phosphatidylinositol 3-kinase
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- UPR
- unfolded protein response
- MEF
- Mouse embryonic fibroblast
- WT
- wild type
- PBS
- phosphate-buffered saline
- siRNA
- small interference RNA
- ChIP
- chromatin immunoprecipitation
- HA
- hemagglutinin
- H3K27me
- histone H3 methylation mark on lysine 27
- CBP
- CREB (cAMP-response element-binding protein)-binding protein.
REFERENCES
- 1.Brown J. M., Wilson W. R. (2004) Nat. Rev. Cancer 4, 437–447 [DOI] [PubMed] [Google Scholar]
- 2.Hockel M., Vaupel P. (2001) Semin. Oncol. 28, 2 Suppl. 8,36–41 [PubMed] [Google Scholar]
- 3.Semenza G. L. (2003) Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 4.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 5.Wykoff C. C., Beasley N. J., Watson P. H., Turner K. J., Pastorek J., Sibtain A., Wilson G. D., Turley H., Talks K. L., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Cancer Res. 60, 7075–7083 [PubMed] [Google Scholar]
- 6.Pastorekova S., Parkkila S., Zavada J. (2006) Adv. Clin Chem. 42, 167–216 [PubMed] [Google Scholar]
- 7.Potter C. P., Harris A. L. (2003) Br. J. Cancer 89, 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svastová E., Hulíková A., Rafajová M., Zat'ovicová M., Gibadulinová A., Casini A., Cecchi A., Scozzafava A., Supuran C. T., Pastorek J., Pastoreková S. (2004) FEBS Lett. 577, 439–445 [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Zaguilán R., Seftor E. A., Seftor R. E., Chu Y. W., Gillies R. J., Hendrix M. J. (1996) Clin. Exp. Metastasis 14, 176–186 [DOI] [PubMed] [Google Scholar]
- 10.Moellering R. E., Black K. C., Krishnamurty C., Baggett B. K., Stafford P., Rain M., Gatenby R. A., Gillies R. J. (2008) Clin. Exp. Metastasis 25, 411–425 [DOI] [PubMed] [Google Scholar]
- 11.Stubbs M., McSheehy P. M., Griffiths J. R., Bashford C. L. (2000) Mol. Med. Today 6, 15–19 [DOI] [PubMed] [Google Scholar]
- 12.Swietach P., Wigfield S., Supuran C. T., Harris A. L., Vaughan-Jones R. D. (2008) BJU Int. 4, Suppl. 101,22–24 [DOI] [PubMed] [Google Scholar]
- 13.Swietach P., Wigfield S., Cobden P., Supuran C. T., Harris A. L., Vaughan-Jones R. D. (2008) J. Biol. Chem. 283, 20473–20483 [DOI] [PubMed] [Google Scholar]
- 14.Supuran C. T. (2008) Nat. Rev. Drug Discov. 7, 168–181 [DOI] [PubMed] [Google Scholar]
- 15.Dubois L., Douma K., Supuran C. T., Chiu R. K., van Zandvoort M. A., Pastoreková S., Scozzafava A., Wouters B. G., Lambin P. (2007) Radiother. Oncol. 83, 367–373 [DOI] [PubMed] [Google Scholar]
- 16.Robertson N., Potter C., Harris A. L. (2004) Cancer Res. 64, 6160–6165 [DOI] [PubMed] [Google Scholar]
- 17.Kaluz S., Kaluzová M., Opavský R., Pastoreková S., Gibadulonová A., Dequiedt F., Kettmann R., Pastorek J. (1999) J. Biol. Chem. 274, 32588–32595 [DOI] [PubMed] [Google Scholar]
- 18.Kaluz S., Kaluzová M., Chrastina A., Olive P. L., Pastoreková S., Pastorek J., Lerman M. I., Stanbridge E. J. (2002) Cancer Res. 62, 4469–4477 [PubMed] [Google Scholar]
- 19.Kopacek J., Barathova M., Dequiedt F., Sepelakova J., Kettmann R., Pastorek J., Pastorekova S. (2005) Biochim. Biophys. Acta 1729, 41–49 [DOI] [PubMed] [Google Scholar]
- 20.Kaluz S., Kaluzová M., Stanbridge E. J. (2006) J. Cell. Biochem. 97, 207–216 [DOI] [PubMed] [Google Scholar]
- 21.Wouters B. G., Koritzinsky M. (2008) Nat. Rev. Cancer 8, 851–864 [DOI] [PubMed] [Google Scholar]
- 22.Koumenis C., Wouters B. G. (2006) Mol. Cancer Res. 4, 423–436 [DOI] [PubMed] [Google Scholar]
- 23.Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J., Scheuner D., Kaufman R. J., Bell J., Ron D., Wouters B. G., Koumenis C. (2005) EMBO J. 24, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman D. E., Chauhan V., Koong A. C. (2005) Mol. Cancer Res. 3, 597–605 [DOI] [PubMed] [Google Scholar]
- 25.Koritzinsky M., Magagnin M. G., van den Beucken T., Seigneuric R., Savelkouls K., Dostie J., Pyronnet S., Kaufman R. J., Weppler S. A., Voncken J. W., Lambin P., Koumenis C., Sonenberg N., Wouters B. G. (2006) EMBO J. 25, 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koumenis C., Naczki C., Koritzinsky M., Rastani S., Diehl A., Sonenberg N., Koromilas A., Wouters B. G. (2002) Mol. Cell. Biol. 22, 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koritzinsky M., Rouschop K. M., van den Beucken T., Magagnin M. G., Savelkouls K., Lambin P., Wouters B. G. (2007) Radiother. Oncol. 83, 353–361 [DOI] [PubMed] [Google Scholar]
- 28.Blais J. D., Filipenko V., Bi M., Harding H. P., Ron D., Koumenis C., Wouters B. G., Bell J. C. (2004) Mol. Cell. Biol. 24, 7469–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Beucken T., Koritzinsky M., Wouters B. G. (2006) Cancer Biol. Ther. 5, 749–755 [DOI] [PubMed] [Google Scholar]
- 30.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. (2001) Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 31.Pastoreková S., Zavadová Z., Kostál M., Babusíková O., Závada J. (1992) Virology 187, 620–626 [DOI] [PubMed] [Google Scholar]
- 32.Bracken A. P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. (2003) EMBO J. 22, 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troost E. G., Bussink J., Kaanders J. H., van Eerd J., Peters J. P., Rijken P. F., Boerman O. C., van der Kogel A. J. (2005) Radiother. Oncol. 76, 194–199 [DOI] [PubMed] [Google Scholar]
- 34.Rijken P. F., Peters J. P., Van der Kogel A. J. (2002) Radiat. Res. 157, 626–632 [DOI] [PubMed] [Google Scholar]
- 35.Donzé O., Jagus R., Koromilas A. E., Hershey J. W., Sonenberg N. (1995) EMBO J. 14, 3828–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 37.Sok J., Wang X. Z., Batchvarova N., Kuroda M., Harding H., Ron D. (1999) Mol. Cell. Biol. 19, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. (1999) Biochem. J. 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein B. E., Meissner A., Lander E. S. (2007) Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 40.Lieskovská J., Opavský R., Záciková L., Glasová M., Pastorek J., Pastoreková S. (1999) Neoplasma 46, 17–24 [PubMed] [Google Scholar]
- 41.Blais J. D., Addison C. L., Edge R., Falls T., Zhao H., Wary K., Koumenis C., Harding H. P., Ron D., Holcik M., Bell J. C. (2006) Mol. Cell. Biol. 26, 9517–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Kazuhiro Nagata K., Heather P., Harding H. P., Ron D. (2004) Genes Dev. 2004 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson A. B., Denko N., Barton M. C. (2008) Mutat. Res. 640, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beyer S., Kristensen M. M., Jensen K. S., Johansen J. V., Staller P. (2008) J. Biol. Chem. 283, 36542–36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollard P. J., Loenarz C., Mole D. R., McDonough M. A., Gleadle J. M., Schofield C. J., Ratcliffe P. J. (2008) Biochem. J. 416, 387–394 [DOI] [PubMed] [Google Scholar]
- 46.Liang G., Hai T. (1997) J. Biol. Chem. 272, 24088–24095 [DOI] [PubMed] [Google Scholar]
- 47.Chérasse Y., Maurin A. C., Chaveroux C., Jousse C., Carraro V., Parry L., Deval C., Chambon C., Fafournoux P., Bruhat A. (2007) Nucleic Acids Res. 35, 5954–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero-Ramirez L., Cao H., Nelson D., Hammond E., Lee A. H., Yoshida H., Mori K., Glimcher L. H., Denko N. C., Giaccia A. J., Le Q. T., Koong A. C. (2004) Cancer Res. 64, 5943–5947 [DOI] [PubMed] [Google Scholar]
- 49.Hochachka P. W., Buck L. T., Doll C. J., Land S. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magagnin M. G., Koritzinsky M., Wouters B. G. (2006) Drug Resist. Updates 9, 185–197 [DOI] [PubMed] [Google Scholar]
- 51.Airley R. E., Loncaster J., Raleigh J. A., Harris A. L., Davidson S. E., Hunter R. D., West C. M., Stratford I. J. (2003) Int. J. Cancer 104, 85–91 [DOI] [PubMed] [Google Scholar]
- 52.Beasley N. J., Wykoff C. C., Watson P. H., Leek R., Turley H., Gatter K., Pastorek J., Cox G. J., Ratcliffe P., Harris A. L. (2001) Cancer Res. 61, 5262–5267 [PubMed] [Google Scholar]
- 53.Jankovic B., Aquino-Parsons C., Raleigh J. A., Stanbridge E. J., Durand R. E., Banath J. P., MacPhail S. H., Olive P. L. (2006) Cytometry B Clin. Cytom. 70, 45–55 [DOI] [PubMed] [Google Scholar]
- 54.Sobhanifar S., Aquino-Parsons C., Stanbridge E. J., Olive P. (2005) Cancer Res. 65, 7259–7266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.