Abstract
Both interleukin-4 (IL-4) and IL-13 can bind to the shared receptor composed of the IL-4 receptor α chain and the IL-13 receptor α1 chain (IL-13Rα1); however, the mechanisms by which these ligands bind to the receptor chains are different, enabling the principal functions of these ligands to be different. We have previously shown that the N-terminal Ig-like domain in IL-13Rα1, called the D1 domain, is the specific and critical binding unit for IL-13. However, it has still remained obscure which amino acid has specific binding capacity to IL-13 and why the D1 domain acts as the binding site for IL-13, but not IL-4. To address these questions, in this study we performed mutational analyses for the D1 domain, combining the structural data to identify the amino acids critical for binding to IL-13. Mutations of Lys-76, Lys-77, or Ile-78 in c′ strand in which the crystal structure showed interaction with IL-13, and those of Trp-65 and Ala-79 adjacent to the interacting site, resulted in significant impairment of IL-13 binding, demonstrating that these amino acids generate the binding site. Furthermore, mutations of Val-35, Leu-38, or Val-42 at the N-terminal β-strand also resulted in loss of IL-13 binding, probably from decreased structural stability. None of the mutations employed here affected IL-4 binding. These results demonstrate that the D1 domain of IL-13Rα1 acts as an affinity converter, through direct cytokine interactions, that allows the shared receptor to respond differentially to IL-4 and IL-13.
Interleukin-4 (IL-4)2 and IL-13 are related cytokines. IL-4 binds to a heterodimeric complex composed of the IL-4R α chain (IL-4Rα) and the common γ chain (γc), or of IL-4Rα and the IL-13R α1 chain (IL-13Rα1), called type I or type II IL-4R, respectively (1, 2). In contrast, IL-13 binds to type II IL-4R, but not type I IL-4R. Therefore, type II IL-4R is also called IL-13R. This means that IL-4 and IL-13 share the same receptor, type II IL-4R·IL-13R, which explains why IL-4 and IL-13 exert similar activities. However, the principal functions of IL-4 and IL-13 are different. As type I IL-4R is mainly expressed on hematopoietic cells, IL-4 acts on these cells, inducing Th2 differentiation in T cells and immunoglobulin class switching to IgE in B cells (1, 3). In contrast, type II IL-4R·IL-13R expresses ubiquitously, including nonhematopoietic cells, and IL-13 plays a central role in the pathogenesis of bronchial asthma by acting on these cells, including epithelial cells and fibroblasts (1, 4). Thus, it can be said that the principal role of IL-4 is an immunoregulatory cytokine, whereas that of IL-13 is an effector cytokine (5).
The assembly mechanism for the binding of either IL-4 or IL-13 to type II IL-4R·IL-13R is unique. IL-4 first binds to IL-4Rα with high affinity (Kd = 1 nm), followed by recruitment of IL-13Rα1 with low affinity. In contrast, IL-13 first binds to IL-13Rα1 with low affinity (Kd = 30–37 nm), and then the complex recruits IL-4Rα, forming a high affinity receptor (Kd = 0.03–0.4 nm (6, 7)). This means that, although both IL-4 and IL-13 use IL-4Rα and IL-13Rα1, the roles of IL-4Rα and IL-13Rα1 as the primary or secondary binding unit are the opposite of those for IL-4 and IL-13. Furthermore, these differences in affinity between the ligand, the primary binding unit, and the secondary binding unit can result in that in nonhematopoietic cells on which IL-131 is expressed more abundantly than IL-4α, the number of the IL-13 receptor complex continues to rise as the IL-13 concentration increases, whereas the formation of the IL-4 receptor complex is saturated at a low IL-4 concentration. This can explain why IL-13 transduces stronger signals than IL-4 in nonhematopoietic cell such as epithelial cells and fibroblasts.
We previously found that the N-terminal Ig-like domain in IL-13Rα1, called the D1 domain, is the specific and critical binding unit for IL-13, but not for IL-4, using the D1 domain-deleted IL-13Rα1 (8). LaPorte et al. recently described the crystal structure of the IL-13·IL-13Rα1·IL-4Rα, showing that the c′ strand of the D1 domain of IL-13Rα1 and the C-D strand of IL-13 generate an antiparallel β-sheet structure (7). Furthermore, this structural analysis showed that the polar bonds between IL-4 and IL-4Rα were diminished in the IL-13·IL-4Rα complex, possibly suggesting why IL-4Rα has high and no affinity with IL-4 and IL-13, respectively. These results confirmed that the unique assembly mechanism of type II IL-4R·IL-13R for IL-4 and IL-13 is dictated by the D1 domain and indicated that the c′ strand in the D1 domain is the binding site of IL-13Rα1 to IL-13. It is thought that IL-13Rα1 has evolved from γc, which does not have the extra Ig domain, acquiring the D1 domain probably from IL-2Rα or IL-15Rα (7, 9). In other words, the acquisition of the D1 domain enables the cells to respond to IL-13 in addition to IL-4. In this sense, the D1 domain appears to be an affinity converter that has evolved differential interactions with IL-4 and IL-13 to respond to the two cytokines distinctly, based on receptor expression levels and cytokine concentration. Thus, the evolution of distinct interactions of D1 with IL-4 versus IL-13 is an unprecedented example of divergent evolution of function of the same structure. Interestingly, in the structural study, it was observed that the c′ strand of the D1 domain of IL-13Rα1 can also generate an antiparallel β-sheet structure with IL-4 that appears similar to that of IL-13 (7), leaving open the question of whether it is energetically important for IL-13 but not IL-4, and whether direct interactions are required.
From these studies, several questions remain unresolved. The structures did not make it clear if this differential effect is indirect, or through direct interaction with the cytokines. Are the c′ contacts with cytokines energetically important and distinct for IL-4 and IL-13? If this is the case, then the second question is which amino acid in the c′ strand has specific binding capacity to IL-13. The third question is why does this portion act as the binding site specific for IL-13, but not IL-4. To address these questions, we took advantage of the mutational approach for the D1 domain, combining data from the structural study, and identified the amino acids critical for binding to IL-13.
MATERIALS AND METHODS
Reagents and Cells
Expression and purification of recombinant human IL-13 was performed as previously described (8, 10). Recombinant human IL-4 was purchased from PeproTech (Rocky Hill, NJ). HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 10 units/ml penicillin G. DND-39 cells transfected with the germ line ϵ promoter-luciferase gene (DND-39/Gϵ cells) were prepared and cultured, as described before (8).
Plasmids and Transfection
Plasmids encoding the wild type of hIL-13Rα1 (pIRES1hyg-FLAG-hIL-13Rα1) were prepared previously (8). The FLAG sequence was attached to the N termini of the D1 domain of hIL-13Rα1 cDNA. The mutated types of IL-13Rα1 were generated by the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using mutation-incorporated oligonucleotides as the primers and pIRES1hyg-FLAG-hIL-13Rα1 as the templates. Plasmids were prepared encoding the wild type of hIL-4Rα (pcDNA3.1-hIL- 4Rα-myc/His). Transient transfection of the plasmids into HEK 293T cells was performed by Lipofectamine 2000 (Invitrogen) or by polyethyleneimine (Polysciences, Inc., Warrington, PA), according to the manufacturer's protocol. The plasmids were transfected into DND-39/Gϵ cells using the Amaxa Nucleofector (Amaxa Biosystems, Cologne, Germany), and stable transfected cells were maintained with a culture medium containing 250 μg/ml hygromycin B (Wako, Osaka, Japan). Expression of the receptors was confirmed by flow cytometry (FACSCalibur, BD Biosciences, San Jose, CA) using anti-FLAG antibody (Ab, Sigma-Aldrich) or R-phycoerythrin-labeled anti-CD124 Ab (BD Biosciences).
Luciferase Assay
The luciferase assay was performed as previously described (8). HEK 293T cells were cotransfected with various types of pIRES1hyg-FLAG-hIL-13Rα1, pME18S-STAT6, and pGL3-N4 × 8 (kind gifts of Dr. T. Sugahara, Asahi Kasei Pharma Corp., Fuji, Japan). pGL3-N4 × 8 has an eight-tandem-lined STAT6-responsible element (TTCNNNNGAA), as a promoter sequence of the firefly luciferase gene. The Renilla-luciferase reporter gene (phRL-Tk, Promega, Madison, WI) was used as an internal control. Forty-eight hours after transfection, HEK 293T cells were stimulated with the indicated concentrations of IL-4 or IL-13 for 6 h, followed by washing once by phosphate-buffered saline, and total cell lysates were applied to the Picagene Dual Luciferase assay kit (TOYO Inc., Tokyo, Japan).
[125I]IL-13 and [125I]IL-4 Binding Assay
Iodination of IL-13 or IL-4 and the binding assay were performed as described previously with slight modifications (8). In brief, 1 μg of IL-13 or IL-4 and 1 mCi of Na125I (MP Biomedicals, Costa, CA) in 50 μl of phosphate-buffered saline were incubated in an IODOGEN-precoated tube (Pierce) at 4 °C for 10 min. Radiolabeled IL-13 or IL-4 was purified by a PD-10 column (Amersham Biosciences) and stored at 4 °C for up to 2 weeks. The concentrations of [125I]IL-13 and [125I]IL-4 were quantified by self-displacement of IL-13Rα2-expressing DND-39 cells or parental DND-39 cells, respectively. After cells were incubated with various concentrations of [125I]IL-13 or [125I]IL-4 at 4 °C for 2.5 h, bound and free ligands were separated by centrifugation through an oil gradient, and their radioactivity was measured. Nonspecific binding was determined by counts in the presence of 100-fold unlabeled IL-13 or IL-4.
Western Blotting and Immunoprecipitation
Western blotting and immunoprecipitation were performed as previously described (8). DND-39/Gϵ cells stably transfected with various kinds of IL-13Rα1 were stimulated with the indicated concentration of IL-4 or IL-13 at 37 °C for 10 min. The solubilized lysates or the immunoprecipitates from the lysates with anti-Tyk2 Ab (Santa Cruz Biotechnology, Santa Cruz, CA) were subjected to SDS-PAGE and transferred to polyvinylidene fluoride membranes. Membranes were incubated with either anti-phosphotyrosyl STAT6 Ab (Cell Signaling Technology, Inc., Beverly, MA), anti-STAT6 Ab (Santa Cruz Biotechnology), anti-phospho-tyrosine Ab (4G10, Upstate Biotechnology, Lake Placid, NY) or anti-Tyk2 Ab, followed by incubation with secondary Abs conjugated to horseradish peroxidase. The signals were visualized with an enhanced chemiluminescence system (ECL, Amersham Biosciences) and LAS-1000PLUS (Fuji Film, Tokyo, Japan).
Calculation of the Contact Area of Amino Acid to IL-13 and IL-4
The accessible surface area for ligand-binding and -unbinding structures of hIL-13Rα1 and the contact area between either IL-13 or IL-4 and hIL-13Rα1 were calculated by using the computer program Areaimol (11) in the CCP4 program suite (12), using atomic coordinates (PDB: 3bpo) of IL-13·IL-13Rα1 (7). In these calculations, default parameters for the van der Waals radii of 1.80, 1.65, 1.60, and 1.85 Å were used for C, N, O, and S, respectively. The 1.4-Å distance as a solvent proof was added to each atom location for accessible surface area calculation.
RESULTS
Truncation of the N-terminal Region of the D1 Domain in IL-13Rα1 to Identify the Critical Region for the IL-13 Signals
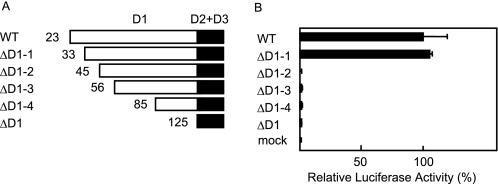
To identify the critical residues in the D1 domain of hIL-13Rα1 for the IL-13 signals, we first generated four kinds of D1 domain-truncated types of hIL-13Rα1 and analyzed their IL-13 responses in HEK 293T cells (hIL-13Rα1ΔD1-(1–4), Fig. 1A). In this experiment, we stimulated hIL-13Rα1-transfected HEK 293T cells with 0.01 ng/ml IL-13, because endogenous IL-13Rα1 cannot transduce IL-13 signals with this concentration of IL-13, as we previously reported (8). hIL-13Rα1ΔD1-1 lacking the N-terminal 32 amino acids showed luciferase activities similar to the wild type, whereas only a little activity was detected in hIL-13Rα1ΔD1–2 lacking the N-terminal 44 amino acids as well as hIL-13Rα1ΔD1–3/4 and hIL-13Rα1ΔD1 (Fig. 1B). The expression of all hIL-13Rα1ΔD1-(1–4) was confirmed by flow cytometry to be at the same level as the wild type and hIL-13Rα1ΔD1 (data not shown). These results suggested that the portion from the 33rd to the 44th amino acid in the a strand of the D1 domain of hIL-13Rα1 was critical for the IL-13 signals.
FIGURE 1.
Identification of the regions critical for the IL-13 signals in the D1 domain of IL-13Rα1. Schematic models of D1 domain-truncated IL-13Rα1 (A) are depicted. The numbers in each truncated type represent the starting amino acid number in A. Either the wild type or the indicated truncated types of IL-13Rα1 were transfected into HEK 293T cells. Then the cells were stimulated with 0.01 ng/ml IL-13 for 6 h. The luciferase activities in the truncated types (B) are shown. The relative luciferase activities of each mutant are calculated from that of the wild type.
Identification of Amino Acid Residues in the N-terminal Region of the D1 Domain That Are Critical for the IL-13 Signals
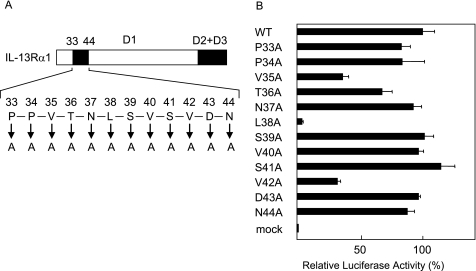
To identify the amino acids critical for the IL-13 signals in the portion from the 33rd to the 44th amino acid in the a strand of the D1 domain, we next substituted each amino acid between the 33rd and 44th amino acids with Ala and analyzed their IL-13 responses in HEK 293T cells (Fig. 2A). Among the Ala-substituted mutants, Leu-38 → Ala significantly diminished the luciferase activities, and Val-35 → Ala and Val-42 → Ala partially impaired these activities, whereas the other mutants showed activities similar to the wild type (Fig. 2B). All of the Ala-substituted mutants were confirmed by flow cytometry to be expressed at the same level as the wild type (data not shown). These results clearly demonstrated that Leu-38, and additionally Val-35 and Val-42, were critical amino acids in the transduction of the IL-13 signals.
FIGURE 2.
Identification of the amino acids critical for the IL-13 signals in the D1 domain of IL-13Rα1. Schematic models of Ala-substituted mutations (A) are depicted. The characters in A represent the amino acids from the 33rd to the 44th amino acid in the D1 domain. Either the wild-type or the indicated mutated types of IL-13Rα1 were transfected into HEK 293T cells, and then the cells were stimulated with 0.01 ng/ml IL-13 for 6 h. The luciferase activities in the substituted mutants (B) are shown. The relative luciferase activities of each mutant are calculated from that of the wild type.
Loss of Binding and Signal Transduction of IL-13 in Val-35 → Ala, Leu-38 → Ala, and Val-42 → Ala of IL-13Rα1
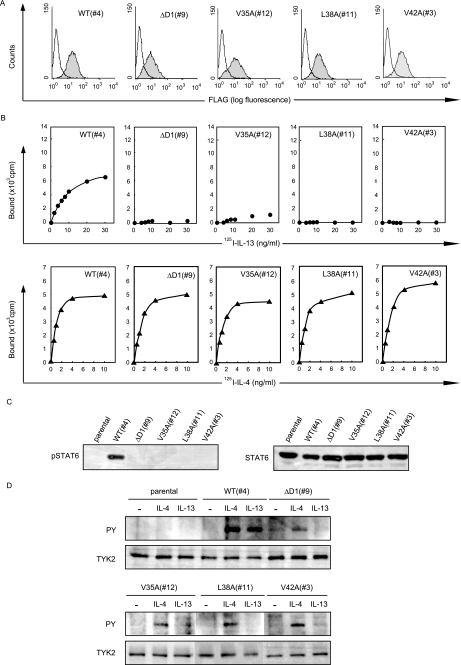
To investigate the importance of Val-35, Leu-38, and Val-42 of hIL-13Rα1 in the binding and signal transduction of IL-13, we generated DND-39/Gϵ cells stably expressing either Val-35 → Ala-, Leu-38 → Ala-, or Val-42 → Ala-harboring IL-13Rα1 and analyzed their abilities to bind to IL-13 and transduce the IL-13 signals. All of the DND-39/Gϵ transfectants used for the assays expressed almost the same level of the receptors (Fig. 3A). Binding to IL-13 was significantly impaired in Val-35 → Ala mutant compared with the wild type (Kd = ∼480 pm). No IL-13 binding was detected in Leu-38 → Ala, or in Val-42 → Ala mutants or D1 domain-deleted hIL-13Rα1, whereas binding activities to IL-4 were invariant in all of the mutants with the Kd of ∼54 pm (Fig. 3B). In accordance with the binding activity to IL-13, STAT6 activation by IL-13 was almost completely impaired in all Val-35 → Ala, Leu-38 → Ala, and Val-42 → Ala mutants (Fig. 3C). We then analyzed activation of Tyk2, a specific event for type II IL-4R·IL-13R (Fig. 3D). In response to IL-13, Leu-38 → Ala mutant completely diminished the activation of Tyk2, whereas both Val-35 → Ala and Val-42 → Ala mutants showed slight activation of Tyk2, compatible with the results of the luciferase activities (Fig. 2B), whereas IL-4 induced invariable bands in these mutants. Taking these results together, Val-35, Leu-38, and Val-42 were identified as crucial for IL-13 binding, subsequently for the IL-13 signals, whereas these amino acids were not involved in the IL-4 binding.
FIGURE 3.
Loss of binding and signal transduction of IL-13, but not IL-4 in Val-35 → Ala, Leu-38 → Ala, and Val-42 → Ala of IL-13Rα1. A, expression of the wild type, D1-deleted type, and the indicated mutated types of IL-13Rα1 on the surface of stable DND-39/Gϵ transfectants as estimated by flow cytometry is shown using anti-FLAG Ab. The shaded and open areas represent the count with or without the first Ab, respectively. B, binding of 125I-labeled IL-13 (closed circles) or IL-4 (closed triangles) to DND-39/Gϵ cells expressing the wild type, D1-deleted type, and the indicated mutated types of IL-13Rα1 is depicted. C and D, activation of STAT6 (C) and Tyk2 (D) using DND-39/Gϵ cells expressing the wild type, D1-deleted type, and the indicated mutated types of IL-13Rα1 is shown. The cells were stimulated with either 0.01 ng/ml of IL-13 (C) or 10 ng/ml of IL-4 or IL-13 (D) for 10 min.
Significance of Val-35, Leu-38, and Val-42 in the Structural Stability of the D1 Domain of IL-13Rα1
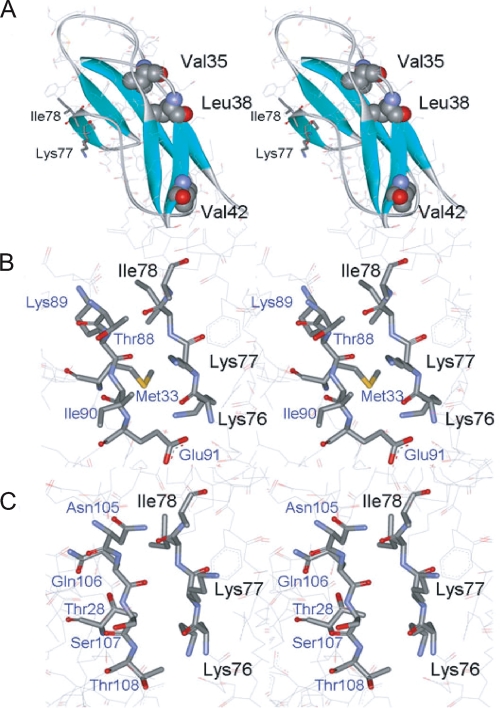
Truncation mutagenesis as well as subsequent Ala-scanning mutagenesis clearly demonstrated that Val-35, Leu-38, and Val-42 are important for IL-13 binding. However, the crystal structure of the D1 domain in hIL-13Rα1 indicated that the side chains of Val-35, Leu-38, and Val-42 are buried between the β-sheets and are not involved in the direct IL-13 binding (7). This may suggest that the hydrophobic character of Val-35, Leu-38, and Val-42 is important for maintaining the structural stability of the D1 domain. To explore this possibility, the accessible surface area of the side chain of each amino acid residue of Val-35, Leu-38, and Val-42 was calculated based on structural information obtained by x-ray crystallography (7). The accessible surface areas for the residues Val-35, Leu-38, and Val-42 were calculated to be 0, 1.2, and 28.9A2, respectively, corresponding to only 0, 1.0, and 26% of the exposed surface area of the side chains, suggesting that both Val-35 and Leu-38 are completely buried (Fig. 4A). These results indicate that the side chains of these hydrophobic residues (Val-35, Leu-38, and Val-42), particularly Leu-38, may contribute to the stability needed to maintain the tertiary structure and consequently the ligand binding activity.
FIGURE 4.
Tertiary structures of mutation sites introduced into the D1 domain of IL-13Rα1. A, location of hydrophobic residues mutated to Ala. Mutated residues that lost IL-13 binding are shown in van der Waals representation. B, interface between the D1 domain and IL-13. The residues critical for IL-13 binding are labeled in black. C, interface between the D1 domain and IL-4. Structures were drawn with WebLab Viewer (Accelrys) using the coordinates 3BPN and 3BPO, determined by LaPorte et al. (7).
Identification of Amino Acids in the c′ Strand of the D1 Domain Critical for Binding to IL-13
The crystal structure showed that the ligand-receptor interaction between IL-13 and the D1 domain is a β-sheet-like interaction created by hydrogen bonding between the main-chain amide groups from Gln-74 to Glu-81 (major contributors are Lys-76, Lys-77, and Ile-78) of the D1 domain and the region from Ala-86 to Glu-91 (major contributors are Asp-87, Lys-89, and Glu-91) of IL-13 (7). The area of the contact region between IL-13 and the D1 domain was calculated to be ∼350 Å2. Hydrophobic interaction from the side chains in this region, particularly Lys-76 (92 Å2), Lys-77 (42 Å2), and Ile-78 (82 Å2), seems to assist the interaction between IL-13 and the D1 domain of receptor (Fig. 4B).
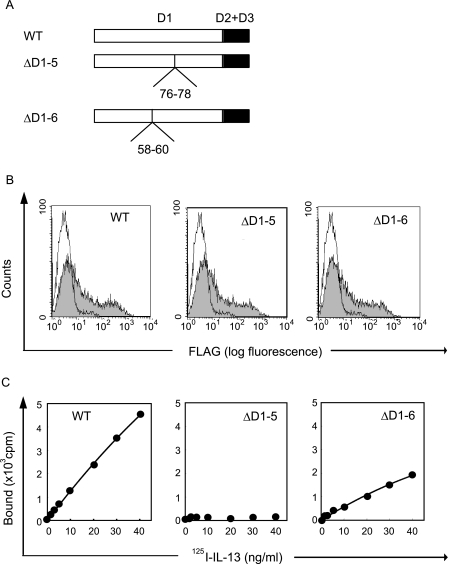
To analyze the significance of Lys-76, Lys-77, and Ile-78 in the c′ strand of the D1 domain, we generated two internal deleted mutants of hIL-13Rα1, hIL-13Rα1ΔD1–5 (deletion of Lys-76, Lys-77, and Ile-78) and hIL-13Rα1ΔD1–6 (deletion of Ala-58, Ser-59, and Ser-60), and analyzed their responses to IL-13 (Fig. 5A). The crystal structure of the IL-13·IL-13Rα1 complex suggested that Ala-58, Ser-59, and Ser-60 would not be involved in the formation of the conformational structure of IL-13Rα1 (18). hIL-13Rα1ΔD1–6 only slightly decreased binding activities compared with the wild type, whereas hIL-13Rα1ΔD1–5 completely diminished the activities (Fig. 5C). The expression of both hIL-13Rα1ΔD1–5 and hIL-13Rα1ΔD1–6 was the same as the wild type (Fig. 5B). These results strongly suggested that three amino acids, Lys-76, Lys-77, and Ile-78 in the c′ strand of hIL-13Rα1, are involved the direct interaction with IL-13.
FIGURE 5.
Importance of Lys-76, Lys-77, and Ile-78 in IL-13Rα1 for binding to IL-13. A, schematic models of two internal deleted mutants of IL-13Rα1 are depicted. The numbers represent the deleted amino acid numbers. The wild type or the indicated internal deleted mutated type of IL-13Rα1 (B and C) was transfected into HEK 293T cells. Expression of IL-13Rα1 (B) on the surface of HEK 293T cells by flow cytometry using anti-FLAG Ab is shown. The shaded and open areas represent the expression of the indicated transfectant or mock transfectant, respectively. Binding of 125I-labeled IL-13 (C) to HEK 293T cells is depicted.
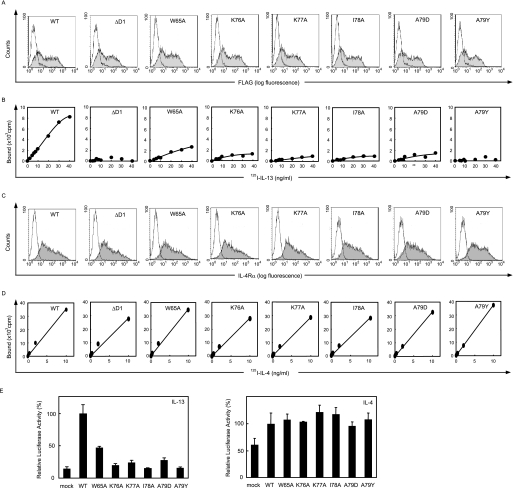
To further identify the important residues in this region, we then substituted Trp-65, Lys-76, Lys-77, and Ile-78 with Ala, and Ala-79 with charged or bulky amino acids, Asp or Tyr, based on the assumption from the crystal structure analysis, and analyzed the binding and signal transduction of IL-13 and IL-4 in these mutants. Lys-76, Lys-77, and Ile-78 contributed the most to the contact with IL-13. Because Trp-65 and Ala-79 of the D1 domain are also located at the interface with IL-13, the substitution of these residues may affect the interaction with IL-13. The Trp-65 → Ala, Lys-76 → Ala, Lys-77 → Ala, Ile-78 → Ala, Ala-79 → Asp, and Ala-79 → Tyr mutants were all expressed at the same level as the wild type, as shown by flow cytometry (Fig. 6A). Binding activity of IL-13 was partially impaired in the Trp-65 → Ala mutant, whereas the Lys-76 → Ala, Lys-77 → Ala, Ile-78 → Ala, Ala-79 → Asp, and Ala-79 → Tyr mutants showed significant impairment of binding activities (Fig. 6B). In contrast, binding activity to IL-4 was not affected in these mutants (Fig. 6, C and D). In accordance with the IL-13 binding, the luciferase activity of IL-13 was slightly decreased in the Trp-65 → Ala mutant. The Lys-76 → Ala, Lys-77 → Ala, Ile-78 → Ala, Ala-79 → Asp, and Ala-79 → Tyr mutants showed significantly decreased activities at almost the same level as the mock transfectant (Fig. 6E). The luciferase activities of IL-4 were again invariant between the wild type and the mutants. It is of note that the substitutions of Ala-79 with two different bulky amino acids, Asp and Tyr, similarly decreased IL-13 luciferase activity, indicating that the bulky side chains, not the specific characteristics of the side chains in Asp and Tyr, contributed to inhibiting the IL-13 signals. Combined with the structural analysis of the IL-13·IL-13Rα1·IL-4Rα complex, these functional studies clearly showed that the hydrophobic part of the side chains composed of Lys-76, Lys-77, and Ile-78, a characteristic feature of the β strand, was the IL-13 recognition site.
FIGURE 6.
Loss of binding and signal transduction of IL-13, but not IL-4 in the mutants of Trp-65, Lys-76, Lys-77, Ile-78, and Ala-79 in IL-13Rα1. The wild type or the indicated mutated type of IL-13Rα1 alone (A and B) or the wild type of IL-4Rα and the wild type or the indicated mutated type of IL-13Rα1 (C and D) were transfected into HEK 293T cells. Expression of IL-13Rα1 (A) or IL-4Rα (C) on the surface of HEK 293T cells by flow cytometry using anti-FLAG Ab or anti-CD124 Ab, respectively, is shown. The shaded and open areas represent the expression of the indicated transfectant or mock transfectant, respectively. Binding of 125I-labeled IL-13 (B) or IL-4 (D) to HEK 293T cells is depicted. E, the luciferase activities by IL-13 or by IL-4 are shown. The cells were stimulated with 0.01 ng/ml IL-13 or IL-4 for 6 h. The relative luciferase activities of each mutant are calculated from that of the wild type.
DISCUSSION
In this study, we identified the amino acids critical for binding to IL-13 using various IL-13Rα1 mutants, combining the data with structural study of the IL-13·IL-13Rα1·IL-4Rα complex. Although structural analysis of the ligand· receptor complex is a powerful approach to identify the critical region for ligand-receptor interaction, this method sometimes gives results for the identified ligand-receptor interaction site that are different from those yielded by the mutational approach. That is, the “structural” and “functional” binding epitopes are different, because it is very difficult to predict the energetics of interactions by inspection of structural data. This information can only be obtained by mutagenesis. For example, structural analysis showed that Phe-108 in IL-13 was strongly involved in receptor interaction at the D3 domain (site II interaction (7)), whereas the substitution of Phe-108 with Ala did not show any measurable loss of function (13). The present mutational analysis solidified the finding by the structural study that three amino acids, Lys-76, Lys-77, and Ile-78, in the c′ strand of the D1 domain of IL-13Rα1 form the recognition site with IL-13. This analysis also showed that three hydrophobic amino acids, Val-35, Leu-38, and Val-42, are critical for binding to IL-13, probably by maintaining the stability of the ternary structure of the D1 domain, which was not shown by the structural study.
It is known that the cytokine receptor homologous region having the WSXWS motif is a characteristic determinant for the interaction between helical cytokines and their receptors (14). However, several cytokine receptors such as gp130 and granulocyte colony-stimulating factor receptor use the extra Ig domain for ligand binding in addition to the cytokine receptor homologous region (15, 16). There are several ligand recognition schemes identified for Ig domains of cytokine receptors. One scheme is generation of a β-sheet-like structure by β strands of ligand and receptor, and another is hydrophobic interaction between ligand and receptor. gp130 uses the N and C termini of the Ig domain for generation of a β-sheet-like structure and hydrophobic interaction, respectively (15), whereas granulocyte colony-stimulating factor receptor binds to the ligand by hydrophobic interaction via N- and C-terminal portions of the Ig domain (16). The present and previous structural studies show that the ligand recognition manner of the D1 domain of IL-13Rα1 is unique in that the middle portion (residues from 65 to 79) of the D1 domain makes a hydrophobic patch in the β-sheet-like structure interfacing with Met-33, Thr-88, Lys-89, Ile-90, and Glu-91 in IL-13 (Fig. 4B).
Overall, from the structures of the IL-4 and IL-13 ternary complexes, it is difficult to understand the exact reason why the IL-13Rα1 D1 interaction with the cytokines is energetically critical for IL-13 binding but not IL-4. In both cases, the c′ strand of the D1 domain of IL-13Rα1 forms an antiparallel β-sheet with the F′-G′ loop of the cytokine in a rather “featureless” interface relatively devoid of deeply embedded and specific pairwise amino acid contacts, or obvious shape complementarity, as one sees, for instance, in the site I and site II interfaces. There is also extensive main-chain contact between the D1 c′ strand and the F′-G′ loop. We speculated from the crystal structures that the reason for the energetic differences could be related to an apparently more polar site III surface in the IL-4 complex (Thr-28, Asn-105, Gln-106, Ser-107, and Thr-108), versus a more hydrophobic surface in the IL-13 site III interface (Met-33, Thr-88, Lys-89, Ile-90, and Glu-91) that could generate binding energy through desolvation (Fig. 4, B and C). In either case, it is clear that even the IL-13 site III interface is weak in isolation, because we cannot see binding of an isolated IL-13Rα1 D1 domain with IL-13 (data not shown). Rather, the site III interaction is cooperative with sites I and II, supporting the notion that the higher energy site III in the IL-13 complex reflects a delicate energetic balance. Here, our mutational data support the assertion that the alkyl side chains of the c′ strand residues contribute binding energy to the interface through apolar van der Waals contacts that are diminished in the IL-4 site III (Fig. 6, B and D).
Because of the importance of IL-13 in the pathogenesis of bronchial asthma, several reagents targeting IL-13 are now being developed as therapeutic agents (1, 17). It is of note that an IL-4 variant (pitrakinra, Aerovant) competitively inhibiting the biological activities of both IL-4 and IL-13 by occupying their receptors, improved asthmatic symptoms in phase 2a studies (18). Our present findings about the specifics of the role of the D1 domain on the IL-13Rα1 signaling will help to develop a new or more efficient antagonist targeting IL-13Rα1. One possibility is to develop a neutralizing Ab or a low-molecular-weight compound targeting the binding site in the D1 domain. Thus, our present findings about the interaction between IL-13 and IL-13Rα1 give us not only basic knowledge about the association mechanism of the cytokine·receptor complex, but also clues useful in developing therapeutic agents.
Acknowledgment
We thank Dr. Dovie R. Wylie for critical review of the manuscript.
This work was supported in part by grant-in-aids for scientific research from the Japan Society for the Promotion of Science.
- IL-4
- interleukin-4
- IL-13R
- IL-13 receptor
- IL-13Rα1
- IL-13Rα1 chain
- IL-4Rα
- IL-4Rα chain
- Ab
- antibody
- STAT6
- signal transducers and activators of transcription 6.
REFERENCES
- 1.Izuhara K., Arima K., Kanaji S., Ohta S., Kanaji T. (2006) Curr. Med. Chem. 13, 2291–2298 [DOI] [PubMed] [Google Scholar]
- 2.Jiang H., Harris M. B., Rothman P. (2000) J. Allergy Clin. Immunol. 105, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 3.Nelms K., Keegan A. D., Zamorano J., Ryan J. J., Paul W. E. (1999) Annu. Rev. Immunol. 17, 701–738 [DOI] [PubMed] [Google Scholar]
- 4.Wills-Karp M. (2004) Immunol. Rev. 202, 175–190 [DOI] [PubMed] [Google Scholar]
- 5.Junttila I. S., Mizukami K., Dickensheets H., Meier-Schellersheim M., Yamane H., Donnelly R. P., Paul W. E. (2008) J. Exp. Med. 205, 2595–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews A. L., Holloway J. W., Puddicombe S. M., Holgate S. T., Davies D. E. (2002) J. Biol. Chem. 277, 46073–46078 [DOI] [PubMed] [Google Scholar]
- 7.LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., Heller N. M., Keegan A. D., Garcia K. C. (2008) Cell 132, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arima K., Sato K., Tanaka G., Kanaji S., Terada T., Honjo E., Kuroki R., Matsuo Y., Izuhara K. (2005) J. Biol. Chem. 280, 24915–24922 [DOI] [PubMed] [Google Scholar]
- 9.Boulay J. L., O'Shea J. J., Paul W. E. (2003) Immunity 19, 159–163 [DOI] [PubMed] [Google Scholar]
- 10.Honjo E., Shoyama Y., Tamada T., Shigematsu H., Hatanaka T., Kanaji S., Arima K., Ito Y., Izuhara K., Kuroki R. (2008) Protein Expr. Purif. 60, 25–30 [DOI] [PubMed] [Google Scholar]
- 11.Lee B., Richards F. M. (1971) J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 12.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 13.Madhankumar A. B., Mintz A., Debinski W. (2002) J. Biol. Chem. 277, 43194–43205 [DOI] [PubMed] [Google Scholar]
- 14.Bazan J. F. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 16.Tamada T., Honjo E., Maeda Y., Okamoto T., Ishibashi M., Tokunaga M., Kuroki R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holgate S. T., Polosa R. (2008) Nat. Rev. Immunol. 8, 218–230 [DOI] [PubMed] [Google Scholar]
- 18.Wenzel S., Wilbraham D., Fuller R., Getz E. B., Longphre M. (2007) Lancet 370, 1422–1431 [DOI] [PubMed] [Google Scholar]