Abstract
Endothelin-1 (ET-1) is a potent vasoconstrictor and co-mitogen for vascular smooth muscle and is implicated in pulmonary vascular remodeling and the development of pulmonary arterial hypertension. Vascular smooth muscle is an important source of ET-1. Here we demonstrate synergistic induction of preproET-1 message RNA and release of mature peptide by a combination of tumor necrosis factor α (TNFα) and interferon γ (IFNγ) in primary human pulmonary artery smooth muscle cells. This induction was prevented by pretreatment with the histone acetyltransferase inhibitor anacardic acid. TNFα induced a rapid and prolonged pattern of nuclear factor (NF)-κB p65 subunit activation and binding to the native preproET-1 promoter. In contrast, IFNγ induced a delayed activation of interferon regulatory factor-1 without any effect on NF-κB p65 nuclear localization or consensus DNA binding. However, we found cooperative p65 binding and histone H4 acetylation at distinct κB sites in the preproET-1 promoter after stimulation with both TNFα and IFNγ. This was associated with enhanced recruitment of RNA polymerase II to the ATG start site and read-through of the ET-1 coding region. Understanding such mechanisms is crucial in determining the key control points in ET-1 release. This has particular relevance to developing novel treatments targeted at the inflammatory component of pulmonary vascular remodeling.
Endothelin-1 is a 21-amino acid peptide which is known to be both a potent vasoconstrictor and mitogen for vascular smooth muscle (1, 2). It is released as a 38-amino acid precursor (Big ET-12) before cleavage to the mature ET-1 form. As such it has been implicated in the pathogenesis of vascular disease and is particularly associated with pulmonary arterial hypertension (3). Indeed, several endothelin receptor antagonists are now approved for the treatment of pulmonary arterial hypertension (4). However, endothelin receptor antagonists as a class are associated with potentially serious side effects (4), making new treatments aimed at blocking ET-1 synthesis an attractive alternative.
Although endothelial cells are thought to be the main source of ET-1 release, several groups including our own have shown that ET-1 can be released from the more numerous vascular smooth muscle cells (5–10). The vascular pathology observed in pulmonary arterial hypertension is propagated by inflammation, and circulating levels of cytokines including tumor necrosis factor α (TNFα) are elevated in patients with pulmonary arterial hypertension (11–15). In many cell types cytokines mediate their biological effects at least in part by the activation of the nuclear factor κB (NF-κB) pathway (16), and a role for NF-κB in pulmonary arterial hypertension has been proposed (17). In addition, we have shown previously that a combination of TNFα and interferon γ (IFNγ) stimulates human pulmonary artery smooth muscle (HPASM) cells to release ET-1 (18). However, the mechanisms underlying this effect are unknown.
The preproET-1 promoter region has been shown experimentally to possess binding sites for nuclear factor (NF)-1 and phorbol ester-sensitive c-Fos and c-Jun complexes (19), acute phase reactant regulatory proteins, and binding sites for AP-1 and GATA-2 (20–22). In addition, binding sites for interferon regulatory factor-1 (IRF-1) and NF-κB are predicted by Transfac analysis (23). The close proximity of the IRF-1 site and one of the NF-κB sites is characteristic of genes that are regulated by the synergistic action of TNFα and IFNγ, such as interleukin-6 (IL-6) and intercellular adhesion molecule-1 (24, 25), although ET-1 has not previously been recognized in this group.
Our aims were, therefore, to investigate the role of NF-κB in ET-1 release by primary HPASM cells. In addition, we were interested in the role of histone acetylation in the epigenetic control of the ET-1 production. Understanding these novel mechanisms will allow a greater understanding of the pathogenesis of vascular remodeling in pulmonary vessels and aid in the development of new treatment strategies aimed at blocking synthesis of ET-1.
EXPERIMENTAL PROCEDURES
Cell Culture
Human pulmonary arteries from healthy segments of lung were obtained from patients undergoing pulmonary resection at the Royal Brompton Hospital. Vessels were dissected clean from adventitia under sterile conditions, and the endothelium was removed mechanically, cut into 3–4-mm2 sections, and cultured as previously described (6). Cells between passages 2 and 9 were either seeded onto 96-well plates or tissue culture flasks.
Measurement of ET-1 and Big-ET-1 Release
Cells were grown to 80% confluence (10,000 cells/well), serum-deprived for 24 h (0.1% BSA) before treatment with either the same medium supplemented with 10% fetal calf serum and/or combinations of TNFα and IFNγ (both at 10 ng/ml), for another 24 h. ET-1 or Big ET-1 in the supernatant was measured by ELISA (R&D systems, Abingdon, UK and Biomedica, Vienna, Austria).
Real-time PCR
Total cellular RNA was extracted as previously described (27), and ET-1 and glyceraldehyde-3-phosphate dehydrogenase mRNA were quantified by real-time PCR using a QuantiTect SYBR Green PCR kit (Qiagen) using a Rotor-Gene 3000 (Corbett Research, N.S.W., Australia) and the ΔCt method according to the manufacturer's instructions. All primer pair details are shown in Table 1.
TABLE 1.
Primer details for all real-time quantitative PCR experiments
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
| Region | Sense primer (5′–3′) | Antisense primer (5′–3′) |
|---|---|---|
| preproET-1 | TCCTCTGCTGGTTCCTGACT | ATGGAAGCCAGTGAAGATGG |
| GAPDH | ACAGTCAGCCGCATCTTCTT | TTGACTCCGACCTTCACCTT |
| ChIP primers | ||
| A | TCTGAGGTGCATTCCTCCTT | AAGAGCTACAGGGGCTGATG |
| A | CCTGTACAGTAACTGCCCTGTG | AGGTGGTGCTGAGAAGGAAA |
| B | CCAGGCTGGACCTGAGAATA | CAGGGACTCCCCCTACTTTT |
| C | GCACACGTCTTGCAAATTCA | CGAGCTCGGGGAAAGTAAAT |
| D | GGTTTGTTGGGGTTCTTCTCCTT | TCCCTTTTCCTCCTGTAGGG |
| E | CTGGCCACGTTTTGTGTTT | GAGAAGTGGGGCAAAATGAA |
| RNA Pol II “run-through” | ||
| ATG | TTGAGGGACCTGAAGCTGTT | CAGAAGTAGACACACTCTTTATCCATC |
| mid-coding | CAGGGCTGAAGACATTATGGA | TCTCCGACCTGGTTTGTCTT |
Immunocytochemistry
HPASM cells were cultured in eight-well chamber slides (3000 cells per well), and immunocytochemistry was performed as previously described (27). Antibodies used were goat anti-p65 antibody (1/50, Santa Cruz Biotechnology, Wiltshire, UK), rhodamine-conjugated anti-goat antibody (1/100, Santa Cruz Biotechnology), rabbit anti-ET-1 antibody (1/25, Peninsula Laboratories, Inc., St. Helens, UK), and fluorescein isothiocyanate-conjugated anti-rabbit antibody (1/20, Dako Ltd, Ely, UK). Stained cells were observed by confocal microscopy, and images were collected using Leica confocal analysis software (TCSNT).
Preparation of Nuclear Extracts
Nuclear extracts were prepared exactly as previously described (27).
Western Blotting
Protein expression in nuclear extracts was determined by Western blot analysis (27) using rabbit anti-p65 (sc-109, Santa Cruz Biotechnology) and goat anti-rabbit antibody conjugated to horseradish peroxidase (Dako).
TransAm NF-κB Binding Assays
NF-κB p65 DNA binding in nuclear extracts was determined using an ELISA-based kit according to the manufacturer's instructions (TransAm, Active Motif, Rixensart, Belgium).
Chromatin Immunoprecipitation (ChIP) Assay
Binding of p65 (sc-372, Santa Cruz Biotechnology), IRF-1 (Santa Cruz Biotechnology), and RNA polymerase II (ab5408, Abcam, Cambridge, UK) to the native preproET-1 promoter and changes in histone acetylation status (06-866, anti-pan acetylated H4 antibody, Upstate, Watford, UK) were measured by ChIP assay exactly as previously described (27), except quantitative PCR was performed with 10 μl of DNA sample in a Rotor-Gene 3000TM Four-Channel Multiplexing System (Corbett Research, Cambridge, UK) using the QuantiTect SYBR Green PCR Kit (Qiagen). Primer details are shown in Table 1.
Materials
Dulbecco's modified Eagle's medium, penicillin, streptomycin, glutamine, onessential amino acids, sodium pyruvate, and fetal calf serum were obtained from Invitrogen. TNFα and IFNγ were obtained from R&D systems. The IκB kinase 2 (IKK-2) inhibitor AS602868 was kindly provided by Dr. Michel Dreano (Serono, Basel, Switzerland). The histone acetyltransferase inhibitor anacardic acid was obtained from Calbiochem. All primers for PCR were constructed by Invitrogen.
Statistics
All data are reported as the means ± S.E. from n experiments unless stated otherwise. Statistical analysis was made by one-way analysis of variance (ANOVA) with post-test analysis or by one-sample t test. p values ≤0.05 were considered significant.
RESULTS
Effect of TNFα and IFNγ on ET-1 Release and mRNA Induction by HPASM Cells
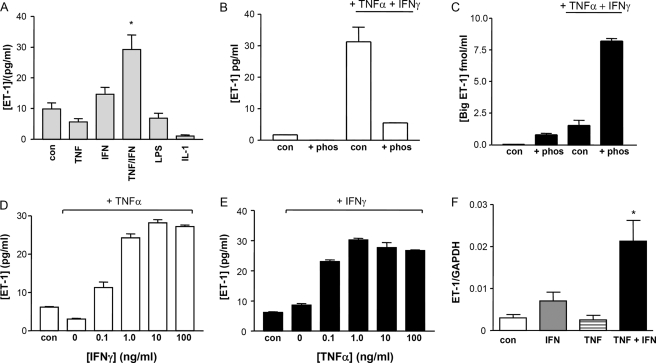
Under basal conditions HPASM cells released 9.8 pg/ml ET-1 into the supernatant after 24 h. Neither TNFα (10 ng/ml) nor INFγ (10 ng/ml) alone significantly increased ET-1 release, confirming our previous data (18). However, when TNFα and INFγ were added together, a synergistic induction of ET-1 production was observed (29.2 versus 9.8 pg/ml p < 0.05, Fig. 1A). In addition, neither lipopolysaccharide nor IL-1β had any significant effect on ET-1 release. The addition of the endothelin converting enzyme inhibitor phosphoramidon (50 μm) dramatically reduced ET-1 release with consequent increases in big ET-1 levels (Figs. 1, B and C, respectively), suggesting de novo protein production. The synergistic effects of TNFα and INFγ were apparent at all concentrations of cytokine tested (Fig. 1, D and E) and reached a plateau at 1–10 ng/ml TNFα and IFNγ. These effects were also seen at the mRNA level. 18 h after stimulation there was a clear synergistic increase in preproET-1 mRNA with both cytokines together compared with either cytokine alone (Fig. 1F, all at 10 ng/ml). Previous studies from our group have suggested that the cytokine-driven increase in preproET-1 mRNA levels in systemic vascular smooth muscle are unlikely to be because of changes in mRNA stability (7).
FIGURE 1.
A, the effect of TNFα and IFNγ on ET-1 expression in HPASM cells. HPASM cells were cultured for 24 h in the presence of TNFα, IFNγ, IL-1β (all at 10 ng/ml), or lipopolysaccharide (LPS, 1 μg/ml). ET-1 released into the supernatant was measured by ELISA. Results are presented as the means ± S.E. from five separate experiments performed in cells from three separate patients. *, p < 0.05 versus control by one way ANOVA. B and C, HPASM cells were cultured in the absence or presence of the endothelin converting enzyme inhibitor phosphoramidon (phos, 50 μm) for 30 min before treatment with medium (con) or TNFα and IFNγ (both at 10 ng/ml). After a further 24-h incubation, supernatants were collected for determination of ET-1 and Big ET-1 (B and C, respectively) by ELISA. Results are presented as the mean from one representative patient repeated in triplicate. D and E, synergistic increase of ET-1 release from HPASM cells treated with 10 ng/ml TNFα and increasing concentrations of IFNγ (D) or 10 ng/ml IFNγ and increasing concentrations of TNFα (E). Results are expressed as the mean from a single representative experiment repeated in triplicate. F, the synergistic effect of TNFα and IFNγ in stimulating expression of preproET-1 mRNA from HPASM cells after 18 h. Results are expressed as the ratio of preproET-1 message to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) message and are expressed as the mean ± S.E. of three independent patient cells. *, p < 0.01 compared with control by one way ANOVA.
NF-κB Activation in HPASM Cells Stimulated with TNFα and IFNγ
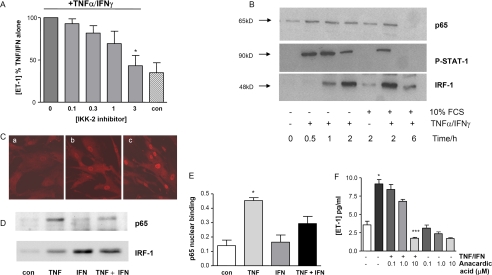
The IKK2 inhibitor AS602868 blocked the ability of TNFα and IFNγ to induce ET-1 expression in these cells in a concentration-dependent manner without affecting basal release (Fig. 2A). This implicates NF-κB as a critical factor in the synergistic induction of ET-1. To further explore the apparent synergy at the level of transcription, we examined nuclear enrichment of NF-κB p65, IRF-1, and phospho-STAT-1. TNFα plus INFγ induced p65 nuclear translocation, which was detectable between 0.5 and 6 h, with a peak signal at 1–2 h (Fig. 2B). In separate studies we found that the IFNγ-regulated transcription factors, IRF-1 and STAT-1, also translocate to the nucleus after treatment of HPASM cells with TNFα and IFNγ (Fig. 2B). As predicted, levels of STAT-1 peaked earlier than those of IRF-1 (data not shown), as STAT-1 is known to regulate IRF-1 production (29). Similarly, by confocal microscopy, p65, which was almost exclusively located in the cytosol of untreated cells that had been serum-deprived for 24 h, appeared enriched in the nucleus of cells stimulated with TNFα plus INFγ (Fig. 2C). Because no STAT binding sites are present in the preproET-1 promoter, we focused hereafter on IRF-1 interactions with NF-κB.
FIGURE 2.
The NF-κB pathway is crucial for TNFα/IFNγ-stimulated ET-1 production. A, HPASM cells were pretreated with the IKK2 inhibitor (AS602868, 0–3 μm) for 30 min before co-treating with TNFα and IFNγ (both at 10 ng/ml) and ET-1 release measured by ELISA. Results are from four donors and are expressed as percentage of ET-1 released from cells treated with TNFα and IFNγ without the inhibitor. *, p = 0.02 by one sample t test. con, control. B, p65, IRF-1, and phospho STAT-1 undergo nuclear translocation when HPASM cells are stimulated with TNFα and IFNγ for various times up to 6 h. Panel B is a representative blot of results from three donors. C, nuclear translocation of p65 assessed by confocal microscopy in the absence of serum (a), cells with 10% fetal calf serum (b), and 10% fetal calf serum with TNFα/IFNγ (c) for 2 h. The confocal images are representative of results from three donors. No enhancement of p65 or IRF-1 nuclear translocation in the presence of both TNFα and IFNγ (both at 10 ng/ml) was determined in nuclear extracts after 2 h (D). The blot is representative of three separate experiments. E, similar experiments were performed, and p65 nuclear binding was determined by TransAm assay. Results are the means ± S.E. from three separate donors (repeated in duplicate). *, p < 0.05 by one way ANOVA for treatment with TNFα versus control. F, concentration-dependent effect of 30 min of pretreatment with the histone acetyltransferase inhibitor anacardic acid on ET-1 expression. *, p < 0.05 versus control unstimulated cells; ###, p < 0.001 versus stimulated cells by Kruskal-Wallis with Dunn's post-test analysis.
NF-κB Consensus DNA Binding in HPASM Cells Treated with TNFα and IFNγ
In separate experiments the effect of TNFα and IFNγ on p65 and IRF-1 nuclear translocation was investigated 2 h post-stimulation. This time point was chosen as this represented the time when there was maximum overlap of nuclear enrichment of p65 and IRF-1. There was no enhanced nuclear import of either p65 or IRF-1 as measured by Western blotting. Interestingly, although IFNγ alone had no effect on nuclear translocation of p65, TNFα caused a small but significant nuclear enrichment of IRF-1 (Fig. 2D). This was confirmed with confocal microscopy (data not shown). Furthermore, TNFα alone significantly enhanced NF-κB p65 DNA binding to a consensus κB element, whereas treatment with IFNγ alone did not enhance NF-κB p65 DNA binding (Fig. 2E). Although cotreatment of HPASMs with TNFα and IFNγ appeared to result in less NF-κB p65 DNA binding to the consensus κB site, this did not reach statistical significance (Fig. 2E). Finally, we demonstrated that the histone acetyltransferase inhibitor anacardic acid suppressed TNFα/IFNγ-stimulated ET-1 expression in a concentration-dependent manner (Fig. 2F), highlighting a key role for histone acetylation in ET-1 induction in these cells.
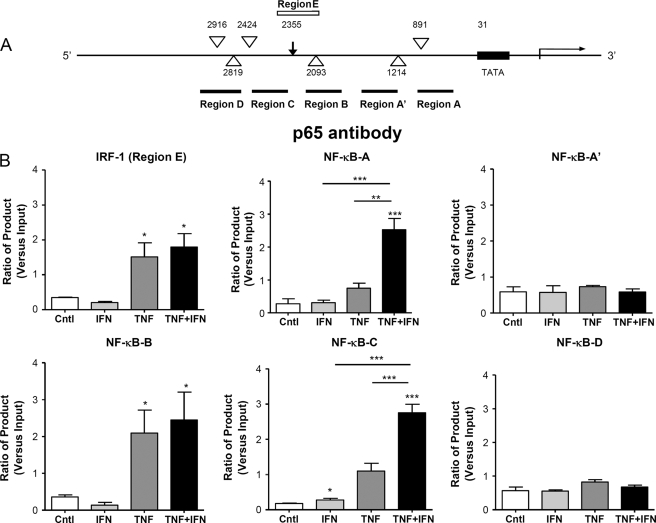
Binding of p65 to the Native PreproET-1 Promoter
ChIP assays were used to determine that binding of p65 to putative NF-κB sites on the native preproET-1 promoter. Using the Transfac MatInspector V2.2 program (23) we were able to identify six possible NF-κB binding sites in the preproET-1 promoter region (Fig. 3A). Of these, two have previously been reported as active sites for NF-κB binding, −2093 bp (30) (Region B) and −891 bp (31) (Region A), using reporter gene assays. Enhanced binding of p65 at regions A and C was observed at 2 h after treatment of cells with both cytokines compared with that seen with either cytokine alone (Fig. 3B). Although there was enhanced p65 DNA binding with TNFα alone in region B compared with IFNγ alone or control, there was no additional enhancement of binding in the presence of both cytokines. There was no evidence of p65 promoter binding under any condition compared with control in regions A′ and D. Interestingly, treatment of cells with TNFα alone resulted in p65 association with the IRF-1 site (Region E), although there was no additional binding after co-stimulation with IFNγ.
FIGURE 3.
Selective recruitment of p65 to the native ET-1 promoter in the presence of TNFα and IFNγ as determined by ChIP assay. The human preproET-1 promoter region was analyzed using the Transfac program for putative NF-κB binding sites (large open arrowhead) and IRF-1 binding sites (arrow) (A). In addition, regions defined by the primers used for the ChIP assay are also shown (black lines for NF-κB and white line for IRF-1). B, HPASM cells were cultured with the cytokines TNFα and IFNγ either alone or in combination (all at 10 ng/ml) for 2 h. ChIP was performed using an antibody directed against p65, and real time PCR was performed using primers directed against the regions indicated. Ct (threshold) values of PCR products were normalized to those of input samples. Results are represented as the mean ± S.E. for three separate donors. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA. Cntl, control.
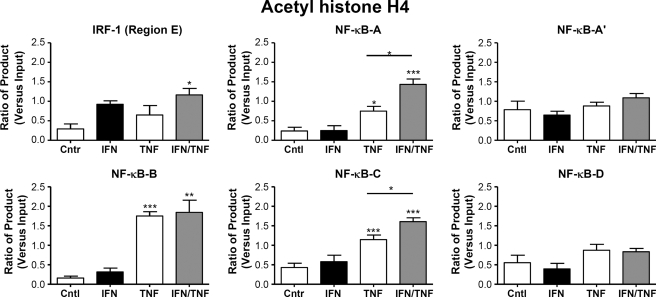
Evidence for Differential Histone Acetylation at NF-κB Sites in the PreproET-1 Promoter Region
Treatment of primary HPASM cells with a combination of TNFα and IFNγ (both at 10 ng/ml) for 2 h resulted in increased acetylation of histone H4 at the κB sites positioned at Region A (−891 bp) and Region C (−2424 bp), compared with that seen with either cytokine alone (Fig. 4). The largest increase in histone H4 acetylation was seen at region B (−2093 bp) after TNFα stimulation. However, this was not further enhanced by the addition of IFNγ. In contrast, there was no H4 acetylation seen at Region A′ (−1214 bp) or Region D (−2819 and −2916 bp). These results were consistent, therefore, with the p65 ChIP experiments. They indicate that although sites within regions A, B, and C all bind p65 and are associated with histone H4 acetylation after TNFα stimulation, only regions A and C demonstrated enhanced binding with the addition of both TNFα and IFNγ. In contrast, regions A′ and D were not acetylated under these conditions despite containing putative NF-κB binding sites reflecting the lack of p65 binding. Although there was evidence of increased acetylation at the IRF-1 site at 2355 bp (Region E) compared with control, there was no difference between cytokine treatments. Interestingly, there was a clear difference between the results for histone acetylation at the NF-κB sites at Region C (−2424 bp) and the IRF-1 site at Region E (−2355 bp), surprisingly distinguishing between binding sites less than 100 bp apart after enzymatic digestion of DNA.
FIGURE 4.
Differential histone H4 acetylation at p65 binding sites in the native ET-1 promoter. HPASM cells were cultured with the cytokines TNFα and IFNγ either alone or in combination (all at 10 ng/ml) for 2 h. ChIP was performed using an antibody directed against acetylhistone H4, and real time PCR was performed using primers directed against the regions indicated in Fig. 3A. Ct (threshold) values of PCR products were normalized to those of input samples. Results are represented as the mean ± S.E. for three separate donors. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA. Cntr, control.
RNA Pol II ChIP
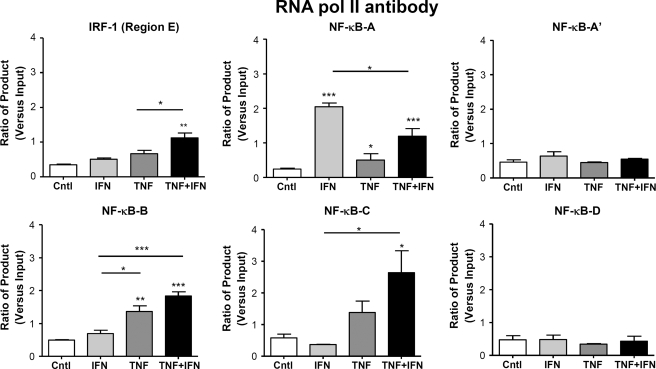
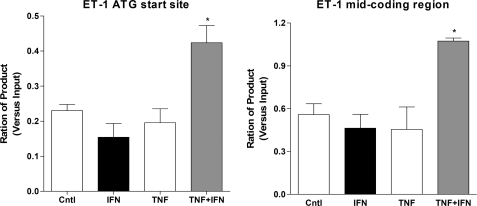
RNA Pol II binding at κB sites in region C and B but not D and A′ was enhanced, consistent with the results for p65 and acetylated histone H4 ChIP experiments. Interestingly in region A, treatment with IFNγ alone enhanced RNA Pol II binding to a greater extent than that seen with TNFα with no additional recruitment seen in the presence of both cytokines. Finally, at the IRF-1 site (region E) there was greatest relative enhancement of RNA Pol II binding after co-stimulation compared with the effect of either cytokine alone, confirming the importance of this site in transcriptional control of ET-1 under these conditions (Fig. 5). Furthermore, RNA Pol II was only recruited to the preproET-1 start site when both cytokines are present, and this coincided with active transcription as measured by RNA Pol II read-through of preproET-1 (Fig. 6).
FIGURE 5.
Differential RNA Pol II association with specific κB sites in the native ET-1 promoter. HPASM cells were cultured with the cytokines TNFα and IFNγ either alone or in combination (all at 10 ng/ml) for 2 h. ChIP was performed using an antibody directed against RNA Pol II, and real time PCR was performed using primers directed against the regions indicated in Fig. 3A. Ct (threshold) values of PCR products were normalized to those of input samples. Results are represented as mean ± S.E. for three separate donors. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA. Cntl, control.
FIGURE 6.
Synergistic association of RNA pol II to the ET-1 start site and coding region. HPASM cells were cultured with the cytokines TNFα and IFNγ either alone or in combination (all at 10 ng/ml) for 2 h. ChIP was performed using an antibody directed against RNA Pol II. Real time PCR was performed using primers covering the ATG start site and a mid coding region. Ct (threshold) values of PCR products were normalized to those of input samples. Results are represented as mean ± S.E. for three separate donors. *, p < 0.05. Cntl, control.
DISCUSSION
Pulmonary vascular smooth muscle cells are an important source of mitogenic factors, which contribute to the development of pulmonary arterial hypertension. The role of cytokines in stimulating vascular smooth muscle cells to produce mitogens such as ET-1 is poorly understood. We demonstrate that enhanced histone acetylation and RNA polymerase II recruitment occurs at selective NF-κB sites in the preproET-1 promoter after cooperative p65 recruitment to these sites in the native promoter and that this is associated with enhanced gene transcription. Furthermore, we show that an additional level of control occurs at the post-transcriptional level in these cells.
In endothelial cells, which were originally considered to be the main source of ET-1 in the vasculature, TNFα, IL-1β, or lipopolysaccharide alone and in combination stimulate ET-1 release and increased expression of preproET-1 mRNA (32–35). In endothelial cells the increase in expression of preproET-1 mRNA is transient. By contrast, in vascular smooth muscle cells, we have previously shown that the preproET-1 mRNA induced by TNFα and IFNγ is both slow and sustained (18). These differences in ET-1 release may be indicative of the physiological versus pathophysiological role of each cell type.
The observation of a dependence on two cytokines for the increased transcription of a gene has previously been described in immune cells (25), and the synergistic action of TNFα and IFNγ is well described and includes control of genes such as intercellular adhesion molecule-1, IL-6, RANTES (regulated on activation normal T cell expressed and secreted), and inducible nitric-oxide synthase (36–39). In this study we extend this regulation for the first time to pulmonary artery smooth muscle cells. Furthermore, this synergistic control of the preproET-1 mRNA expression is a novel observation. As in immune cells, this type of regulation may be seen as a protective function in pulmonary artery smooth muscle, only allowing synthesis of ET-1 under severe stress or with multiple inflammatory hits. Thereby this observation has important implications for vascular remodeling, where the role of inflammation is being increasingly recognized (5).
Where TNFα and INFγ act in synergy to induce genes, the TNFα-associated transcription factor is invariably NF-κB (25). Indeed, we were able to demonstrate that inhibition of IKK2 by AS602868 was able to attenuate TNFα/IFNγ-induced ET-1 expression in these cells. IFNγ-induced transcription factors include IRF-1 as well as STAT-1 and AP-1 (41). Stimulation of HPASM cells with TNFα and IFNγ was associated with p65 nuclear translocation; however, there was no increase in nuclear translocation when the cytokines were combined, and we demonstrated similar findings with IRF-1. In addition, we observed an increase in nuclear enrichment with IRF-1 when HPASM cells were treated with TNFα alone. Stimulation of IRF-1 synthesis by TNFα has been shown previously (29). However, treatment of HPASM cells with TNF-α alone is still insufficient for preproET-1 gene transcription, suggesting that the amount of IRF-1 induced by TNFα is insufficient to enable formation of a pre-initiation complex and subsequent ET-1 transcription. Alternatively, other transcription factors such as AP-1 may play a predominant role. We were unable to demonstrate a STAT-1 binding site in the preproET-1 promoter region, which limits the potential role for this transcription factor in interacting with p65 and precluded further investigation. Further experiments will need to be performed to investigate the importance of the AP-1 site at −108 bp and the effect of c-Jun NH2-terminal kinase (JNK) inhibition.
Using immunocytochemistry, Western blotting, and DNA binding assays, we were unable to show enhanced p65 activation in the presence of IFNγ. In fact, there appeared to be a decrease in consensus p65 binding in the presence of TNFα and IFNγ. This observation has been reported before in relation to the regulation of IL-6, IL-8, and eotaxin (42). Although this may explain regulation of genes inhibited by the combination of TNFα and IFNγ, it clearly does not explain the situation where there is a synergistic increase in gene regulation such as for inducible nitric-oxide synthase and IL-6. Furthermore, within the situation of total cell reduction in p65 binding and histone acetylation, there was clearly no loss of p65 binding and indeed enhanced histone acetylation and RNA Pol II binding to specific κB sites in the preproET-1 promoter.
Using p65 ChIP analysis we were able to show that in the presence of TNFα and IFNγ there was selective recruitment to specific κB sites in the native preproET-1 promoter. We could show that three of the five regions (containing six κB sites) investigated were active. Furthermore, there was clearly enhanced binding at regions A (−891bp) and C (−1214 bp) with both cytokines but not at region B (−2093 bp), where there was no increase compared with TNFα alone. We were also able to show tethering of p65 at the IRF-1 binding site in the ET-1 promoter in the presence of TNFα alone but no enhanced binding with the addition of IFNγ. Therefore, our results extend those of Quehenberger et al. (30), who detected increased binding using electrophoretic mobility shift assay and a specific probe for the −2093 binding site for p65 (corresponding to our region B) in bovine endothelial cells. In fact, although we also show p65 binding to this site in Region B, we also reveal the potentially more important sites in Regions A and C with regard to enhancement of ET-1 transcription in the presence of TNFα and IFNγ. As far as we are aware, this is the first report of this powerful technique in primary human vascular smooth muscle cells and the first to describe direct interactions of p65 with the native preproET-1 promoter.
To further investigate potential mechanisms, we examined the effect of TNFα and IFNγ alone and in combination on histone H4 acetylation at the relevant sites within the native ET-1 promoter. Histone acetylation is an epigenetic control mechanism involved in the regulation of gene transcription (44). We were able to demonstrate enhanced histone H4 acetylation at distinct NF-κB sites in the preproET-1 promoter region under conditions of TNFα and IFNγ stimulation compared with the cytokines alone. In agreement with the p65 ChIP binding data, most histone acetylation was associated with regions A, B, and C, suggesting that these are the most transcriptionally active NF-κB binding sites. However, as with the p65 ChIP data, it was only at the sites in Regions A and C where enhanced histone H4 acetylation with both cytokines was demonstrated. Histone H4 acetylation was increased in all conditions at the IRF-1 site compared with control, but there was no difference between these conditions, unlike at the NF-κB sites. Surprisingly, the resolution of the ChIP assay here using enzymatic digested DNA was such that it could distinguish between an active and inactive site (Region C and E), less than 100 bp apart. Finally, the importance of histone acetylation was confirmed by the action of the histone acetyltransferase inhibitor anacardic acid.
RNA Pol II ChIP confirmed the importance of the NF-κB sites in regions B and C and confirmed the redundancy of the sites in regions A′ and D. The greatest relative increase in binding occurred at the IRF-1 binding site, indicating its importance in transcription. The most unexpected result was at the NF-κB binding site in region A. Here, we found that IFNγ alone resulted in most RNA Pol II association. However, taken together, these ChIP experiments confirm a complex interaction of key NF-κB sites together with a single IRF-1 site in close proximity. This may reflect the formation of an enhanceosome (in the presence of both cytokines) which allows RNA Pol II to bind to the ATG start site and drive active preproET-1 transcription as demonstrated by RNA Pol II association with the preproET-1 coding region. This would need to be confirmed using addition experiments including chromosome conformation capture analysis to demonstrate looping out at the ET-1 locus. Alternatively, as the RNA Pol II antibody used measures both phosphorylated and non-phosphorylated forms, we could merely be reflecting localization of distinct hypo- and hyperphosphorylated forms at distinct sites performing separate functions. The unphosphorylated form of RNA Pol II binds to the pre-initiation complex through interaction with the general transcription factor TBP and the Mediator complex. This complex tethers RNA Pol II to the TGFβ1 promoter and integrates the signals from p65 and IRF-1 to the general transcription machinery (45). Subsequent RNA Pol II Ser-5 phosphorylation then allows release from the Mediator complex and transcription to start as demonstrated by the read-through of the coding sequence. Further temporal analysis of phosphorylated forms of RNA Pol II across the TGFβ1 promoter and coding sequence may resolve this.
There are several limitations to this study. First, the ChIP studies in particular involved small n numbers (n = 3–4). However, we chose to study only primary human cells from the pulmonary vasculature of patients, making the results more meaningful. We were not able to reproduce the synergy in ET-1 release or p65 DNA binding with TNFα and IFNγ in a commercially available human pulmonary artery smooth muscle cell line (CC-2581, Lonza), which also limited the potential to perform further mechanistic studies. The number of cells available was, therefore, limited, especially for techniques such as ChIP, which demand large numbers of cells. In addition, the use of limited numbers of primary cells prevented the optimization of the time course for histone acetylation at the ET-1 promoter. Initial studies in cells from a single patient demonstrate that enhanced p65 binding occurs at the NF-κB-A site at 1 h post-stimulation before measurable pan H4 acetylation (data not shown). Second, studying individual patients inevitably will lead to individual variations, and we believe that is why the error margins are quite large in some of the experiments. It is possible that other IFNγ regulated transcription factors/sites are important such as AP-1, but these were not studied here. The formation of enhanceosomes has been demonstrated elegantly for virus-induced IFN-β expression and is essential for maximal and sustained transcription but needs to be formally demonstrated here (46). Further experiments including optimization of the time-course for chromatin modification and remodeling events may provide further information underlying the synergistic transcription of ET-1.
Understanding basic mechanisms of induction of genes such as ET-1 are crucial to elucidating the pathogenesis of vascular remodeling. In particular, the development of assays such as the ChIP assay allows the study of native transcriptional control not only in human cells but also in a tissue-specific manner. This model may allow the development of therapeutic strategies that are themselves tissue-specific, such as NF-κB inhibition (47) or by the use of sequence-specific artificial zinc finger proteins (48) to target vascular smooth muscle. Indeed, methods directed at transcription factors, such as antisense oligonucleotides against NF-κB, have already been shown to be of therapeutic benefit in vascular disease (40).
Footnotes
- ET-1
- endothelin-1
- TNFα
- tumor necrosis factor α
- IFN
- interferon γ
- IL
- interleukin
- ELISA
- enzyme-linked immunosorbent assay
- STAT
- signal transducers and activators of transcription
- Pol
- polymerase
- HPASM
- human pulmonary artery smooth muscle
- IRF-1
- interferon regulatory factor-1
- ANOVA
- analysis of variance
- ChIP
- chromatin immunoprecipitation
- AP-1
- activator protein 1.
REFERENCES
- 1.Bobik A., Grooms A., Millar J. A., Mitchell A., Grinpukel S. (1990) Am. J. Physiol. 258, C408–C415 [DOI] [PubMed] [Google Scholar]
- 2.Yanagisawa M., Kurihara H., Kimura S., Tomobe Y., Kobayashi M., Mitsui Y., Yazaki Y., Goto K., Masaki T. (1988) Nature 332, 411–415 [DOI] [PubMed] [Google Scholar]
- 3.Stewart D. J., Levy R. D., Cernacek P., Langleben D. (1991) Ann. Intern. Med. 114, 464–469 [DOI] [PubMed] [Google Scholar]
- 4.Langleben D. (2007) Clin. Chest. Med. 28, 117–125 [DOI] [PubMed] [Google Scholar]
- 5.Wort S. J., Woods M., Warner T. D., Evans T. W., Mitchell J. A. (2001) Am. J. Respir. Cell Mol. Biol. 25, 104–110 [DOI] [PubMed] [Google Scholar]
- 6.Wort S. J., Mitchell J. A., Woods M., Evans T. W., Warner T. D. (2000) J. Cardiovasc. Pharmacol. 36, S410–413 [DOI] [PubMed] [Google Scholar]
- 7.Woods M., Wood E. G., Bardswell S. C., Bishop-Bailey D., Barker S., Wort S. J., Mitchell J. A., Warner T. D. (2003) Mol. Pharmacol. 64, 923–931 [DOI] [PubMed] [Google Scholar]
- 8.Tchekneva E., Quertermous T., Christman B. W., Lawrence M. L., Meyrick B. (1998) J. Clin. Invest. 101, 1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchekneva E., Lawrence M. L., Meyrick B. (2000) Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L813–L821 [DOI] [PubMed] [Google Scholar]
- 10.Upton P. D., Wharton J., Coppock H., Davie N., Yang X., Yacoub M. H., Ghatei M. A., Polak J. M., Bloom S. R., Smith D. M., Morrell N. W. (2001) Am. J. Respir. Cell Mol. Biol. 24, 170–178 [DOI] [PubMed] [Google Scholar]
- 11.Caslin A. W., Heath D., Madden B., Yacoub M., Gosney J. R., Smith P. (1990) Histopathology 16, 9–19 [DOI] [PubMed] [Google Scholar]
- 12.Humbert M., Monti G., Brenot F., Sitbon O., Portier A., Grangeot-Keros L., Duroux P., Galanaud P., Simonneau G., Emilie D. (1995) Am. J. Respir. Crit. Care Med. 151, 1628–1631 [DOI] [PubMed] [Google Scholar]
- 13.Kubo K., Hanaoka M., Hayano T., Miyahara T., Hachiya T., Hayasaka M., Koizumi T., Fujimoto K., Kobayashi T., Honda T. (1998) Respir. Physiol. 111, 301–310 [DOI] [PubMed] [Google Scholar]
- 14.Lesprit P., Godeau B., Authier F. J., Soubrier M., Zuber M., Larroche C., Viard J. P., Wechsler B., Gherardi R. (1998) Am. J. Respir. Crit. Care Med. 157, 907–911 [DOI] [PubMed] [Google Scholar]
- 15.Voelkel N. F., Tuder R. M. (1995) Eur. Respir. J. 8, 2129–2138 [DOI] [PubMed] [Google Scholar]
- 16.Collins T., Read M. A., Neish A. S., Whitley M. Z., Thanos D., Maniatis T. (1995) FASEB J. 9, 899–909 [PubMed] [Google Scholar]
- 17.Aziz S. M., Toborek M., Hennig B., Endean E., Lipke D. W. (1997) Cell Biol. Int. 21, 801–812 [DOI] [PubMed] [Google Scholar]
- 18.Wort S. J., Woods M., Warner T. D., Evans T. W., Mitchell J. A. (2002) Mol. Pharmacol. 62, 1147–1153 [DOI] [PubMed] [Google Scholar]
- 19.Lee M. E., Bloch K. D., Clifford J. A., Quertermous T. (1990) J. Biol. Chem. 265, 10446–10450 [PubMed] [Google Scholar]
- 20.Dorfman D. M., Wilson D. B., Bruns G. A., Orkin S. H. (1992) J. Biol. Chem. 267, 1279–1285 [PubMed] [Google Scholar]
- 21.Inoue A., Yanagisawa M., Takuwa Y., Mitsui Y., Kobayashi M., Masaki T. (1989) J. Biol. Chem. 264, 14954–14959 [PubMed] [Google Scholar]
- 22.Lee M. E., Dhadly M. S., Temizer D. H., Clifford J. A., Yoshizumi M., Quertermous T. (1991) J. Biol. Chem. 266, 19034–19039 [PubMed] [Google Scholar]
- 23.Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhäuser R., Prüss M., Schacherer F., Thiele S., Urbach S. (2001) Nucleic Acids Res. 29, 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tau G., Rothman P. (1999) Allergy 54, 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paludan S. R. (2000) J. Leukoc. Biol. 67, 18–25 [DOI] [PubMed] [Google Scholar]
- 26.Deleted in proof
- 27.Ito K., Jazrawi E., Cosio B., Barnes P. J., Adcock I. M. (2001) J. Biol. Chem. 276, 30208–30215 [DOI] [PubMed] [Google Scholar]
- 28.Deleted in proof
- 29.Pine R. (1997) Nucleic Acids Res. 25, 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quehenberger P., Bierhaus A., Fasching P., Muellner C., Klevesath M., Hong M., Stier G., Sattler M., Schleicher E., Speiser W., Nawroth P. P. (2000) Diabetes 49, 1561–1570 [DOI] [PubMed] [Google Scholar]
- 31.Fujita M., Shannon J. M., Irvin C. G., Fagan K. A., Cool C., Augustin A., Mason R. J. (2001) Am. J. Physiol. Lung Cell. Mol. Physiol. 280, L39–L49 [DOI] [PubMed] [Google Scholar]
- 32.Marsden P. A., Brenner B. M. (1992) Am. J. Physiol. 262, C854–C861 [DOI] [PubMed] [Google Scholar]
- 33.Yoshizumi M., Kurihara H., Morita T., Yamashita T., Oh-hashi Y., Sugiyama T., Takaku F., Yanagisawa M., Masaki T., Yazaki Y. (1990) Biochem. Biophys. Res. Commun. 166, 324–329 [DOI] [PubMed] [Google Scholar]
- 34.Sugiura M., Inagami T., Kon V. (1989) Biochem. Biophys. Res. Commun. 161, 1220–1227 [DOI] [PubMed] [Google Scholar]
- 35.Molet S., Furukawa K., Maghazechi A., Hamid Q., Giaid A. (2000) J. Allergy Clin. Immunol. 105, 333–338 [DOI] [PubMed] [Google Scholar]
- 36.Jahnke A., Johnson J. P. (1995) Immunobiology 193, 305–314 [DOI] [PubMed] [Google Scholar]
- 37.Kamijo R., Harada H., Matsuyama T., Bosland M., Gerecitano J., Shapiro D., Le J., Koh S. I., Kimura T., Green S. J. (1994) Science 263, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 38.Sancéau J., Kaisho T., Hirano T., Wietzerbin J. (1995) J. Biol. Chem. 270, 27920–27931 [DOI] [PubMed] [Google Scholar]
- 39.Casola A., Henderson A., Liu T., Garofalo R. P., Brasier A. R. (2002) Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L1280–L1290 [DOI] [PubMed] [Google Scholar]
- 40.Sawa Y., Morishita R., Suzuki K., Kagisaki K., Kaneda Y., Maeda K., Kadoba K., Matsuda H. (1997) Circulation 96, Suppl. 9, II– 280–284 [PubMed] [Google Scholar]
- 41.Aaronson D. S., Horvath C. M. (2002) Science 296, 1653–1655 [DOI] [PubMed] [Google Scholar]
- 42.Keslacy S., Tliba O., Baidouri H., Amrani Y. (2007) Mol. Pharmacol. 71, 609–618 [DOI] [PubMed] [Google Scholar]
- 43.Deleted in proof [Google Scholar]
- 44.Adcock I. M., Ford P., Ito K., Barnes P. J. (2006) Respir. Res. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egloff S., Murphy S. (2008) Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 46.Kim T. K., Maniatis T. (1997) Mol. Cell 1, 119–129 [DOI] [PubMed] [Google Scholar]
- 47.Karin M. (2005) Proc. Am. Thorac. Soc. 2, 386–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Negi S., Imanishi M., Matsumoto M., Sugiura Y. (2008) Chemistry 14, 3236–3249 [DOI] [PubMed] [Google Scholar]