Abstract
In response to DNA damage, eukaryotic cells activate a series of DNA damage-dependent pathways that serve to arrest cell cycle progression and remove DNA damage. Coordination of cell cycle arrest and damage repair is critical for maintenance of genomic stability. However, this process is still poorly understood. Nucleotide excision repair (NER) and the ATR-dependent cell cycle checkpoint are the major pathways responsible for repair of UV-induced DNA damage. Here we show that ATR physically interacts with the NER factor Xeroderma pigmentosum group A (XPA). Using a mass spectrometry-based protein footprinting method, we found that ATR interacts with a helix-turn-helix motif in the minimal DNA-binding domain of XPA where an ATR phosphorylation site (serine 196) is located. XPA-deficient cells complemented with XPA containing a point mutation of S196A displayed a reduced repair efficiency of cyclobutane pyrimidine dimers as compared with cells complemented with wild-type XPA, although no effect was observed for repair of (6-4) photoproducts. This suggests that the ATR-dependent phosphorylation of XPA may promote NER repair of persistent DNA damage. In addition, a K188A point mutation of XPA that disrupts the ATR-XPA interaction inhibits the nuclear import of XPA after UV irradiation and, thus, significantly reduced DNA repair efficiency. By contrast, the S196A mutation has no effect on XPA nuclear translocation. Taken together, our results suggest that the ATR-XPA interaction mediated by the helix-turn-helix motif of XPA plays an important role in DNA-damage responses to promote cell survival and genomic stability after UV irradiation.
The genomes of all living cells are under constant attack from both endogenous and exogenous agents that may lead to genome instability. The nucleotide excision repair pathway (NER)3 is the primary mechanism in cells for the removal of bulky DNA lesions induced by exogenous agents such as UV radiation and a variety of genotoxic chemicals (1). In eukaryotic cells NER requires more than 25 proteins to perform the DNA damage recognition, excision, and DNA synthesis steps necessary to remove the lesion and restore the integrity of DNA (2, 3). In humans, defects in NER lead to the clinical disorder Xeroderma pigmentosum (XP) that is characterized by increased sensitivity to UV light and a predisposition to development of skin cancer (4, 5).
Xeroderma pigmentosum group A protein (XPA) is one of eight factors found to be deficient in XP disorder (2, 3, 6). XPA is a 32-kDa zinc metalloprotein that is believed to verify the damage site after initial recognition of the presence of a lesion, stabilize repair intermediates, and play a role in recruiting other NER factors (7–13). XPA is an indispensable factor for both the transcription-coupled repair and global genome NER pathways. Given its central role in NER, patients with XPA deficiency display the most severe XP phenotypes (2, 3). In addition, XPA has also been implicated to play a role in laminopathy-induced premature aging syndromes (14, 15).
The DNA damage checkpoint pathways serve to monitor genomic integrity and to coordinate multiple cellular pathways to ensure efficient repair of DNA damage (16). The ATM (ataxia-telangiectasia mutated) and ATR (ATM and RAD3-related)-mediated checkpoint pathways represent two major DNA damage-dependent checkpoints. Both ATM and ATR are protein kinases belonging to the phosphoinositide 3-kinase-like kinase family. These pathways are composed of a series of DNA damage sensors, signal mediators and transducers, and downstream effector molecules (1, 16, 17). The ATR-dependent checkpoint pathway serves to sense replication stress and responds primarily to DNA damage typically generated by UV irradiation (1, 18–20). ATR is targeted to the sites of elongated RPA-coated single-strand DNA generated when DNA replication forks stall because of DNA damage. This event is mediated by interactions between RPA and the ATR interaction protein ATRIP (18). Upon sensing DNA damage, ATR initiates a complex signaling cascade via phosphorylation of downstream protein substrates, which ultimately leads to cell cycle arrest (20, 21).
Previous studies have implied a role for the ATR-mediated checkpoint pathway in regulation of the NER pathway (17, 22, 23). In particular, ATR kinase activity may participate in the regulation of global genome NER uniquely during the S-phase of the cell cycle. Additionally, XPA has been defined as a direct ATR target for phosphorylation and cytoplasm-to-nucleus redistribution in response to UV-C irradiation (22). XPA−/− cells complemented with recombinant phosphorylation-deficient XPA protein displayed an increased sensitivity to UV-C irradiation compared with cells complemented with wild-type XPA (22). In addition, ATR directed the nuclear import of XPA in both a dose-dependent and time-dependent manner for regulation of NER activity (17).
Although there is growing evidence that the ATR-dependent checkpoint pathway coordinates with NER via an ATR-XPA interaction to promote DNA repair, how the interaction occurs and its significance have not been defined. Furthermore, the molecular basis of the ATR-XPA interaction remains to be elucidated. There is no structural information available for the ATR kinase or a model for how it binds to target proteins. In this study we investigated the molecular basis for the ATR-XPA interaction. Using our mass spectrometric protein footprinting technique, we identified an α-helix in the XPA minimum DNA-binding domain that mediates the XPA-ATR interaction. In addition, we demonstrate that regulation of XPA activity by ATR via ATR-XPA interaction is required for promoting repair of UV-C induced DNA damage by NER.
EXPERIMENTAL PROCEDURES
Cell Lines and Tissue Culture
XPA−/− cells (GM04429) were obtained from Coriell Cell Repositories (Camden, NJ) and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. XPA-complemented cells were generated by stably transfecting GM04429 cells with pcDNA3.1 vectors (Invitrogen) containing either wild-type or mutated XPA cDNA with the indicated point mutations as described previously (22). U2OS stably transfected with the doxycycline-inducible FLAG-tagged ATR expression construct were a generous gift from Dr. Paul Nghiem (University of Washington Medical Center) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 0.2 mg/ml neomycin, and 0.2 mg/ml hygromycin. All cell lines were grown at 37 °C, 5% CO2. UV-C irradiation was performed using a 254-nm lamp at a fluence of 1.3 J/m2/s. For time course analysis cells were incubate at 37 °C, 5% CO2 for the indicated amounts of time.
FLAG-ATR Immunoprecipitation and ATR-XPA Complex Formation
U2OS-FLAG-ATR cells were grown overnight in 10-cm tissue culture dishes in Dulbecco's modified Eagle's medium supplemented with doxycycline (5 μg/ml). Cells were harvested by scraping and resuspended in lysis buffer (50 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1× protease inhibitor mixture (Roche Applied Science)). Clarified lysates were immunoprecipitated using monoclonal mouse-anti-FLAG M2 antibody (Sigma) and captured with Protein G-coated Sepharose beads (Amersham Biosciences). Beads were rinsed with wash buffer A (50 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.05% NP-20) before washing with high salt buffer (50 mm HEPES-KOH, pH 7.4, 1 m NaCl, 1 mm EDTA, 0.05% Nonidet P-40). Immunoprecipitates then were equilibrated in the XPA-ATR binding buffer (50 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm ATP), and purified His6-XPA was added and incubated at room temperature for 30 min. Unbound XPA was washed away using buffer B (50 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm ATP, 0.05% Nonidet P-40). The resulting complex was modified by the addition of NHS-biotin (1 mm final concentration) for 30 min. The reactions were quenched with 10 mm lysine in its free form. The interacting proteins were separated by SDS-PAGE and visualized by Coomassie stain. The XPA bands were excised from the gel and subjected to in-gel trypsin hydrolysis as described previously (24, 25).
Mass Spectrometric Analysis
The tryptic peptide fragments were analyzed with matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry using an Axima-CRF instrument (Shimadzu Scientific Instruments). Samples were ionized with an α-cyano-4-hydroxycinnamic acid matrix. To identify XPA peptide peaks, the experimental data were compared with the theoretical values obtained with Protein Prospector Version 4.0.6. The experimental measurements indicated the mass accuracy of 0.1–0.01%. Modified lysine residues were assigned by identifying the peptide peaks formed upon the NHS-biotin treatment of the protein. The experimental mass/charge data for modified peptides were then compared with the predicted theoretical values considering that each modification adds 226 Da to the affected Lys and renders the residue resistant to tryptic hydrolysis. For accurate quantitative analysis of the modified peptide peaks, at least two unmodified proteolytic peptide peaks were used as internal controls. A protection was considered to be significant when the intensity of the given modified peptide peak derived from NHS-biotin-treated free protein was reduced at least 10-fold in the context of the protein-protein complex. A modified peptide peak was considered unprotected when the intensities of the given peptide obtained from free protein and protein-protein complexes were within ±20% of each other. The data were reproducibly compiled and analyzed from at least four independent experimental groups.
Coimmunoprecipitation
Coimmunoprecipitation experiments were conducted by lysing cells with lysis buffer A and adding dilution buffer to reduce the final NaCl concentration to 150 mm. One mg of total protein was immunoprecipitated using 2 μg of monoclonal mouse-anti-XPA (Clone 12FA, Kamiya Biochemical). Samples were resolved on a 4–12% gradient SDS-PAGE for Western blot analysis.
Immunofluorescent DNA Repair Assay
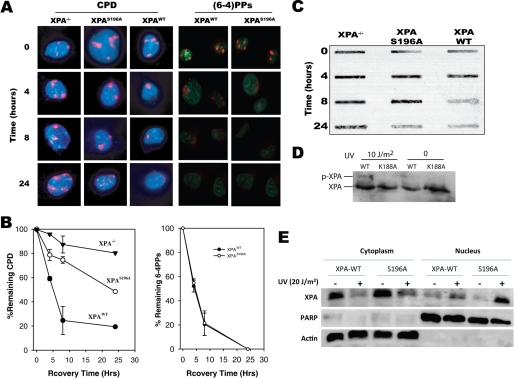
For immunofluorescence microscopy, cells were grown on coverslips and UV-C-irradiated through 3- or 5-μm polycarbonate isopore filters (Millipore) and allowed to recover for the indicated amounts of time. Cells were fixed with 100% methanol and treated with 1 m HCl to denature the DNA. Cyclobutane pyrimidine dimers (CPD) or (6-4) photoproducts ((6-4)PPs) were detected with monoclonal mouse-anti-CPD (TDM-2, MBL International Corporation) or anti-(6-4)PPs (D195–1, MBL) and donkey-anti-mouse Alexa Fluor 568 antibodies (Molecular Probes). Coverslips were mounted in ProLong Antifade with DAPI (Molecular Probes) and visualized using 100× magnification. Data were recorded under single-blind conditions in which the individual performing the microscopy did not know the identity of the samples. Repair of CPD or (6-4)PPs damage was quantified as a percentage of DAPI-stained nuclei containing at least one well defined CPD or (6-4)PPs focus by overlaying the anti-CPD or anti-(6-4)PPs and DAPI images. At least 50 nuclei per time point were counted for damage repair quantification. Samples then were normalized, with time point 0 h representing 100% nuclei containing CPD or (6-4)PPs foci. Images were analyzed using Photoshop CS.
Slot-blot DNA Damage Repair Assay
Cells were seeded at 1 × 106 cells per 10-cm tissue culture dish and allowed to grow for 48 h before UV-C irradiation. Cells were allowed to recover for the indicated amounts of time, and genomic DNA was purified using the PureLink Genomic DNA kit (Invitrogen). Purified DNA was quantified by measuring the A260 nm, and samples were diluted to 0.2 μg/ml in a final volume of 200 μl of TE buffer containing 10 mm Tris-HCl, pH 7.4, and 1 mm EDTA. Samples were denatured by incubating at 90 °C for 10 min then rapidly chilled on ice for 10 min before adding an equal volume of 2 m ammonium acetate. Samples were filter-immobilized on a nylon membrane and probed using monoclonal mouse-anti-CPD.
Subcellular Fractionation
The subcellular protein fractionation was performed using the Proteo JETTM cytoplasmic and nuclear protein extraction kit (Fermentas) and by following the procedures as suggested by the manufacturer. Briefly, 10 volumes of cell lysis buffer (with protease inhibitors) were added to 1 volume of packed cells. After vortexing for 10 s and incubation on ice for 10 min, cytoplasmic proteins were separated from nuclei by centrifugation at 500 × g for 7 min. Isolated nuclei were washed once with 500 μl of the nuclei washing buffer and then collected by centrifugation. The collected nuclear pellets were resuspended in ice-cold nuclear storage buffer, and volume of the nuclear lysis reagents were added to the mixtures to lysis the nuclei by shaking for 15 min at 4 °C. Then nuclear lysate was collected after rinsing by centrifugation at 20,000 × g for 12 min.
Computational Modeling
An initial model of phosphorylated XPA was generated from the PDB coordinates 1D4U (26) using the Biopolymer module of Insight II (Accelrys, Inc., San Diego, CA). Native and phosphorylated XPA structures were subsequently minimized in Amber 9 (27). Electrostatic surfaces were calculated using the APBS plug-in PyMOL (28–30); atomic charge and radius assignments were prepared for the electrostatic calculation using PDB 2PQR (31).
RESULTS
Protein Purification and ATR-XPA Complex Formation
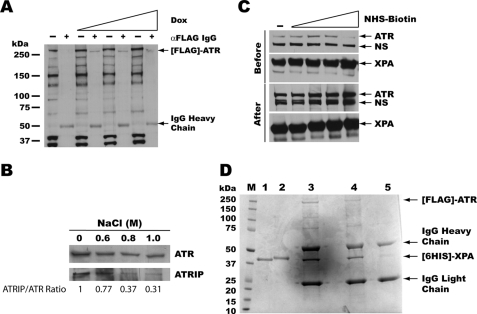
Recombinant XPA containing an N-terminal His6 tag was purified from baculovirus-infected insect cells as described previously (10). Recombinant ATR protein containing an N-terminal FLAG tag was purified from U2OS cells expressing ATR cDNA under the control of a tetracycline-inducible promoter (32). To obtain the highest yield of ATR, FLAG-ATR expression was induced by tetracycline derivative doxycycline at varying concentrations of 1, 2, and 5 μg/ml (Fig. 1A). Whole cell lysates were obtained followed by immunoprecipitation with FLAG antibody. Analysis of the samples with and without immunoprecipitation by Western blotting of a 3–8% gradient SDS gel shows that the amount of ATR increases with doxycycline concentration and a single band of ATR present only in the doxycycline-induced cell lysates (Fig. 1A). Based on these results, we chose 5 μg/ml doxycycline to induce FLAG-ATR expression. To obtain relatively pure ATR for XPA binding, the immunoprecipitated FLAG-ATR was washed with a high salt-containing buffer to remove proteins associated with the FLAG-ATR. Because ATR forms a tight complex with ATRIP in vivo (33), the presence of ATRIP coimmunoprecipitated with ATR was monitored with increasing concentrations of NaCl to test the efficiency of the high salt wash. Fig. 1B illustrates that as the salt concentration increases to 1 m, the levels of ATRIP co-immunoprecipitated with FLAG-ATR decreases significantly, as detected by Western blotting. Although about 30% ATRIP remained after the 1 m NaCl wash, the reduction of tightly bound ATRIP shows the efficiency of the wash procedure. We also found that further increasing the salt concentration above 1 m interfered with the antibody-antigen interaction. The high salt-washed FLAG-ATR could then be incubated with purified recombinant XPA to investigate complex formation.
FIGURE 1.
XPA-ATR complex preparation. A, U2OS-ATR cells were treated with increasing amounts of doxycycline for 24 h to induce expression of the FLAG-ATR construct. Then, cell lysates were mixed with anti-FLAG IgG, and immunoprecipitated proteins were analyzed by Western blot using anti-ATR monoclonal IgG. Western blot analysis indicates that as the concentration of doxycycline increases, the amount of FLAG-ATR expression also increases, whereas immunoprecipitation of the FLAG-tagged protein results in a single full-length protein. B, FLAG-ATR protein was immunoprecipitated and washed with increasing concentrations of NaCl solution to remove bound proteins. ATRIP forms a tight complex with ATR and is efficiently removed by washing with 1 m NaCl. The ATRIP/ATR ratios were normalized to the ratio at zero salt concentration. C, recombinant His6-XPA was immobilized on nickel-nitrilotriacetic acid beads and modified with increasing amounts of NHS-biotin (0, 0.1, 0.25, 0.5, and 1 mm) before or after the addition of FLAG-ATR lysates. XPA modified with 1 mm NHS-biotin before the addition of FLAG-ATR lysate prevents formation of the XPA-ATR complex; however, modification after complex formation does not affect the protein-protein interaction. A nonspecific (NS) protein band was also observed during the immunoprecipitation even though no XPA was added to the beads, indicating that the protein interacts with the nickel matrix and not with XPA or ATR (data not shown). Because of the nonspecific nature of the band, it was used as a loading control for these experiments. D, FLAG-ATR purified by immunoprecipitation was mixed with His6-XPA (lane 3 and 4) and modified with NHS-biotin (lane 4). As control, anti-FLAG antibody was mixed with the recombinant XPA in the absence of ATR protein (lane 5). Protein bands were excised and digested with trypsin for mass spectrometry analysis.
Chemical Modification of XPA and XPA-ATR Complex
Previous studies demonstrated that ATR can directly interact with XPA (22). To map the XPA sites involved in the interaction with ATR, we used our mass spectrometry-based protein footprinting approach (24, 25, 34, 35). The method enables a comparison of surface topologies of the free protein versus the protein-protein complex using small chemical modifiers. We chose to use a primary amine-selective reagent, NHS-biotin, for probing the ATR-XPA interactions given that lysines are one of the most abundant residues in these proteins. Furthermore, a similar experimental approach applied to other protein-protein interactions consistently identified physiologically important interactions (24, 25, 34, 35).
For meaningful footprinting experiments, it was essential to establish mild modification conditions under which the integrity of the functional protein-protein complexes would be preserved. For this purpose we carefully optimized the concentrations of NHS-biotin in the reaction mixtures. We immobilized recombinant His6-XPA on nickel-nitrilotriacetic acid beads and modified the protein with increasing amounts of NHS-biotin before or after the addition of doxycycline-induced U2OS cell lysates to assay the effects of biotinylation on XPA-ATR complex formation. Fig. 1C shows that biotinylation of XPA before the addition of U2OS lysate prevents the coimmunoprecipitation of FLAG-ATR at concentrations greater than 1 mg/ml. These data suggest that at least one surface lysine in XPA is essential for interactions with ATR. Of note, the identical treatment of the preformed the XPA-ATR complex did not disturb the protein-protein complex, indicating the modification conditions were sufficiently mild. These findings defined optimal experimental conditions for subsequent mass spectrometry footprinting of the ATR-XPA complex.
Mass Spectrometric Footprinting of the XPA-ATR Complex
To form the protein-protein complex, purified His6-XPA was added in excess to FLAG-ATR immobilized on Protein-G beads and incubated in the binding buffer (see “Experimental Procedures”). For control experiments, XPA was also incubated with the Protein-G beads that were preincubated with anti-FLAG-treated cell lysates from un-induced U2OS cells under the same experimental conditions. Unbound XPA was then washed away, and the resulting samples were treated with NHS-biotin. In parallel experiments, free XPA was also subjected to the NHS-biotin treatment. The modified proteins were then resolved by SDS-PAGE. Fig. 1D shows a Coomassie Blue-stained gel. The unmodified and modified forms of free XPA are shown in lanes 1 and 2, respectively. Four major bands were observed for the unmodified (lane 3) and NHS-biotin treated (lane 4) XPA-ATR complexes corresponding to ATR, XPA, and the heavy and light chains of the FLAG antibody. The XPA bands were excised from the gel and subjected to in-gel trypsin proteolysis to generate small peptide fragments amenable for MALDI-TOF analysis.
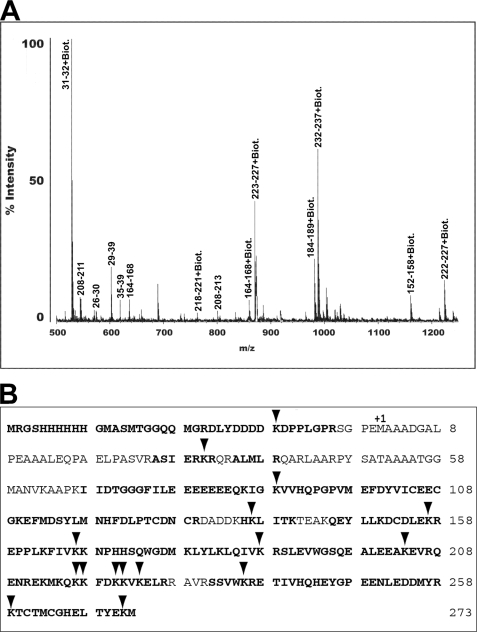
Fig. 2A depicts a representative mass spectrum for tryptic fragments of the modified XPA protein. Monoisotopic resolution of the peaks were obtained, allowing us to reliably identify the tryptic fragments of XPA. Fig. 2B illustrates the peptide fragments (bold sequences) and biotinylated lysines identified by MALDI-TOF analysis.
FIGURE 2.
MALDI-TOF analysis of biotin-modified XPA. A, a typical MALDI-TOF mass spectrum of peptide fragments resulting from trypsin digestion of biotin modified XPA. B, summary of MALDI-TOF results in the context of the XPA primary structure. The sequence of N-terminal His6 tag added for purification of the recombinant protein is shown. The His6 tag residue numbering begins with −1 and continues backward to the N terminus. The initial methionine residue of the XPA sequence is labeled +1. Amino acid sequences corresponding to tryptic peptide fragments detected by MALDI-TOF are depicted in bold. The lysine residues affected by NHS-biotin treatment are indicated by arrows.
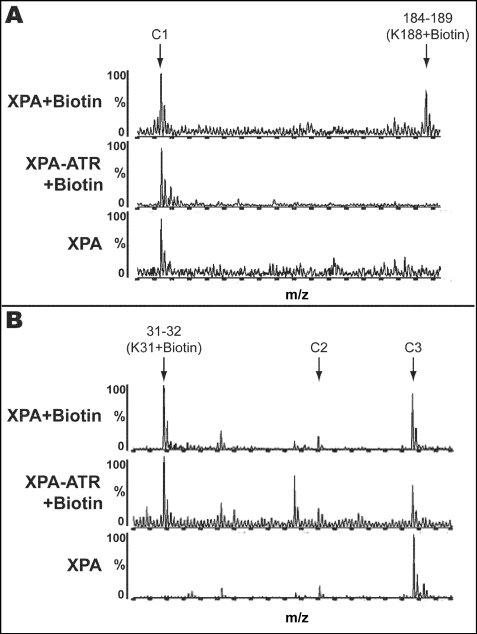
To identify the XPA surface lysine(s) interacting with ATR, we compared the modification profiles of free XPA and the XPA-ATR complex. Fig. 3 depicts representative MALDI-TOF fragment profiles used for comparison. Fig. 3A demonstrates that Lys-188 was readily susceptible to modification in free XPA but was inaccessible to NHS-biotin in the XPA-ATR complex. These results indicate that Lys-188 is surface-exposed in free XPA and becomes shielded upon ATR binding. In contrast, Fig. 3B illustrates an m/z peak corresponding to biotinylated XPA fragment 31–32 (K31+biotin) present in the spectra for both the free XPA and the XPA-ATR complex. These data suggest that surface topology of Lys-31 is not affected by the bound ATR. Peaks C1, C2, and C3 are unmodified peptide peaks of XPA and provide internal reference. The lysine footprinting results for the XPA-ATR complex are summarized in Table 1.
FIGURE 3.
MALDI-TOF analysis of lysine protection in the XPA-ATR complex. A, top and middle spectra show free XPA and the XPA-ATR complex treated with NHS-biotin. The bottom spectrum shows untreated free XPA. The peak corresponding to XPA tryptic peptide fragment amino acids 184–189 containing a single modified Lys at position 188 is indicated. This peak is detected in free XPA samples and is significantly diminished in the XPA-ATR complex. B, unlike Lys-188, lysine residue 31 is modified in both free XPA and the XPA-ATR complex. Peaks C1, C2, and C3 are unmodified peptide fragments of XPA and serve as internal controls.
TABLE 1.
Summary of MALDI-TOF analysis of biotinylated XPA peptides
Presented are tryptic digest fragments containing biotinylated lysine residues. Lysine residues protected from modification in the presence of ATR are indicated by a +, whereas residues not protected by ATR are indicated by a −. The asterisk indicates that the biotinylated fragment is located in the His tag. The intensity of the biotinylated peptide peak 184–189 (Lys-188 + biotin) was at least 10-fold higher for free XPA treated with NHS-biotin than for XPA-ATR complex modified under identical conditions. Intensities of other biotinylated XPA peptide fragments in the absence and presence of ATR varied within ±20%.
| Fragment | Modified Lys | Protection |
|---|---|---|
| (−18)–(−5)* | −12* | − |
| 31–32 | 31 | − |
| 87–110 | 89 | − |
| 136–141 | 137 | − |
| 152–158 | 157 | − |
| 164–168 | 167 | − |
| 184–189 | 188 | + |
| 190–207 | 204 | − |
| 218–221 | 218 | − |
| 216–221 | 217, 218 | − |
| 219–224 | 221, 222 | − |
| 222–227 | 222, 224 | − |
| 223–227 | 224 | − |
| 232–237 | 236 | − |
| 259–273 | 259 | − |
| 260–273 | 272 | − |
Lys-188 and the XPA-ATR Interaction
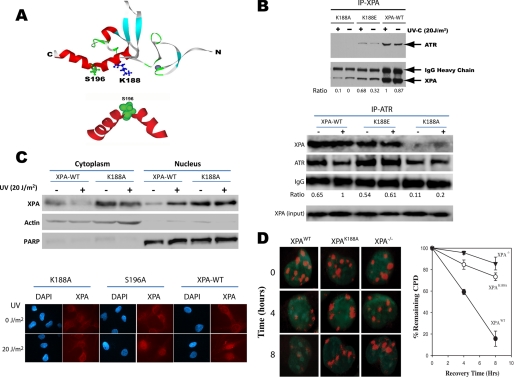
We identified 16 lysine residues biotinylated in XPA, of which one residue, Lys-188, was protected from modification in the presence of ATR (Table 1). Fig. 4A is a view of the minimum DNA-binding domain structure of XPA as determined by NMR spectroscopy (PBD code 1D4U) (26). ATR phosphorylates XPA at serine 196 (shown in green), which is located in the turn of a helix-turn-helix motif, a part of the proposed DNA-binding cleft. Protected lysine residue Lys-188 (shown in blue) is also located in the helix-turn-helix motif and is oriented in nearly the same plane as Ser-196. This is consistent with the fact that phosphorylation requires binding of ATR to XPA.
FIGURE 4.
Residue Lys-188 is required for XPA-ATR complex formation, XPA nuclear import upon UV damage, and NER. A, ribbon diagram of XPA minimum DNA-binding domain (PDB code 1D4U), indicating that the identified lysine residue Lys-188 (shown in blue) is located in the helix-turn-helix motif-containing ATR phosphorylation site serine 196. B, point mutations were generated in the pcDNA-XPA expression construct generating alanine (K188A) and glutamic acid (K188E) substitutions. The mutated constructs as well as wild-type XPA were stably expressed in XPA−/− cells, and their effects on the XPA-ATR interaction were investigated by coimmunoprecipitation (IP). The K188A mutant protein was unable to coimmunoprecipitate ATR or vice versa from lysates generated from UV-irradiated or un-irradiated cells. The K188E mutant maintained the interaction between XPA and ATR and exhibited a similar UV-induced pattern as seen for XPA-WT. The relative amounts of the co-immunoprecipitated ATR were estimated by its ratio to those of the immunoprecipitated protein that were normalized to the loading control IgG. C, XPA cells complemented with wild-type XPA and XPA-K188A were subjected to subcellular fractionation and immunofluorescence microscopy analysis. The specificity of the fractionation assay is demonstrated by the presence and absence of cytoplasm-specific and nucleus-specific proteins β-actin and PARP (poly(ADP-ribose) polymerase), respectively, in the cytosol and nucleus. D, cells were irradiated with UV of 50 J/m2 through isopore filters to induce localized DNA damage and then fixed at the indicated times for immunofluorescence analysis with anti-CPD antibody. Nuclei containing at least one well defined CPD focus were counted as a percentage of total DAPI-stained nuclei and plotted versus time post-irradiation. At least 50 DAPI-stained nuclei were randomly chosen for the quantification at each time point.
Given that Lys-188 is shielded from the solvent in the presence of ATR, site-directed mutagenesis of Lys-188 was performed to investigate its possible role in XPA-ATR complex formation. Thus, pcDNA3.1 expression constructs were generated in which Lys-188 was changed to either alanine (K188A) or glutamic acid (K188E), and the vectors were stably expressed in XPA−/− cells. Coimmunoprecipitation assays were performed to examine the effects of the Lys-188 mutations on ATR binding to XPA. As shown in Fig. 4B, anti-XPA antibody efficiently co-immunoprecipitated ATR from whole cell lysates generated from XPA−/− cells complemented with wild-type XPA protein. Consistent with previous observations, UV-C irradiation increased the affinity of ATR for XPA but was unnecessary for the interaction (22). Interestingly, XPA-K188E protein also was able to co-immunoprecipitate ATR in a similar pattern to that observed for the wild-type protein, although its affinity seemed slightly reduced. The K188A mutation, however, completely abolished the XPA-ATR interaction in both the irradiated and un-irradiated cells. These results were further confirmed by the reversed coimmunoprecipitation in which XPA was coimmunoprecipitated by anti-ATR antibody (Fig. 4B).
Effect of K188A Mutation on Nuclear Import of XPA upon UV Irradiation and DNA Damage Repair
We previously reported that after DNA damage, XPA translocates from the cytoplasm to the nucleus in an ATR-dependent manner (17). Thus, an interesting question is whether the ATR-XPA interaction is required for XPA nuclear import. To address this question, a cellular fractionation assay was performed using XPA−/− cells expressing recombinant wild-type XPA (XPA-WT) or XPA-K188A mutant. After UV-C irradiation (20 J/m2), wild-type XPA is re-distributed from the cytoplasm to the nucleus. However, UV-C irradiation does not affect the cellular distribution of the XPA-K188A protein (Fig. 4C). These results suggest that the K188A mutation negatively affects the ability of ATR to induce nuclear accumulation of XPA in response to DNA damage. The results were further confirmed by an immunofluorescence microscopy assay in which the subcellular localization of XPA was directly visualized (Fig. 4C). Interestingly, the phosphorylation of XPA at Ser-196 appears to have no effect on XPA nuclear import. All these suggest that physical interaction between XPA and ATR, but not Ser-196 phosphorylation, may play a role in DNA damage-induced XPA nuclear import. It should be noted that although the total levels of XPA appeared to be different in cells expressing XPA-WT and mutant, the effect of total XPA level on the UV-induced subcellular translocation patterns of XPA should be very minimal. To determine the dependence of NER on the Lys-188-mediated ATR-XPA interaction, XPA-WT or XPA-K188A mutant cells were cultured on glass coverslips and irradiated through isopore filters to generate localized DNA damage. Damage foci were visualized by immunofluorescence microscopy and measured as a function of repair time. As shown in Fig. 4D, foci removal is impaired in XPA−/− and XPA-K188A cells compared with XPA-WT cells, suggesting that the mutation largely abrogated the repair.
XPA Phosphorylation and DNA Damage Repair
Another possible role of ATR binding to XPA in cells is to phosphorylate the NER protein. The ATR-dependent phosphorylation of XPA previously has been shown to play a role in cell survival after UV-C irradiation (22). Given the unique role of XPA in NER, we hypothesized that XPA phosphorylation may play a role in promoting removal of UV photoproducts. Previous experiments in which ATR kinase activity was inhibited by small interfering RNA knockdown reduced the repair rate of (6-4) photoproducts ((6-4)PPs) (17); however, most of the lesions were removed within several hours post-irradiation, which appears to be earlier than XPA phosphorylation (22). This suggests that the phosphorylation may not be involved in (6-4)PP repair. We, therefore, reason that XPA phosphorylation might play a role in promoting the removal of persistent lesions, such as CPDs.
To test the notion, repair of UV-C-induced photoproducts was monitored by immunofluorescence microscopy in XPA−/− cells complemented with either wild-type XPA or XPA in which serine 196 was replaced by alanine (S196A). As shown in Fig. 5A, CPD and (6-4)PP foci (stained in red), formed in the nuclei of cells complemented with XPA-WT protein, decrease in size and frequency as the recovery time increases up to 24 h. However, CPD foci are more persistent in nuclei from XPA-S916A complemented cells. Nuclei from cells transfected with empty vector alone show little change in CPD focus size or frequency across time. These results suggest that CPD lesions are repaired at a slower rate in cells complemented with XPA-S196A compared with XPA-WT protein (Fig. 5, A and B). In agreement with previous results, no substantial difference in repair rate was observed for (6-4)PPs between cells complemented with wild-type and phosphorylation-deficient XPA (Fig. 5B). In a parallel DNA repair assay (Fig. 5C), XPA-complemented cells were irradiated with UV-C (20 J/m2) followed by extraction of the genomic DNA. Equal amounts of purified DNA were then immobilized on nylon membrane and probed with antibody specific for CPDs. Consistently, CPD lesions were removed more efficiently in cells expressing XPA-WT compared with cells expressing phosphorylation-deficient XPA-S196A protein, whereas little repair occurred in XPA cells transfected with empty vector. These results suggest that XPA phosphorylation promotes the repair of persistent UV-induced photolesions. Also importantly, the phosphorylation is dependent on the ATR-XPA interaction as demonstrated in Fig. 5D in which the XPA phosphorylation was abolished in cells expressing XPA-K188A mutant. Taken together, these results support the observation on the effects of XPA-K188A mutation on CPD repair in Fig. 4D.
FIGURE 5.
Effects of XPA phosphorylation on repair of cyclobutane pyrimidine dimers. Recombinant pcDNA constructs containing wild-type XPA or XPA-S196A cDNA were stably expressed in XPA−/− cells. A, cells were grown on coverslips and UV-irradiated at 20 J/m2 through isopore filters to induce localized DNA damage followed by immunofluorescence staining with anti-(6-4)PPs and anti-CPD antibodies at the indicated time points. B, nuclei containing at least one DNA damage focus were counted as a percentage of total nuclei and plotted versus time post-irradiation. At least 50 DAPI-stained nuclei were randomly counted for each time point. C, genomic DNA was isolated from cells complemented with wild-type or phosphorylation-deficient XPA after UV-C irradiation. The DNA was then immobilized on nylon membranes, and total CPDs were detected using mouse-anti-CPD. D, cells expressing recombinants XPA-WT and XPA-K188A, respectively, were UV-irradiated and then subjected to Western blot analysis of phosphorylated and intact XPA. E, subcellular distribution of XPA-WT and XPA-S196A after UV-C irradiation was determined by cellular fractionation. β-Actin and PARP (poly(ADP-ribose) polymerase), cytoplasm- and nucleus-specific proteins, respectively, demonstrate the specificity of the assay and serve as loading controls.
To determine whether the reduced repair of CPD lesions by XPA-S196A was because of reduced nuclear translocation, we performed a cellular fractionation assay to determine the subcellular distribution of XPA-S196A in response to UV-C irradiation. As demonstrated in Fig. 5E, phosphorylation-deficient XPA-S196A is imported into the nucleus with the same efficiency as wild-type XPA, suggesting that XPA translocation is not affected by the phosphorylation. The result is consistent with that determined by the immunofluorescence analysis (Fig. 4C). Taken together, XPA phosphorylation is likely involved in a mechanism independent of subcellular protein redistribution to promote repair of persistent DNA lesions.
DISCUSSION
Our previous work has demonstrated that XPA is phosphorylated by the DNA damage checkpoint kinase ATR in response to UV irradiation at serine residue 196 located in the minimum DNA-binding domain of XPA (17). In addition, ATR directed the UV-induced subcellular redistribution of XPA from the cytosol to the nucleus in both a dose-dependent and time-dependent manner (22). Given the central role of XPA in nucleotide excision repair, an interaction between XPA and ATR may represent a novel regulatory mechanism for NER to be modulated by DNA damage checkpoints. The dependence of NER on ATR is further supported by a recent report by Auclair et al. (23).
In this study we have employed our protein footprinting approach to map the interaction sites of ATR and XPA. We probed the surface topology of XPA in complex with ATR and identified one lysine residue, Lys-188, which is involved in the ATR-XPA interaction. Lys-188 is located in the N-terminal helix of the helix-turn-helix motif containing the ATR phosphorylation site Ser-196. In contrast, lysine residue 204 located in the C-terminal helix of the helix-turn-helix motif is not protected from modification by ATR, suggesting that only the N-terminal helix is directly involved in the ATR-XPA interaction. Mutating Lys-188 to alanine effectively abolished the interaction between XPA and ATR, whereas the K188E substitution had a relatively modest effect on the complex formation. One explanation for this observation is that ATR is able to remodel its XPA binding surface and effectively bind the K188E protein but not the K188A. A second explanation is that Lys-188 is not directly in the binding site, but the alanine and glutamic acid mutations to this residue differentially alter the stability of the N-terminal α-helix that mediates the XPA-ATR interaction. Lysine residues are often involved in the formation of salt bridges that stabilize helical secondary structures. Moreover, the presence of acidic residues in the N terminus are also known to stabilize the α-helices (36–38). The substitution of alanine at Lys-188 might destabilize the N-terminal helix of the XPA helix-turn-helix motif, whereas substitution with glutamic acid may result in an equally or possibly more stable helix (39, 40). In summary, these results strongly imply a key role for the N-terminal α-helix of the XPA helix-turn-helix motif in the interaction with ATR and suggest that Lys-188 mediates the effects on this interaction indirectly by modulating the stability of the helix rather than through direct interaction with ATR. It should be noted, however, that because ∼30% ATRIP remained bound to the ATR assayed in this study, the involvement of ATRIP in the ATR-XPA interaction is also possible.
The significance of the ATR-XPA interaction has been shown by its requirement for XPA nuclear import in response to UV irradiation of the cells (Fig. 4). Because XPA is an indispensable factor for NER and the nuclear availability of XPA is critical for NER, it is reasonable to expect that NER may depend on ATR-XPA interaction. Indeed our result indicates that the nucleotide excision repair of UV-induced photolesions requires the ATR-XPA interaction in cells. There are at least two possible scenarios in which XPA nuclear translocation and, thus, NER could depend on ATR-XPA interaction. First, the interaction could occur in the nucleus, which reduces the nuclear concentration of free XPA. The depletion of free XPA in the nucleus disrupts the concentration balance across the nuclear membrane and subsequently allows more XPA to be imported into the nucleus driven by favorable free energy. In the second scenario, XPA could form a complex with ATR in the cytoplasm and then be imported into the nucleus across the nuclear membrane in the complex form.
It is obvious that ATR-XPA complex formation is also required for the phosphorylation of XPA. Phosphorylation of XPA at serine 196 has previously been shown to moderately promote cell survival after UV-C irradiation (22). Given that the only known function for XPA is in NER, we assayed UV-C-induced photoproducts removal to investigate the role of XPA phosphorylation in NER. We found that XPA−/− cells complemented with XPA-S196A displayed a slower repair rate for CPD photoproducts when compared with wild-type XPA. By contrast, no effect has been observed for repair of (6-4) photoproducts. It has been well established that CPD is much more persistent DNA-damaging than (6-4) photoproducts (41). Taken together, these results suggest that XPA phosphorylation may play a role in stimulating NER activity for removal of persistent DNA damage through an as yet undetermined mechanism. This also appears consistent with our previous observation that phosphorylated XPA represents only a small portion of the cellular pool of XPA (22).
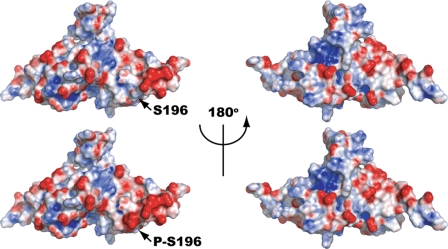
We hypothesized that phosphorylation of XPA may modulate NER activity by altering the affinity of XPA for the damage site. Indeed, phosphorylated XPA was shown to associate more tightly with UV-damaged chromatin in cells than the wild-type XPA (22). Previous NMR data generated by docking the XPA minimum DNA-binding domain to a ssDNA 9-mer projected a DNA-binding cleft consisting of a series of basic residues aligned across the minimal DNA-binding domain (42). We generated a computational model of the XPA minimal DNA-binding domain to obtain insight into how phosphorylation of serine 196 could affect the surface charge distribution of the protein. Fig. 6 illustrates that Ser-196 phosphorylation induces an accumulation of negative charge along the surface of the helix-turn-helix motif. However, our computational model did not predict any significant change in the positive charge distribution of the DNA-binding cleft after Ser-196 phosphorylation. Although we did not observe any change in surface charge distribution for the proposed DNA-binding cleft, the current structural model of the XPA-DNA interaction is based on XPA interacting with an ssDNA substrate. It is well documented that XPA binds damaged dsDNA, and moreover, our previous study indicates that XPA interacts most efficiently with a substrate containing a ssDNA-dsDNA junction. Thus, additional stabilizing contacts between DNA and XPA may be present that were not identified in the NMR study. The need for further analysis of the XPA interaction with ssDNA-dsDNA junction substrates is highlighted by multiple reports supporting the interaction as the most probable DNA structure in which XPA would be presented in vivo. Indeed, the recruitment of XPF-ERCC1 complex by XPA after strand opening by TFIIH supports a model of XPA binding at or near the ssDNA-dsDNA junction in the pre-incision complex (3, 9, 43–45).
FIGURE 6.
Modeling analysis of the effects on surface charge distribution from phosphorylation of XPA at serine residue 196. A surface representation of the DNA-binding domain of XPA (PDB code 1D4U) is shown at the left with the electrostatic field mapped in red (negative) and blue (positive). A computation model of the DNA-binding domain of XPA phosphorylated at serine 196 is shown at the right with the same mapping of the electrostatic field. The circle highlights the region of the protein near serine 196 where a large net increase in negative charge is associated with phosphorylation.
Another possible mechanism for increased chromatin association after Ser-196 phosphorylation could be related to the increased affinity of XPA with respect to its binding partners. Our electrostatic model clearly indicates that phosphorylation of Ser-196 induces significant accumulation of negative charge on the helix-turn-helix motif. This motif, in addition to DNA binding, is located in close proximity to both the RPA70 and TFIIH interaction domains on XPA (8). Increasing the XPA affinity for RPA70 and/or TFIIH could, therefore, modulate the association of XPA with the damage site in a manner independent of its affinity for DNA.
This work was supported, in whole or in part, by National Institutes of Health Grants CA86927 and AG031503 (to Y. Z.), AI077341 (to M. K.), and GM065484 and CA092584 (to W. J. C.).
- NER
- nucleotide excision repair
- ATM
- ataxia-telangiectasia mutated
- ATR
- ATM and RAD3-related
- ATRIP
- ATR interaction protein
- XPA
- Xeroderma pigmentosum (XP) group A
- CPD
- cyclobutane pyrimidine dimers
- (6-4)PPs
- (6-4) photoproducts
- NHS-biotin
- N-hydroxysuccinimidobiotin
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- DAPI
- 4,6-diamidino-2-phenylindole
- WT
- wild type
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- RPA
- replication protein A.
REFERENCES
- 1.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 2.Park C. J., Choi B. S. (2006) FEBS J. 273, 1600–1608 [DOI] [PubMed] [Google Scholar]
- 3.Riedl T., Hanaoka F., Egly J. M. (2003) EMBO J. 22, 5293–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer K. H., Lee M. M., Andrews A. D., Lambert W. C. (1994) Arch. Dermatol. 130, 1018–1021 [PubMed] [Google Scholar]
- 5.Kraemer K. H., Lee M. M., Scotto J. (1987) Arch. Dermatol. 123, 241–250 [DOI] [PubMed] [Google Scholar]
- 6.Lehmann A. R. (2003) Biochimie 85, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 7.Batty D. P., Wood R. D. (2000) Gene 241, 193–204 [DOI] [PubMed] [Google Scholar]
- 8.Cleaver J. E., States J. C. (1997) Biochem. J. 328, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzder S. N., Sommers C. H., Prakash L., Prakash S. (2006) Mol. Cell. Biol. 26, 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z., Roginskaya M., Colis L. C., Basu A. K., Shell S. M., Liu Y., Musich P. R., Harris C. M., Harris T. M., Zou Y. (2006) Biochemistry 45, 15921–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shell S. M., Zou Y. (2008) Adv. Exp. Med. Biol. 637, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S. H., Kim D. K. (1995) J. Biol. Chem. 270, 12801–12807 [DOI] [PubMed] [Google Scholar]
- 13.Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C. J., Edwards A. M., Chazin W. J. (2000) Cell 103, 449–456 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Wang Y., Rusinol A. E., Sinensky M. S., Liu J., Shell S. M., Zou Y. (2008) FASEB J. 22, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musich P. R., Zou Y. (2009) Aging 1, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 17.Wu X., Shell S. M., Liu Y., Zou Y. (2007) Oncogene 26, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 19.Abraham R. T. (2004) DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 20.Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 21.Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003) Nat. Cell Biol. 5, 255–260 [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Shell S. M., Yang Z., Zou Y. (2006) Cancer Res. 66, 2997–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auclair Y., Rouget R., Affar el B., Drobetsky E. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17896–17901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Kvaratskhelia M., Hess S., Qu Y., Zou Y. (2005) J. Biol. Chem. 280, 32775–32783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shell S. M., Hess S., Kvaratskhelia M., Zou Y. (2005) Biochemistry 44, 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchko G. W., Daughdrill G. W., de Lorimier R., Rao B. K., Isern N. G., Lingbeck J. M., Taylor J. S., Wold M. S., Gochin M., Spicer L. D., Lowry D. F., Kennedy M. A. (1999) Biochemistry 38, 15116–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., III, Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Kollman P. A. (2006) AMBER 9, University of California, San Francisco [Google Scholar]
- 28.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 30.Lerner M., Carlson H. A. (2006) APBS Plugin for PyMOL, University of Michigan, Ann Arbor, MI [Google Scholar]
- 31.Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nghiem P., Park P. K., Kim Y., Vaziri C., Schreiber S. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9092–9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortez D., Guntuku S., Qin J., Elledge S. J. (2001) Science 294, 1713–1716 [DOI] [PubMed] [Google Scholar]
- 34.Shkriabai N., Datta S. A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. (2006) Biochemistry 45, 4077–4083 [DOI] [PubMed] [Google Scholar]
- 35.Zhao Z., McKee C. J., Kessl J. J., Santos W. L., Daigle J. E., Engelman A., Verdine G., Kvaratskhelia M. (2008) J. Biol. Chem. 283, 5632–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aurora R., Creamer T. P., Srinivasan R., Rose G. D. (1997) J. Biol. Chem. 272, 1413–1416 [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker K. R., Kim P. S., Brems D. N., Marqusee S., York E. J., Chaiken I. M., Stewart J. M., Baldwin R. L. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2349–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker K. R., Kim P. S., York E. J., Stewart J. M., Baldwin R. L. (1987) Nature 326, 563–567 [DOI] [PubMed] [Google Scholar]
- 39.Blagdon D. E., Goodman M. (1975) Biopolymers 14, 241–245 [DOI] [PubMed] [Google Scholar]
- 40.Chou P. Y., Fasman G. D. (1978) Adv. Enzymol. Relat. Areas Mol. Biol. 47, 45–148 [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi N., Katsumi S., Imoto K., Nakagawa A., Miyagawa S., Furumura M., Mori T. (2001) Pigment Cell Res. 14, 94–102 [DOI] [PubMed] [Google Scholar]
- 42.Buchko G. W., Tung C. S., McAteer K., Isern N. G., Spicer L. D., Kennedy M. A. (2001) Nucleic Acids Res. 29, 2635–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croteau D. L., Peng Y., Van Houten B. (2008) DNA Repair 7, 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripsianes K., Folkers G. E., Zheng C., Das D., Grinstead J. S., Kaptein R., Boelens R. (2007) Nucleic Acids Res. 35, 5789–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsodikov O. V., Ivanov D., Orelli B., Staresincic L., Shoshani I., Oberman R., Schärer O. D., Wagner G., Ellenberger T. (2007) EMBO J. 26, 4768–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]