Abstract
Type I interferons (IFNs) bind IFNAR receptors and activate Jak kinases and Stat transcription factors to stimulate the transcription of genes downstream from IFN-stimulated response elements. In this study, we analyze the role of protein palmitoylation, a reversible post-translational lipid modification, in the functional properties of IFNAR. We report that pharmacological inhibition of protein palmitoylation results in severe defects of IFN receptor endocytosis and signaling. We generated mutants of the IFNAR1 subunit of the type I IFN receptor, in which each or both of the two cysteines present in the cytoplasmic domain are replaced by alanines. We show that cysteine 463 of IFNAR1, the more proximal of the two cytoplasmic cysteines, is palmitoylated. A thorough microscopic and biochemical analysis of the palmitoylation-deficient IFNAR1 mutant revealed that IFNAR1 palmitoylation is not required for receptor endocytosis, intracellular distribution, or stability at the cell surface. However, the lack of IFNAR1 palmitoylation affects selectively the activation of Stat2, which results in a lack of efficient Stat1 activation and nuclear translocation and IFN-α-activated gene transcription. Thus, receptor palmitoylation is a previously undescribed mechanism of regulating signaling activity by type I IFNs in the Jak/Stat pathway.
Type I interferons (IFN7 α/β) are potent cellular mediators essential for several key cell functions, including immunomodulatory, antiviral, and antiproliferative activities. These pleiotropic effects occur through the transcriptional regulation of many IFN-stimulated genes (ISGs) (1). IFN signal transduction relies mainly on the activation of the Janus tyrosine kinase (Jak)/signal-transducing activators of transcription (Stat) pathways, although several other signaling cascades have also been associated with IFN-regulated transcription (2, 3). In general, the binding of type I IFNs to the cell surface receptor IFNAR1 and IFNAR2 subunits induces tyrosine phosphorylation in trans of the IFNAR-associated Jak kinases (Tyk2 with IFNAR1 and Jak1 with IFNAR2), which in turn leads to IFNAR tyrosine phosphorylation. Several members of the Stat family can be activated by type I IFNs, and Stat1 and Stat2 are the main downstream effectors of the type I IFN transcriptional response. Upon IFN-α stimulation, cytosolic Stat2 is recruited to the activated IFNAR complex where it becomes tyrosine-phosphorylated by the receptor-associated Jak kinases. Stat2 activation is a key event in IFN-α signaling because it is required for the indirect recruitment, through binding to Stat2, of Stat1 to IFNAR1 and its activation. There is some debate as to whether cytosolic Stat2 is preferentially recruited to IFNAR1 or to IFNAR2 (4–9). Whereas the SH2 domain of Stat2 binds to a region surrounding the phosphorylated tyrosine 466 of IFNAR1, Stat2 can bind to IFNAR2 whether it is tyrosine-phosphorylated or not. This series of sequential tyrosine phosphorylations precedes the translocation of the IFN-stimulated gene factor 3 complex to the nucleus where it activates gene transcription from promoters containing an IFN-stimulated response element (ISRE) (10, 11).
Recent data indicate that signal transduction through the Jak/Stat pathway cannot fully account for the diversity and complexity of the biological response elicited by type I IFNs (12), and that other factors, for example receptor configuration and alternate signaling pathways, have to be considered (13). We recently showed that endocytosis plays an important role in the control of IFN-α signaling and biological activity (14). Little is known about the potential links between membrane trafficking and the control of the Jak/Stat signaling pathway, and the contribution of IFNAR trafficking to IFN signaling is just beginning to be appreciated (15). To study this poorly investigated aspect of IFN signaling, we examined the role, if any, of receptor palmitoylation. Palmitoylation is a reversible lipid modification involving the specific attachment of a saturated fatty acid chain to cysteines via a thioester bond. Palmitoylation is among the most prevalent post-translational modifications found on the cytoplasmic face of transmembrane proteins. Various functions have been proposed for protein palmitoylation, although the precise mechanisms by which it works remain to be established (16–18). Palmitoylation controls the stability of several proteins, including CCR5, yeast SNAREs, the anthrax toxin receptor, and the neutral sphingomyelinase 2, by preventing their ubiquitination and thereby their targeting to lysosomal degradation (19–22). In hematopoietic cells and lymphocytes, palmitoylation regulates signal transduction by promoting the association of the signaling molecules with lipid microdomains at the plasma membrane and by regulating protein-protein interactions (23). The chemokine receptor CCR5 and Fas are receptors whose palmitoylation is required for the induction of efficient signaling (24, 25). Finally, palmitoylation is involved in various trafficking events, including export from the endoplasmic reticulum and the Golgi apparatus, and recycling to the plasma membrane (25–27).
We investigated whether palmitoylation contributes to IFNAR1 trafficking and IFN-α-induced signaling. We report that IFNAR1 is palmitoylated on cysteine 463, and although this modification has no major effect on IFNAR1 cellular trafficking, it strongly affects Jak/Stat signaling and the gene transcription induced by IFN-α.
EXPERIMENTAL PROCEDURES
IFNs and Antibodies
Recombinant human IFN-α2b (specific activity of 108 units/mg) from (Biosidus, Argentina) was kindly provided by J. Wietzerbin. Mouse anti-IFNAR1 mAb 34F10 and 64G12 and mouse anti-IFNAR2 mAbs 8F11 and 10E10 were described previously (13). Mouse anti-IFNAR1 mAb AA3 and EA12 were the kind gifts from Biogen Inc. (Boston). Rabbit anti-phospho-Stat1 (Tyr-701), anti-phospho-Tyk2 (Tyr-1054/1055), anti-phospho-Jak1 (Tyr-1022/1023), anti-Stat1, and anti-Tyk2 pAb were from Cell Signaling Technology. Rabbit anti-Stat2 and rabbit anti-phospho-Stat2 (Tyr-689) were from Upstate. Rabbit anti-Lamp2, goat anti-EEA1, goat anti-calnexin, and rabbit anti-Rab6 were from Santa Cruz Biotechnology. Biotinylated anti-phosphotyrosine (RC20) was from BD Biosciences. Secondary antibodies were goat Alexa 488-conjugated anti-mouse pAb, goat Cy3-conjugated anti-mouse pAb, goat Cy3-conjugated anti-rabbit pAb, donkey Cy3-conjugated anti-goat pAb, donkey horseradish peroxidase-conjugated anti-mouse pAb, and donkey horseradish peroxidase-conjugated anti-rabbit pAb (Jackson ImmunoResearch). Streptavidin-horseradish peroxidase was from Roche Applied Science.
Parental Plasmids and Mutagenesis
Wild-type human IFNAR1 was expressed in pEFIREShyg derived from pIREShyg as described previously (28). Mutagenesis of the IFNAR1 receptor chain was performed using the QuickChange site-directed mutagenesis kit (Stratagene, Amsterdam). pcDNA3.1(+)-IFNAR1-YFP was a kind gift from Dr. Jacob Piehler. It was modified by changing the cytomegalovirus promoter by an EF-1α promoter. pRC-CMV-Tyk2-VSV plasmid was a kind gift from Dr. Sandra Pellegrini. Renilla luciferase sequence was amplified from phRluc-C1 plasmid (Clontech) and was inserted in pRC-CMV-Tyk2-VSV plasmid in 5′ of the Tyk2 sequence by PCR. Stat2-GFP and IFNGR1-YFP plasmids were gifts from H. Hauser and J. L. Casanova, respectively. The accuracy of all cDNA was confirmed by DNA sequencing.
Cell Culture and Transfection
L929R2 murine fibroblasts stably expressing human IFNAR2 (28) were transfected by wild-type or mutated forms of human IFNAR1 using FuGENE 6 (Roche Applied Science). Mixed populations of transfected cells were selected under hygromycin selective pressure. The generated cell lines (L929R1R2) were cultured in Dulbecco's modified Eagle's essential medium supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% penicillin/streptomycin, 400 μg/ml hygromycin, and 1.5 mg/ml geneticin. CHO cells were cultured as above except for Dulbecco's modified Eagle's essential medium/F-12 growth medium and without selection antibiotics.
Drug Treatment
Palmitoylation was inhibited by incubating the cells with 200 μm 2-bromopalmitate for 1 h at 37 °C before starting the experiments. Chemical removal of palmitoylation was performed by treating cell extracts with 1 m hydroxylamine, pH 7, for 1 h at room temperature. Protein synthesis was inhibited by a 1-h treatment with 50 μm cycloheximide at 37 °C. All drugs were from Sigma.
Immunoprecipitations
40 × 106 cells were detached with phosphate-buffered saline/EDTA 2 mm and lysed 30 min in immunoprecipitation buffer (1% Triton X-100, 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, and a mixture of protease and phosphatase inhibitors (Sigma) with or without 10 mm N-ethylmaleimide). Cell lysates were centrifuged for 30 min at 14,000 × g, and supernatants were incubated overnight at 4 °C on protein G-Sepharose beads (Amersham Biosciences) with 2.5 μg of EA12 mAb for IFNAR1 or 2.5 μg of 8F11 mAb for IFNAR2. After washing the beads, samples were boiled for 5 min before being analyzed by Western blotting using 64G12 mAb for detecting IFNAR1 or 10E10 mAb for detecting IFNAR2.
Metabolic Labeling with [3H]Palmitate
Cells were first starved for 1 h in serum-free medium, then incubated for 4 h at 37 °C in Dulbecco's modified Eagle's essential medium containing 0.2% bovine serum albumin with 200 μCi/ml [9,10-3H]palmitic acid (American Radiolabeled Chemicals), washed, and immunoprecipitated for IFNAR1 or IFNAR2. After fixation (25% isopropyl alcohol, 65% H2O, and 10% acetic acid), gels were incubated for 30 min in enhanced Amplify NAMP100 (GE Healthcare), dried, and exposed for 3 weeks to Hyperfilm MP (GE Healthcare).
Immunofluorescence Microscopy
For analysis of IFNAR1 endocytosis, cells grown on coverslips were incubated on ice with the 34F10 antibody for 30 min. Cells were then incubated at 37 °C for 30 min, washed, fixed using 4% paraformaldehyde, and permeabilized with saponin treatment, and endocytosed antibody-IFNAR1 complexes were revealed with Cy3-conjugated anti-mouse antibody. Cells were imaged with an epifluorescent Leica microscope. For analysis of IFNAR1 intracellular co-localization experiments, cells were first fixed, then permeabilized, and incubated simultaneously with 34F10 and either anti-EEA1, anti-Rab6, anti-Rab11 or anti-Lamp2 antibodies as indicated. Secondary Alexa488-conjugated anti-mouse antibody was used to reveal IFNAR1, and Cy3-conjugated anti-goat antibody was used to reveal EEA1, and Cy3-conjugated anti-rabbit antibody was used to reveal Rab6, Rab11, and Lamp2. Cells were imaged with a confocal Leica microscope.
IFN-induced Activation of Tyk2, Jak1, and Stat
Cells were treated with or without 1000 units/ml IFN-α2b at 37 °C for the indicated times. For biochemical analysis, cells were washed with phosphate-buffered saline at 4 °C and lysed in Lysis Buffer (1% Triton X-100, 50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA, and a mixture of proteases and phosphatases inhibitors (Sigma)). After centrifugation for 10 min at 15,000 × g, lysates were resolved on SDS-PAGE and analyzed by Western blot/ECL for activated Tyk2 and Jak1 using anti-pTyk2 and anti-pJak1 or activated Stat1 and Stat2 using anti-pStat1 and anti-pStat2 antibodies. Anti-Jak1 and anti-Stat1 antibodies were used to determine the total amount of Jak1 and Stat1. For immunofluorescent analysis of pStat1 and pStat2 nuclear translocation, cells grown on coverslips were treated with IFN-α2b at 37 °C for the indicated time and fixed with cold methanol at −20 °C for 10 min. pStat1 and pStat2 were then stained by successive incubations with anti-phospho-Stat1, anti-phospho-Stat2, and Cy3-conjugated anti-rabbit antibodies.
Flow Cytometry
Cells were detached with phosphate-buffered saline/EDTA 2 mm and stained with AA3 antibody for 40 min in FACS buffer (phosphate-buffered saline supplemented with 3% fetal calf serum and 0.05% sodium azide) on ice. Goat anti-mouse Alexa 488-conjugated antibody (Jackson ImmunoResearch) was used as secondary antibody. Dead cells were excluded by gating on forward/side light scatter. Events corresponding to 2 × 104 gated cells were accumulated per sample. Flow cytometry was performed on a FACSCalibur machine, and data were analyzed by CellQuest software (BD Biosciences).
Luciferase Reporter Assay
L929R1R2 cells were transfected with an ISG54-luciferase construct kindly provided by S. Pellegrini using Lipofectamine 2000 (Invitrogen). After 48 h, cells were treated or not (control) with 1000 units/ml IFN-α2b for 8 h. Luciferase activity was quantified in cell lysates using a luminometer (Lumat LB9501, Berthold, Wildbald, Germany), and results were reported to the quantity of proteins as quantified with the Bradford method. Results were expressed as fold increase over the basal activity without IFN stimulation.
BRET Saturation Assays
5 × 106 CHO cells were transiently transfected with 0.1 μg of the DNA construct coding for BRET donor (RLuc-Tyk2) and increasing (0.05–1.5 μg) amounts of the BRET acceptor plasmid (IFNAR1 CC-YFP, IFNAR1 AC-YFP, or IFNGR1-YFP) and 0.1 μg of the DNA construct coding for IFNAR2 using GeneJuice Reagent (Novagen). A total amount of transfected DNA was maintained constant using an appropriate quantity of pcDNA3 (Invitrogen). 48 h after transfection, the luciferase substrate, coelenterazine h (Interchim), was added at a final concentration of 5 μm to 1 × 105 cells. Luminescence and fluorescence were measured simultaneously using the MithrasTM fluorescence-luminescence detector (Berthold). Cells expressing BRET donors alone were used to determine background. Filter sets were 485 ± 10 nm for luciferase emission and 530 ± 12.5 nm for YFP emission. BRET ratios were calculated as described previously (29).
RESULTS
IFNAR1 Endocytosis and IFN-α-induced Jak/Stat Signaling Depend on Palmitoylation Events
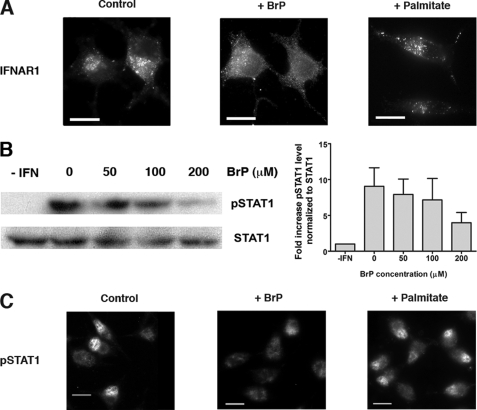
We recently showed that IFNAR1 uptake at the plasma membrane proceeds through classical clathrin- and dynamin-dependent endocytosis; inhibition of IFNAR1 endocytosis by either small interfering RNA-mediated knockdown of clathrin or inactivation of the GTPase dynamin by the dominant negative mutant K44A inhibits both activation of the Jak/Stat signaling pathway and the antiviral and antiproliferative activities otherwise promoted by IFN-α (14). Based on previous studies of the contribution of palmitoylation to the regulation of membrane trafficking (19, 30) and cell signaling pathways (23–25), we tested whether palmitoylation is involved in IFNAR1 endocytosis and IFN-α signaling. We first analyzed the effects of 2-bromopalmitate, a drug that blocks general protein palmitoylation (Fig. 1A) (31). IFNAR1 is rapidly endocytosed, and within 30 min is found in the recycling compartment as shown by co-localization with the small GTPAse Rab11 (data not shown) and as established previously (14). However, preincubation of cells with 200 μm 2-bromopalmitate strongly inhibited IFNAR1 endocytosis, and few IFNAR1 subunits were detected in the recycling compartment (Fig. 1A). The inhibition appears to be specific to 2-bromopalmitate because palmitate had no effect; this implicates palmitoylation in IFNAR1 endocytosis. We next tested the effect of 2-bromopalmitate on the Jak/Stat signaling pathway activated by IFN-α because we have shown this process to be dependent on IFNAR1 endocytosis (14). Upon IFN-α binding, the Tyk2 and Jak1 kinases associated with IFNAR are activated by tyrosine phosphorylation, resulting in the phosphorylation of tyrosine 701 of Stat1. In L929R1R2 cells, tyrosine phosphorylation of Stat1 (pStat1) was complete after 10 min of stimulation with 1000 units/ml IFN-α (Fig. 1B). Treatment with 2-bromopalmitate inhibited tyrosine phosphorylation of Stat1 in a dose-dependent manner with a maximum effect at 200 μm. We also analyzed later steps of the Jak/Stat signaling pathway by testing for the translocation of pStat1 in the nucleus (Fig. 1C). pStat1 accumulated in the nuclei of control or palmitate-treated cells after 30 min of IFN-α stimulation but not in the nuclei of cells treated with 200 μm 2-bromopalmitate (Fig. 1C). Thus, IFNAR1 endocytosis and IFN-α-dependent Jak/Stat signaling require protein palmitoylation.
FIGURE 1.
IFNAR1 endocytosis and IFN-α-induced Jak/Stat signaling depend on palmitoylation events. A, effect of the palmitoylation inhibitor 2-bromopalmitate (BrP)on IFNAR1 endocytosis. L929R1R2 cells, pretreated or not for 1 h with 200 μm 2-bromopalmitate or 200 μm palmitate, were incubated with anti-IFNAR1 34F10 mAb as described under “Experimental Procedures.” Cells were incubated at 37 °C for 30 min with 1000 units/ml IFN-α2b, and IFNAR1 endocytosis was detected by visualizing internalized IFNAR1-antibody complexes after an acid wash. Scale bar, 20 μm. Results are representative of at least four independent experiments. B, effect of 2-bromopalmitate on IFN-α-induced Stat1 tyrosine phosphorylation. L929R1R2 cells were pretreated for 1 h with the indicated concentrations of 2-bromopalmitate and stimulated with 1000 units/ml IFN-α for 10 min at 37 °C. Total lysates were analyzed by Western blot/ECL to detect Stat1 and phosphorylated Stat1 (pStat1). Quantification of pStat1 phosphorylation was made using ImageJ software, and the results correspond to the ratio between pStat1 amount and tubulin amount normalized to the control without 2-bromopalmitate. Results represent mean and S.D. of three independent experiments. C, effect of 2-bromopalmitate on IFN-α-induced pStat1 nuclear translocation. L929R1R2 cells were pretreated or not for 1 h with 200 μm 2-bromopalmitate or 200 μm palmitate and stimulated with 1000 units/ml IFN-α2b for 30 min at 37 °C. Cells were fixed, and the nuclear localization of phosphorylated Stat1 was detected by immunofluorescence. Results are representative of at least three independent experiments. Scale bar, 20 μm.
IFNAR1 and IFNAR2 Subunits of IFNAR Are Palmitoylated
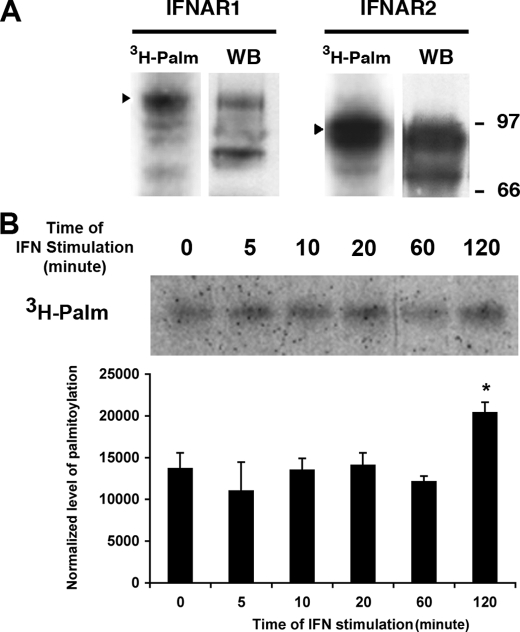
We analyzed whether the type I IFN receptor subunits IFNAR1 and IFNAR2 were themselves palmitoylated. IFNAR1 and IFNAR2 were immunoprecipitated from lysates of [3H]palmitic acid-labeled L929R1R2 cells. Radiolabeled bands corresponding to both IFNAR1 and IFNAR2 were detected (Fig. 2A), with a stronger signal for IFNAR2. The difference of band intensity was probably due to the higher level of IFNAR2 than IFNAR1 expression in this cell line, as shown on the corresponding whole cell extract (28). A similar experiment with a Jurkat cell line expressing a FLAG-tagged IFNAR1 showed that IFNAR1 was palmitoylated in this human lymphoid cell line also (data not shown). The two IFN-α receptor subunits are required for IFN-α activity; however, IFNAR1 is essential for IFN-α-induced signaling, and IFNAR2 is more involved in IFN-α binding; we therefore focused our analysis on IFNAR1 (32).
FIGURE 2.
IFNAR1 and IFNAR2 subunits of the IFN-α receptor are palmitoylated. A, L929R1R2 cells were incubated for 4 h with [3H]palmitate before immunoprecipitation of either IFNAR1 or IFNAR2 as indicated. Immunoprecipitates were analyzed by autoradiography (3H-Palm) or Western blot/ECL (WB) with specific antibodies to detect IFNAR1 and IFNAR2. B, L929R1R2 cells were pretreated or not with 1000 units/ml IFN-α for the indicated times 2 h after the addition of [3H]palmitate in the medium. After 4 h of incubation in [3H]palmitate, IFNAR1 was immunoprecipitated, and samples were analyzed by autoradiography. Quantification was made using ImageJ software, and means ± S.D. of three independent experiments are presented. Results correspond to the ratio between the amount of palmitoylation and IFNAR1 amount normalized to the control without IFN. A Student's t test was performed and revealed that the differences between the normalized level of palmitoylation are not significant except for t = 120 min, n = 3 (for t = 5 min, p = 0.3; for t = 10 min, p = 0.5; for t = 20 min, p = 0.4; for t = 60 min, p = 0.6; for t = 120 min, p < 0.01).
Several studies have reported an inducible cycle of palmitoylation and depalmitoylation. For example, stimulation of the β-adrenergic receptor by isoproterenol increases the binding of [3H]palmitic acid reflecting a higher turnover of bound palmitate, whereas it induces the depalmitoylation of the Gα subunits associated with the receptor (33, 34). The association of the CD19-CD21-CD81 complex with the activated B cell antigen receptor induces palmitoylation of CD81, thereby enhancing the stability of this association (35). Also, 17β-estradiol reduces the palmitoylation of the estradiol receptor in a time- and dose-dependent manner (36). This prompted us to investigate whether IFN treatment regulates the turnover of palmitate on IFNAR1. Stimulation of the cells with IFN-α from 5 min to 1 h prior to and during radiolabeling with [3H]palmitic acid had no effect on the quantity of palmitate bound to IFNAR1 (Fig. 2B); however, after 2 h of stimulation there was an increase of palmitate incorporation indicating that IFNAR1 palmitoylation turnover can be regulated by IFN-α stimulation.
IFNAR1 Is Palmitoylated on Cysteine 463
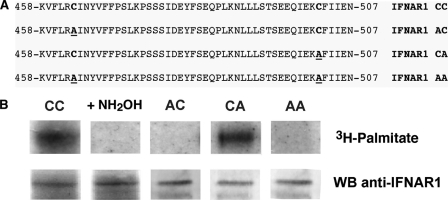
We found that IFNAR1 palmitoylation occurs on cysteine residues via a thioester bond because hydroxylamine, which cleaves the thioester bond, removed the [3H]palmitic acid incorporated in IFNAR1 (Fig. 3B). The cytoplasmic domain of IFNAR1 contains only two cysteines, at positions 463 and 502, that are likely to be palmitoylated. To determine which cysteines are palmitoylated, we mutated each of them to an alanine in single (AC and CA) and double (AA) mutants (Fig. 3A). We transfected L929R2 cells with either the wild-type form of human IFNAR1 or each of the three mutants. Mixed populations of transfected cells were generated, and mutant cell lines expressing levels of cell surface IFNAR1 and IFNAR2 similar to that in cells expressing the wild-type subunits, as determined by flow cytometry, were used for metabolic labeling with [3H]palmitic acid. The CA mutant incorporated as much palmitate as the wild-type CC (Fig. 3B); in contrast, the AC mutant, carrying the cysteine 463 to alanine mutation, did not show any radioactive signal. Similarly, the double mutant AA showed no incorporation of [3H]palmitic acid. Thus, IFNAR1 is palmitoylated on cysteine 463 but not on cysteine 502. This is in agreement with previous reports that transmembrane proteins are mostly palmitoylated on cysteine residues in close proximity to the plasma membrane and near stretches of hydrophobic acids (37), an environment corresponding to that of the cysteine 463.
FIGURE 3.
IFNAR1 is palmitoylated on cysteine 463. A, amino acid sequence of the 458–507 part of the carboxyl-terminal cytoplasmic tail of human IFNAR1. The two cytoplasmic cysteines in positions 463 and 502 are highlighted in boldface type. Cys-463 and Cys-502 were mutated into alanine individually or in combination, and the corresponding mutants are named AC, CA, and AA as indicated (mutations are underlined). B, L929R2 cells stably transfected with wild-type IFNAR1 (CC) or with the indicated mutants were incubated for 4 h with [3H]palmitate. IFNAR1 was immunoprecipitated from cell lysates, and immune complexes (treated or not with 1 m NH2OH for 1 h) were separated by SDS-PAGE. The IFNAR1 immunoprecipitate was split in two before being analyzed either by Western blotting (WB) (lower panel) or by autoradiography (upper panel).
IFNAR1 Palmitoylation Is Not Required for Receptor Endocytosis or Intracellular Distribution
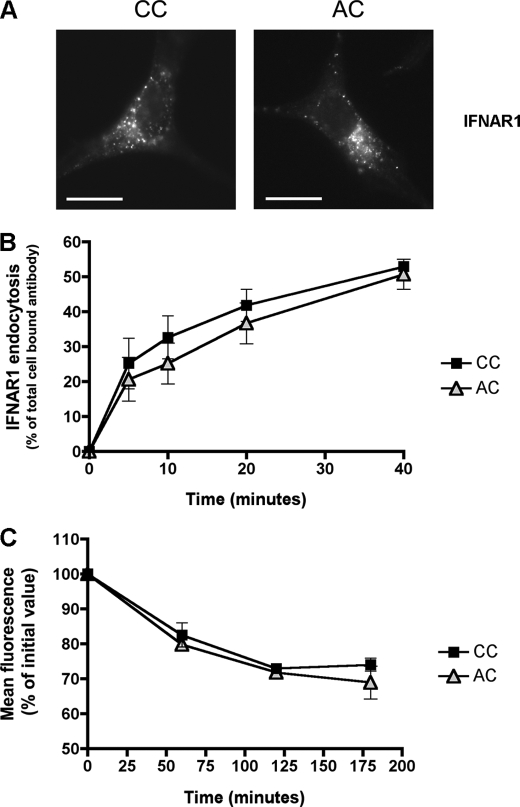
Next, we examined the effect of IFNAR1 palmitoylation on the functional properties of IFNAR. For several proteins, a lack of palmitoylation results in various trafficking defects, including reduced export from the Golgi apparatus, accumulation in lysosomes, and defective recycling to the plasma membrane (19, 20, 25, 27). Therefore, we investigated whether IFNAR1 palmitoylation regulates any of the steps of IFNAR1 intracellular trafficking. We first tested whether IFNAR1 palmitoylation was required for IFNAR1 uptake from the plasma membrane. Indeed, a lack of protein palmitoylation has been associated with abnormalities in endocytosis in a number of cases (19, 30). The endocytic characteristics of IFNAR1 were investigated as described previously (14). Cells were incubated at 4 °C with an anti-IFNAR1 antibody, and the uptake of the IFNAR1-antibody complexes was examined by immunofluorescence after 30 min of endocytosis at 37 °C. The patterns of IFNAR1 uptake of the wild-type (CC) and the nonpalmitoylated (AC) mutant were similar (Fig. 4A), suggesting that IFNAR1 endocytosis was not dependent on IFNAR1 palmitoylation. The endocytosis of the AC mutant was, however, inhibited by 2-bromopalmitate confirming that IFNAR1 trafficking depends on palmitoylation events other than IFNAR1 itself (data not shown). This was further confirmed by flow cytometry measurement of IFNAR1 uptake; the rate and extent of IFNAR1 endocytosis at early and later time points were identical for the wild-type and the AC mutant cell lines (Fig. 4B).
FIGURE 4.
IFNAR1 palmitoylation is not involved in IFNAR1 endocytosis and IFNAR1 half-life at the cell surface. A, endocytic pattern of the wild-type and AC IFNAR1 subunits. L929R2 cells stably transfected with wild-type IFNAR1 or the AC IFNAR1 mutant were processed for endocytosis as described in Fig. 1. Scale bar, 20 μm. B, FACS analysis of IFNAR1 endocytosis. L929R2 cells expressing either wild-type IFNAR1 or the IFNAR1 AC mutant were labeled with AA3 anti-IFNAR1 mAb prior endocytosis at 37 °C with 1000 units/ml IFN-α2b for the indicated times. Remaining cell surface IFNAR1-AA3 complexes were quantified by flow cytometry. Results are expressed as the disappearance of total cell-bound antibody from the cell surface. Each value is the mean ± S.D. of triplicate experiments. C, cell surface down-modulation of wild-type and AC IFNAR1 subunits. The cell surface expression of IFNAR1 was measured by flow cytometry in L929R2 cells expressing either wild-type or AC IFNAR1 subunits in the presence of cycloheximide and 1000 units/ml IFN-α2b for the indicated times.
There are reports that a loss of palmitoylation may favor ubiquitination resulting in increased lysosomal degradation. Tlg1 was the first described example of a yeast protein for which palmitoylation prevents interaction with the ubiquitin ligase Tul1, and thereby prevents its ubiquitination and targeting to the vacuole (21). Palmitoylation inhibits ubiquitination of the anthrax receptor by preventing its association with lipid rafts (19). Recently, Fuchs and co-workers (38, 39) have identified the SCFβTrcp as an E3 ubiquitin ligase that mediates IFNAR1 ubiquitination and degradation. We therefore investigated whether the palmitoylation-deficient IFNAR1 mutant would show increased degradation as a result of enhanced ubiquitination. We measured the half-life of IFNAR1 at the surface of cells stimulated with IFN-α in the presence of cycloheximide to prevent the neosynthesis of IFNAR1 receptors. The kinetics of disappearance of cell surface IFNAR1 subunits was measured by flow cytometry in both cell lines; after 180 min of IFN stimulation, there was a 30% decrease in the amount of IFNAR1 subunits present at the cell surface of both cell lines (one expressing the wild-type and the other the nonpalmitoylated AC mutant IFNAR1). Thus, IFNAR1 was not down-modulated further by the absence of IFNAR1 palmitoylation (Fig. 4C). These different experiments demonstrate that IFNAR1 palmitoylation is not required for IFNAR1 endocytosis, intracellular distribution, or stability at the plasma membrane.
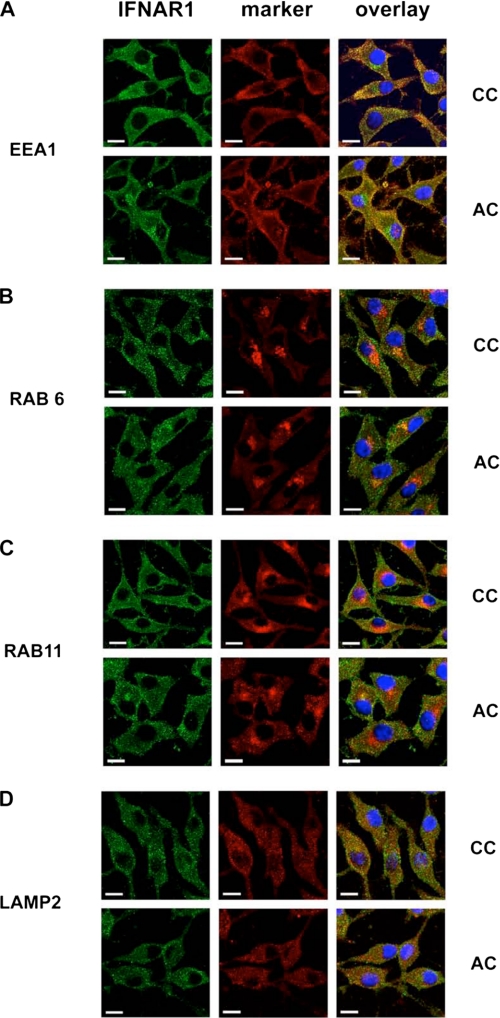
We next examined the intracellular distribution of wild-type and palmitoylation-deficient IFNAR1 by co-localization studies with markers of various intracellular compartments, including early endosomes (EEA1), the Golgi apparatus (Rab6), recycling endosomes (Rab11), and lysosomes (Lamp2). The wild-type and the AC mutant displayed the same pattern of intracellular distribution and both localized mostly in early endosomes at steady state (Fig. 5). The finding that the intracellular distribution of IFNAR1 was not modified by the absence of palmitoylation is in agreement with the lack of effect on both IFNAR1 endocytosis and stability at the plasma membrane, and it further confirms that IFNAR1 palmitoylation does not play a major role in the secretory or in the intracellular trafficking pathways followed by IFNAR1.
FIGURE 5.
Similar intracellular distribution of wild-type IFNAR1 and palmitoylation-deficient AC IFNAR1 mutant. L929R2 cells stably transfected with wild-type IFNAR1 or the AC mutant were fixed, permeabilized, and co-labeled with anti-IFNAR1 34F10 mAb and antibodies directed either against the early endosome marker EEA1 (A), the Golgi apparatus marker Rab6 (B), the recycling endosome marker Rab11 (C), or the lysosomal marker Lamp2 (D). Secondary antibodies coupled with Alexa 488 (green) and with Cy3 (red) were used to reveal IFNAR1 and cellular markers, respectively. 4′,6-Diamidino-2-phenylindole was added to detect nuclei. Cells were imaged with a confocal Leica microscope. Scale bar, 15 μm.
IFNAR1 Palmitoylation Is Required for the Efficient Activation and Nuclear Translocation of Stat2 and Stat1 and the Gene Transcription Induced by IFN-α
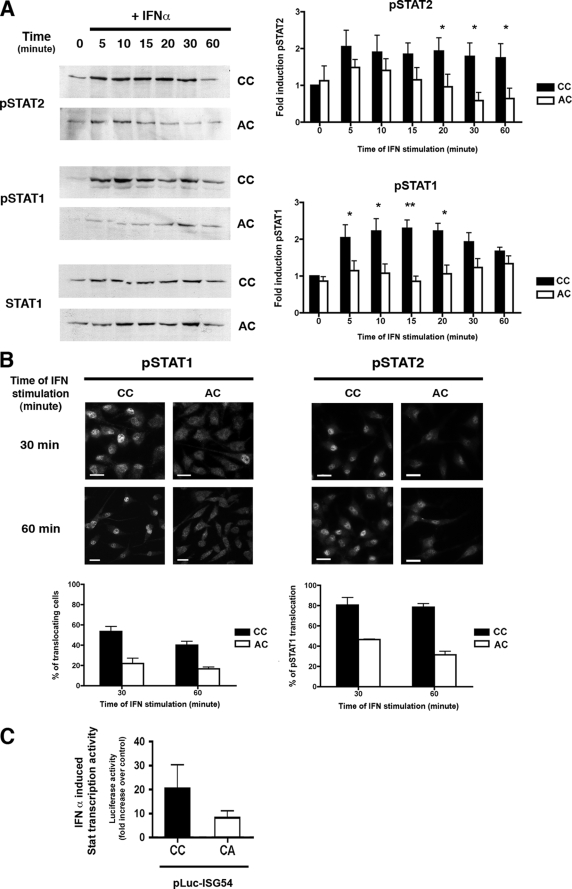
Our initial experiment with 2-bromopalmitate indicated that protein palmitoylation was required for IFN-α-induced Jak/Stat signaling, namely Stat activation and nuclear translocation. We tested whether palmitoylation of IFNAR1 itself was necessary for IFN-α-induced signaling. IFN-α binding to its receptor triggers the activation by tyrosine phosphorylation of the IFNAR1-bound Tyk2 and IFNAR2-bound Jak1 kinases. This activation leads to phosphorylation of tyrosine 466 on IFNAR1, which allows the indirect recruitment of Stat1 to IFNAR1 through binding to Stat2. On IFNAR1 recruitment, Stat1 and Stat2 become tyrosine-phosphorylated on tyrosines 701 and 690, respectively. In cells expressing wild-type IFNAR1, Stat1 and Stat2 tyrosine phosphorylation was detected within 5 min of IFN-α stimulation, was maximal after 10 min, continued for up to 30 min, and decreased thereafter (Fig. 6A). In contrast, there was a strong decrease of Stat2 activation in cells expressing the AC mutant. In these cells, the effect was even more pronounced on Stat1 with no detectable tyrosine phosphorylation of Stat1 along the time course of IFN-α stimulation. Later steps of the Jak/Stat signaling pathway were also investigated with the translocation of activated Stat1 and Stat2 to the nucleus. Cells expressing wild-type IFNAR1 showed a substantial, albeit heterogeneous, translocation of pStat1 to the nucleus, whereas the amount of pStat1 translocation in cells expressing the AC IFNAR1 mutant was about 60% lower (Fig. 6B). Similar results were observed for the nuclear translocation of activated Stat2. Importantly, Stat1 and Stat2 nuclear translocation was still strongly impaired at 60 min indicating that the lack of IFNAR1 palmitoylation prevents rather than delays Stat nuclear translocation.
FIGURE 6.
IFNAR1 palmitoylation is required for the activation of the Stat signaling pathway by IFN-α. A, IFN-α-induced Stat1 and Stat2 tyrosine phosphorylation in L929R2 cells expressing either the wild-type or the AC IFNAR1 subunits. Cells were stimulated or not with 1000 units/ml IFN-α for the indicated times. Cell lysates were analyzed by Western blotting against tyrosine-phosphorylated Stat1 (pStat1) or tyrosine-phosphorylated Stat2 (pStat2) or Stat1 as indicated. The figure shown is representative of three independent experiments. Quantification was made using ImageJ software, and mean ± S.D. of three independent experiments are presented. Results correspond to the ratio between the amount of pStat1 or pStat2 and Stat1 amount normalized to the control without IFN. Significant differences were evaluated by using a Student's t test: *, p < 0.05; **, p < 0.01. B, IFN-α-induced pStat1 and pStat2 nuclear translocation in L929R2 cells expressing either the wild-type or the AC IFNAR1 subunits. Upper part, pStat1 and pStat2 nuclear translocation was analyzed by immunofluorescence as described under “Experimental Procedures” after 30 or 60 min of treatment with 1000 units/ml IFN-α (scale bar, 20 μm). Lower part, the percentage of cells with pStat1 translocation was quantified by counting (n = 100). Results are the mean ± S.D. of three independent experiments. C, IFN-α induced gene transcription is strongly reduced in the palmitoylation-deficient IFNAR1 AC mutant. IFN-α-induced gene transcription was analyzed using the luciferase reporter gene. L929R2 cells stably expressing either the wild-type or the AC IFNAR1 subunits were transfected by the ISG-54 luciferase construct and stimulated with 1000 units/ml IFN-α, and luciferase activity was quantified by measurement of luminescence reported to the quantity of proteins, as described under “Experimental Procedures.” Results are the mean ± S.D. of three independent experiments and are expressed as fold increase of normalized luciferase activity over the basal luciferase activity without IFN-α stimulation.
Finally, we analyzed whether the Jak/Stat signaling defects found in the AC IFNAR1 mutant affect the transcriptional activity induced by IFN-α. The transcription of type I interferon-stimulated genes depends on the binding of the IFN-stimulated gene factor 3 complex to ISRE. We analyzed the transcription of ISRE-containing genes by transfecting cells expressing wild-type or palmitoylation-deficient IFNAR1 with an ISRE-luciferase reporter construct, and we measured the luciferase activity 8 h after IFN-α stimulation (14). The transcriptional activity induced by IFN-α in cells expressing the AC IFNAR1 mutant was more than 60% lower than that in the wild-type controls (Fig. 6C). These findings are consistent with those for Stat1/Stat2 activation and nuclear translocation, and they confirm the major role of IFNAR1 palmitoylation in IFN-α signaling.
IFN-α-induced Signaling Events Upstream of Stat2 and Stat1 Activation Are Not Affected by the Lack of IFNAR1 Palmitoylation
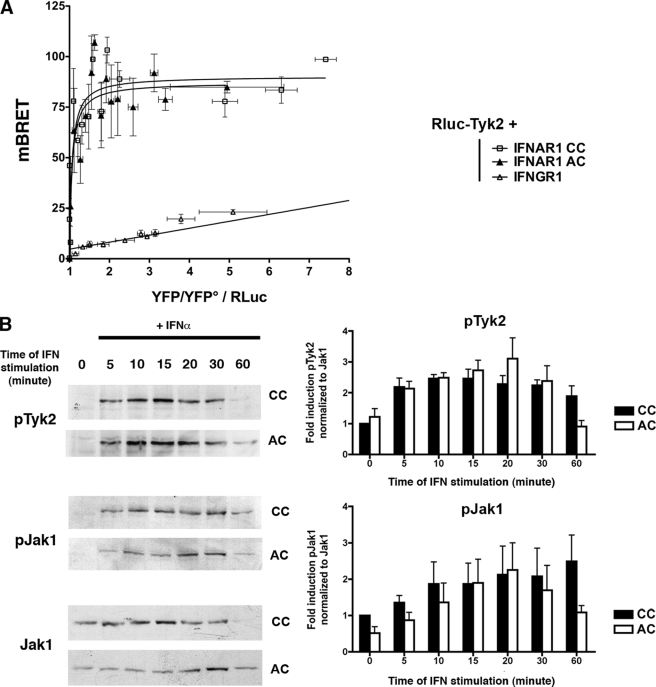
We next sought the precise step where IFNAR1 palmitoylation was required for bona fide IFN-α signaling. Because phosphorylation of tyrosine 466 on IFNAR1 appears to be a critical event for Stat1 and Stat2 activation, we searched for potential defects in the activity of the IFNAR1-associated tyrosine kinases. The tyrosine kinase Tyk2 is stably associated with IFNAR1 independently from IFN-α and becomes rapidly tyrosine-phosphorylated upon IFN-α treatment (7, 40). Interestingly, it has been shown that the Tyk2-binding site spans residues 465–511 of the IFNAR1 cytoplasmic domain (40). We therefore tested whether the palmitoylation of cysteine 463, which is only two amino acids away from the Tyk2-binding site, could affect the association of Tyk2 with IFNAR1 and thereby the tyrosine phosphorylation of Stat1 and Stat2. Because of the poor sensitivity of immunoprecipitation experiments, we set up a BRET assay to measure the interaction between Tyk2 and IFNAR1 (41). Fig. 7A shows saturation experiments conducted in cells expressing the Tyk2 kinase fused to Renilla luciferase (RLuc-Tyk2) as a BRET donor, and increasing concentrations of both IFNAR1 CC and IFNAR1 AC fused to the yellow variant of enhanced green fluorescent protein (YFP) as the BRET acceptors. BRET measurements displayed hyperbolic curves characterized by similar BRET50 (1.059 ± 0.037 and 1.061 ± 0.028) and BRETmax values (90.23 ± 7.87 and 87.18 ± 4.9) for CC and AC IFNAR1, respectively. These data are consistent with a similar propensity of Tyk2 to constitutively interact with IFNAR1 whether it is palmitoylated or not on cysteine 463. To validate this interaction, control experiments were conducted with the IFNGR1 subunit of the IFN-γ receptor, which does not interact with Tyk2 (42). When IFNGR1-YFP was used instead of IFNAR1-YFP as a BRET acceptor, a nonspecific bystander linear plot was obtained confirming the lack of interaction between Tyk2 and IFNGR1.
FIGURE 7.
IFNAR1 palmitoylation does not affect Tyk2 association to IFNAR1 or Jak kinases activation. A, Tyk2 is constitutively associated with IFNAR1 independently of its palmitoylation status. BRET saturation curves were obtained by measuring BRET in CHO cells expressing fixed quantities of BRET donor (Rluc-Tyk2) and increasing amounts of BRET acceptors (carboxyl-terminally YFP-tagged IFNAR1 CC, IFNAR1 AC, or IFNGR1). Relative amounts of BRET acceptor are expressed as the ratio between the fluorescence of the acceptor over the luciferase activity of the donor. YFP° corresponds to background fluorescence in cells expressing the BRET donor alone. BRET ratio values were from 26 individual transfections and were grouped as a function of the amount of BRET acceptor (mean ± S.D.; n = 4). B, Jak kinases are still fully activated even in absence of IFNAR1 palmitoylation. L929R2 cells stably transfected with wild-type IFNAR1 or the AC mutant were stimulated or not with 1000 units/ml IFN-α for the indicated times. Cell lysates were analyzed by Western blotting against tyrosine-phosphorylated Tyk2 (pTyk2), tyrosine-phosphorylated Jak1 (pJak1), and Jak1 as indicated. The figure shown is representative of three independent experiments. Quantification was made using ImageJ software, and mean ± S.D. of at least three independent experiments are presented. Results correspond to the ratio between the amount of pTyk2 or pJak1 and Jak1 amount normalized to the control without IFN. Significant differences were evaluated by using a Student's t test.
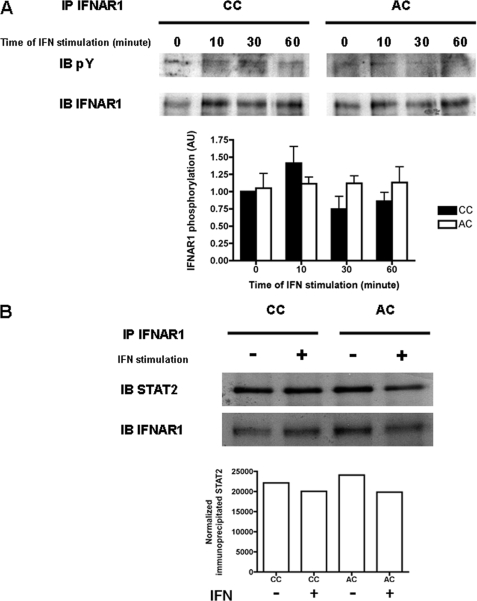
It has been proposed that the heterodimerization of the IFNAR1/IFNAR2 subunits induced by IFN-α leads to the activation by auto- and trans-phosphorylation of Jak1 and Tyk2 (42). We next examined the activation of the Tyk2 kinase itself upon IFN-α stimulation. Tyk2 tyrosine phosphorylation (pTyk2) was detected within 5 min of IFN-α stimulation, was maximal after 10 min, and decreased thereafter (Fig. 7B). No significant difference could be observed for the pattern of Tyk2 activation in cells expressing either IFNAR1 CC or IFNAR1 AC in agreement with the BRET results above. We also examined the activation of the Jak1 tyrosine kinase, which is selectively associated with the IFNAR2 subunit of the IFN-α receptor. Again, the pattern of Jak1 tyrosine phosphorylation did not show any major difference between wild-type and palmitoylation-deficient IFNAR1 (Fig. 7B). The finding that Tyk2 and Jak1 activation was not altered strongly suggests that IFNAR1 palmitoylation does not play a role in the heterodimerization of IFNAR1 and IFNAR2 subunits. In agreement with these data, the pattern of IFNAR1 tyrosine phosphorylation was similar in IFNAR1 CC and IFNAR1 AC expressing cells (Fig. 8A). Among the tyrosine residues that are phosphorylated by IFN-α, tyrosine 466 appears critical because it allows the binding of the SH2 domain of Stat2 in vitro (4, 8, 49). Even though IFNAR1 was properly tyrosine-phosphorylated, the proximity of cysteine 463 with tyrosine 466 led us to test the hypothesis that the lack of cysteine 463 palmitoylation could interfere with Stat2 recruitment to IFNAR1, and thereby decrease its level of activation by Tyk2. Therefore, we sought to detect the amount of Stat2 that was present on IFNAR1 after IFN-α stimulation. The weak and transient nature of this interaction has made this analysis difficult in vivo in cells expressing endogenous levels of Stat2 and IFNAR1. Several groups have tried to circumvent this limitation by overexpression or by in vitro approaches; however, contradictory results have been obtained (4–9). Unfortunately, our attempts to measure this interaction by BRET failed, and we could detect Stat2 bound to IFNAR1 only in transiently transfected cells (Fig. 8B). In agreement with published experiments (7), we found that Stat2 was bound to IFNAR1 independently from IFN-α stimulation. The lack of IFNAR1 palmitoylation, however, did not interfere with the measurable amount of Stat2 recruited to IFNAR1 under our experimental conditions. Altogether, these data indicate that IFNAR1 palmitoylation is specifically required for the efficient tyrosine phosphorylation of Stat2 and Stat1 and that none of the molecular events preceding this single signaling step are affected.
FIGURE 8.
IFNAR1 phosphorylation and Stat2 docking to IFNAR1 are not affected by the palmitoylation of the receptor. A, IFNAR1 total phosphorylation is not influenced by the level of IFNAR1 palmitoylation. L929R2 cells stably transfected with wild-type IFNAR1 or the AC mutant were stimulated or not with 1000 units/ml IFN-α for the indicated times. IFNAR1 was immunoprecipitated (IP) as described under “Experimental Procedures,” and phosphorylation levels were analyzed by Western blotting against phosphorylated tyrosine (pY) or the IFNAR1 receptor as indicated. Quantification was made using ImageJ software, and means ± S.D. of three independent experiments are presented. Results correspond to the ratio between the amount of phosphorylated tyrosine and IFNAR1 amount normalized to the control without IFN. IB, immunoblot. B, Stat2 is associated with IFNAR1 CC and IFNAR1 AC receptors. CHO cells were transiently transfected with IFNAR1 CC or AC, IFNAR2, and Stat2-GFP constructs. 48 h after transfection, cells were stimulated or not with 1000 units/ml IFN-α for 10 min. IFNAR1 was immunoprecipitated as described under “Experimental Procedures,” and Stat2 association was analyzed by Western blotting against Stat2 or IFNAR1 as indicated. Quantification was made using ImageJ software. Results correspond to the ratio between the amount of Stat2 and IFNAR1 amount normalized to the control without IFN and are representative of three independent experiments.
DISCUSSION
Our study demonstrates that the IFN-α receptor subunits IFNAR1 and IFNAR2 are palmitoylated. Cysteine 463, the more proximal of the two cysteines present in the cytoplasmic domain of IFNAR1, is palmitoylated, which is consistent with the preferential linkage of palmitate to cysteines that are next to the cytoplasmic end of the transmembrane domain, which in the case of IFNAR1 encompasses the amino acids 437–457. Accordingly, a palmitoylation site prediction program computing several palmitoylation sites gave a higher probability for the palmitoylation of cysteine 463 than that of cysteine 502 (43).
Because IFNAR1 is essential in IFN-α signaling, we investigated the effects of IFNAR1 palmitoylation in the functions of the IFN-α receptor by examining several established consequences of this common post-translational lipid modification of proteins. Protein palmitoylation has effects on a broad range of activities that include protein trafficking, membrane association, protein stability, protein aggregation into lipid microdomains, and regulation of signaling (44). We already showed that upon IFN-α binding, IFNAR1 subunits do not cluster within lipid microdomains at the plasma membrane, ruling out a role for IFNAR1 palmitoylation in this process (14).
IFNAR1 endocytosis was strongly inhibited by the treatment with 2-bromopalmitate (a general inhibitor of palmitoylation), but the palmitoylation-deficient IFNAR1 mutant did not show any obvious abnormality of endocytosis or intracellular distribution. However, 2-bromopalmitate inhibited the endocytosis of the palmitoylation-deficient IFNAR1 mutant indicating that palmitoylation of proteins other than the receptor itself is involved in IFNAR1 trafficking. It is interesting to consider the potential role of the second IFN-α receptor subunit, IFNAR2, that we also show to be palmitoylated. IFNAR2 is the subunit that presents the highest affinity for IFN-α, and the current model is that the primary interaction at the cell surface is between IFN-α and IFNAR2, with the IFNAR2-bound IFN-α complex interacting with IFNAR1 being a subsequent event (45, 46). However, another study suggests that IFNAR1 and IFNAR2 can also be found pre-associated at the plasma membrane prior to IFN binding (28), a model also described for the two IFN-γ receptor chains (47). In any case, it is possible that nonpalmitoylated IFNAR1 displays normal trafficking properties as a result of its association with IFNAR2. Thus, IFNAR2, by associating with IFNAR1, may drive IFNAR1 uptake and control its intracellular distribution, explaining the differences observed between 2-bromopalmitate treatment and the palmitoylation-deficient IFNAR1 mutant. The IFNAR2 subunit bears two cysteines that are likely to be palmitoylated, and their mutation will clarify this point.
The precise mechanisms by which palmitoylation affects the behavior of transmembrane proteins is currently not predictable (16). These proteins are already strongly embedded in the membrane through their transmembrane domains, and it seems unlikely that palmitoylation of a single cysteine residue is required to anchor them. It is therefore plausible that palmitoylation is not required for IFNAR1 trafficking as has been recently shown for other transmembrane protein; Snc1 and Syn8 are two examples of SNARE proteins that are palmitoylated in yeast, and nonpalmitoylated mutants of these proteins are sorted normally (21). It is interesting that, like IFNAR1, Snc1 and Syn8 are palmitoylated on a single cysteine close to the transmembrane domain. MUC1 is another example of a transmembrane protein whose palmitoylation is not required for its endocytosis and its stability at the cell surface (27).
Protein palmitoylation may be an important device for regulating signaling in a versatile manner because it is the only lipid modification of proteins that can be either transient or permanent. In agreement with the effect of 2-bromopalmitate, we found that the absence of palmitoylation on IFNAR1 cysteine 463 had a major effect on the activation of Stat1 and Stat2 by IFN-α and on downstream signaling events. Thus, the nuclear translocation of Stat1 and Stat2 and the transcriptional activity after IFN-α stimulation, in cells expressing the nonpalmitoylated form of IFNAR1, were all substantially lower than the wild type. Another means by which palmitoylation may control signaling, other than affecting membrane trafficking, is through the regulation of lipid-protein or protein-protein interactions, in the same way as other reversible post-translational modifications such as tyrosine phosphorylation and GTP/GDP cycling. It is therefore possible that IFNAR1 palmitoylation contributes to the recruitment of signaling proteins to the IFNAR1 subunit and stabilization of the association. The presence of palmitate may affect the spatial conformation of IFNAR1 in the vicinity of the transmembrane domain, thereby reversibly regulating the association with downstream effectors. Our finding that the tyrosine phosphorylation of Stat1 was rapidly and strongly inhibited in cells expressing the palmitoylation-deficient IFNAR1 mutant (Fig. 8A) led us to investigate the impact of cysteine 463 palmitoylation on the molecular events involved in this process. A combination of in vitro and in vivo experiments clearly established the key role that Stat2 plays in the activation of Stat1 (4, 5, 7–9). Indeed, cytosolic Stat2 binds first to IFNAR and becomes tyrosine-phosphorylated by the Jak kinases associated with the receptor. The phosphorylation of the tyrosine 690 on Stat2 serves as a docking site for the SH2 domain of Stat1, which in turn becomes phosphorylated on tyrosine 701 upon IFNAR1 recruitment. Although there is debate on which IFNAR subunit recruits Stat2, it appears that tyrosine phosphorylation of Stat2 is a prerequisite for both Stat1 receptor binding and activation. Our results clearly indicating that IFNAR1 palmitoylation is required for the efficient tyrosine phosphorylation of Stat2, and we focused our analysis on the mechanisms of Stat2 activation. We first tested the hypothesis of a defect in kinase activation. Tyk2 was a likely candidate because it is constitutively associated with IFNAR1 on a stretch of amino acids close to the palmitoylated cysteine (40), and it has been recently shown that the accessibility of Tyk2 to IFNAR1 depends on post-translational modifications of IFNAR1 (48). We probed this interaction using BRET, which is more sensitive than classical immunoprecipitation assays. Our results, using BRET, measure for the first time a strong interaction between Tyk2 and IFNAR1, confirming previous data obtained in vitro (40). However, this interaction was not altered by the lack of cysteine 463 palmitoylation. In agreement with these data, the pattern of Tyk2 activation was similar in cells expressing control and palmitoylation-deficient IFNAR1 subunits. We also examined the activity of the IFNAR2-associated Jak1 kinase and found no difference. Finally, under our experimental conditions, the absence of palmitoylated cysteine 463 did not alter the measurable amount of Stat2 recruited to IFNAR1. In conclusion, our results are consistent with the idea that IFNAR1 palmitoylation is exclusively required for the efficient tyrosine phosphorylation of Stat2 upon IFNAR1 recruitment. As a result, Stat1 is not fully recruited to Stat2 and cannot be tyrosine-phosphorylated as demonstrated by the strong decrease of Stat1 tyrosine phosphorylation in these cells. Although Tyk2 appears to be active and correctly bound to IFNAR1, it is still possible that the presence of palmitate on cysteine 463 confers a structural conformation to IFNAR1 that is required for the efficient phosphorylation of Stat2, in agreement with the established role of palmitoylation in protein-protein interaction. As mentioned above, it is also possible that the overexpression approach failed to detect a decrease of endogenous Stat2 binding to IFNAR1. Alternatively, it is possible that the conformational change induced by the lack of palmitoylation still allows the recruitment of Stat2 but prevents its efficient phosphorylation by Tyk2. Further studies, including crystallographic analyses, will be required to discriminate between these different possibilities. This aspect of signaling control by palmitoylation-induced conformational change was described for the interaction between the kinase Fyn and the T cell receptor (50). Similarly, palmitoylation of one critical cysteine of the prostanoid thromboxane A2 receptor is specifically required for efficient coupling of the phospholipase Cβ to the Gq subunit (51).
In this study, we have demonstrated that palmitoylation of a single cysteine (cysteine 463) adjacent to the transmembrane domain of the IFN-α receptor subunit IFNAR1 is critical for full extent of activation of the Jak/Stat signaling pathway by type I IFNs. This is consistent with the essential role of Stat2 and IFNAR1 in IFN-α-induced signaling. We have shown that IFNAR endocytosis was essential for the activation of the Jak/Stat signaling cascade and the antiviral and antiproliferative effects of IFN-α (14). Ubiquitination of IFNAR1 is important for the regulation of IFNAR1 internalization and degradation (38, 52), and several studies have documented a relationship between ubiquitination and palmitoylation of transmembrane proteins (44). Thus, it appears that IFNAR1 palmitoylation and ubiquitination may contribute, through the control of downstream Jak/Stat signaling and IFNAR trafficking, respectively, to orchestrating the complexity and selectivity of the signaling and pleiotropic cellular effects of IFNs. This novel aspect of the regulation of the Jak/Stat signaling pathway may have important therapeutic implications because dysfunctions of this pathway have been reported in several cancers (53). It would therefore be potentially valuable to test whether the recently developed inhibitors of human palmitoylacyltransferases and ubiquitin ligases (54, 55) can modulate the antiviral, immune, and antitumoral activities of IFNs.
Acknowledgments
We thank Florence Niedergang for helpful discussions and Winfried Römer for help with the confocal microscope and reading of the manuscript. We are grateful to Stefano Marullo for free access to the BRET station in the laboratory.
This work was supported in part by Association pour la Recherche sur le Cancer Grant 3143 and Agence Nationale de la Recherche Grant BLAN0211.
- IFN
- interferon
- BRET
- bioluminescence resonance energy transfer
- ISG
- IFN-stimulated gene
- ISRE
- IFN-stimulated response element
- mAb
- monoclonal antibody
- pAb
- polyclonal antibody
- CHO
- Chinese hamster ovary
- SNARE
- soluble NSF attachment protein receptor
- SH2
- Src homology 2
- YFP
- yellow fluorescent protein
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 2.Kalvakolanu D. V. (2003) Pharmacol. Ther. 100, 1–29 [DOI] [PubMed] [Google Scholar]
- 3.Parmar S., Platanias L. C. (2003) Curr. Opin. Oncol. 15, 431–439 [DOI] [PubMed] [Google Scholar]
- 4.Li X., Leung S., Kerr I. M., Stark G. R. (1997) Mol. Cell. Biol. 17, 2048–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadeau O. W., Domanski P., Usacheva A., Uddin S., Platanias L. C., Pitha P., Raz R., Levy D., Majchrzak B., Fish E., Colamonici O. R. (1999) J. Biol. Chem. 274, 4045–4052 [DOI] [PubMed] [Google Scholar]
- 6.Nguyen V. P., Saleh A. Z., Arch A. E., Yan H., Piazza F., Kim J., Krolewski J. J. (2002) J. Biol. Chem. 277, 9713–9721 [DOI] [PubMed] [Google Scholar]
- 7.Abramovich C., Shulman L. M., Ratovitski E., Harroch S., Tovey M., Eid P., Revel M. (1994) EMBO J. 13, 5871–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan K., Singh B., Krolewski J. J. (1998) J. Biol. Chem. 273, 19495–19501 [DOI] [PubMed] [Google Scholar]
- 9.Uddin S., Chamdin A., Platanias L. C. (1995) J. Biol. Chem. 270, 24627–24630 [DOI] [PubMed] [Google Scholar]
- 10.Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. (1998) Annu. Rev. Biochem. 67, 227–264 [DOI] [PubMed] [Google Scholar]
- 11.de Weerd N. A., Samarajiwa S. A., Hertzog P. J. (2007) J. Biol. Chem. 282, 20053–20057 [DOI] [PubMed] [Google Scholar]
- 12.van Boxel-Dezaire A. H., Rani M. R., Stark G. R. (2006) Immunity 25, 361–372 [DOI] [PubMed] [Google Scholar]
- 13.Baychelier F., Nardeux P. C., Cajean-Feroldi C., Ermonval M., Guymarho J., Tovey M. G., Eid P. (2007) Cell. Signal. 19, 2080–2087 [DOI] [PubMed] [Google Scholar]
- 14.Marchetti M., Monier M. N., Fradagrada A., Mitchell K., Baychelier F., Eid P., Johannes L., Lamaze C. (2006) Mol. Biol. Cell 17, 2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claudinon J., Monier M. N., Lamaze C. (2007) Biochimie 89, 735–743 [DOI] [PubMed] [Google Scholar]
- 16.Charollais J., Van Der Goot F. G. (2009) Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 17.Nadolski M. J., Linder M. E. (2007) FEBS J. 274, 5202–5210 [DOI] [PubMed] [Google Scholar]
- 18.Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 19.Abrami L., Leppla S. H., van der Goot F. G. (2006) J. Cell Biol. 172, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tani M., Hannun Y. A. (2007) J. Biol. Chem. 282, 10047–10056 [DOI] [PubMed] [Google Scholar]
- 21.Valdez-Taubas J., Pelham H. (2005) EMBO J. 24, 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Percherancier Y., Planchenault T., Valenzuela-Fernandez A., Virelizier J. L., Arenzana-Seisdedos F., Bachelerie F. (2001) J. Biol. Chem. 276, 31936–31944 [DOI] [PubMed] [Google Scholar]
- 23.Flaumenhaft R., Sim D. S. (2005) Hematology 10, 511–519 [DOI] [PubMed] [Google Scholar]
- 24.Chakrabandhu K., Hérincs Z., Huault S., Dost B., Peng L., Conchonaud F., Marguet D., He H. T., Hueber A. O. (2007) EMBO J. 26, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanpain C., Wittamer V., Vanderwinden J. M., Boom A., Renneboog B., Lee B., Le Poul E., El Asmar L., Govaerts C., Vassart G., Doms R. W., Parmentier M. (2001) J. Biol. Chem. 276, 23795–23804 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi T., Rumbaugh G., Huganir R. L. (2005) Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 27.Kinlough C. L., McMahan R. J., Poland P. A., Bruns J. B., Harkleroad K. L., Stremple R. J., Kashlan O. B., Weixel K. M., Weisz O. A., Hughey R. P. (2006) J. Biol. Chem. 281, 12112–12122 [DOI] [PubMed] [Google Scholar]
- 28.Cajean-Feroldi C., Nosal F., Nardeux P. C., Gallet X., Guymarho J., Baychelier F., Sempé P., Tovey M. G., Escary J. L., Eid P. (2004) Biochemistry 43, 12498–12512 [DOI] [PubMed] [Google Scholar]
- 29.Boularan C., Scott M. G., Bourougaa K., Bellal M., Esteve E., Thuret A., Benmerah A., Tramier M., Coppey-Moisan M., Labbé-Jullié C., Fåhraeus R., Marullo S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18061–18066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez E., Gironès N., Davis R. J. (1990) J. Biol. Chem. 265, 16644–16655 [PubMed] [Google Scholar]
- 31.Webb Y., Hermida-Matsumoto L., Resh M. D. (2000) J. Biol. Chem. 275, 261–270 [DOI] [PubMed] [Google Scholar]
- 32.Pestka S., Krause C. D., Walter M. R. (2004) Immunol. Rev. 202, 8–32 [DOI] [PubMed] [Google Scholar]
- 33.Mumby S. M., Kleuss C., Gilman A. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2800–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouillac B., Caron M., Bonin H., Dennis M., Bouvier M. (1992) J. Biol. Chem. 267, 21733–21737 [PubMed] [Google Scholar]
- 35.Cherukuri A., Carter R. H., Brooks S., Bornmann W., Finn R., Dowd C. S., Pierce S. K. (2004) J. Biol. Chem. 279, 31973–31982 [DOI] [PubMed] [Google Scholar]
- 36.Acconcia F., Ascenzi P., Bocedi A., Spisni E., Tomasi V., Trentalance A., Visca P., Marino M. (2005) Mol. Biol. Cell 16, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bijlmakers M. J., Marsh M. (2003) Trends Cell Biol. 13, 32–42 [DOI] [PubMed] [Google Scholar]
- 38.Kumar K. G., Barriere H., Carbone C. J., Liu J., Swaminathan G., Xu P., Li Y., Baker D. P., Peng J., Lukacs G. L., Fuchs S. Y. (2007) J. Cell Biol. 179, 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar K. G., Tang W., Ravindranath A. K., Clark W. A., Croze E., Fuchs S. Y. (2003) EMBO J. 22, 5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colamonici O. R., Uyttendaele H., Domanski P., Yan H., Krolewski J. J. (1994) J. Biol. Chem. 269, 3518–3522 [PubMed] [Google Scholar]
- 41.Boute N., Jockers R., Issad T. (2002) Trends Pharmacol. Sci. 23, 351–354 [DOI] [PubMed] [Google Scholar]
- 42.Müller M., Briscoe J., Laxton C., Guschin D., Ziemiecki A., Silvennoinen O., Harpur A. G., Barbieri G., Witthuhn B. A., Schindler C., et al. (1993) Nature 366, 129–135 [DOI] [PubMed] [Google Scholar]
- 43.Zhou F., Xue Y., Yao X., Xu Y. (2006) Bioinformatics 22, 894–896 [DOI] [PubMed] [Google Scholar]
- 44.Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 45.Lamken P., Gavutis M., Peters I., Van der Heyden J., Uzé G., Piehler J. (2005) J. Mol. Biol. 350, 476–488 [DOI] [PubMed] [Google Scholar]
- 46.Kumaran J., Wei L., Kotra L. P., Fish E. N. (2007) FASEB J. 21, 3288–3296 [DOI] [PubMed] [Google Scholar]
- 47.Krause C. D., Mei E., Xie J., Jia Y., Bopp M. A., Hochstrasser R. M., Pestka S. (2002) Mol. Cell. Proteomics 1, 805–815 [DOI] [PubMed] [Google Scholar]
- 48.Kumar K. G., Varghese B., Banerjee A., Baker D. P., Constantinescu S. N., Pellegrini S., Fuchs S. Y. (2008) J. Biol. Chem. 283, 18566–18572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H., Krishnan K., Greenlund A. C., Gupta S., Lim J. T., Schreiber R. D., Schindler C. W., Krolewski J. J. (1996) EMBO J. 15, 1064–1074 [PMC free article] [PubMed] [Google Scholar]
- 50.van't Hof W., Resh M. D. (1999) J. Cell Biol. 145, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid H. M., Kinsella B. T. (2007) Cell. Signal. 19, 1056–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar K. G., Krolewski J. J., Fuchs S. Y. (2004) J. Biol. Chem. 279, 46614–46620 [DOI] [PubMed] [Google Scholar]
- 53.Calò V., Migliavacca M., Bazan V., Macaluso M., Buscemi M., Gebbia N., Russo A. (2003) J. Cell. Physiol. 197, 157–168 [DOI] [PubMed] [Google Scholar]
- 54.Bernassola F., Karin M., Ciechanover A., Melino G. (2008) Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 55.Ducker C. E., Griffel L. K., Smith R. A., Keller S. N., Zhuang Y., Xia Z., Diller J. D., Smith C. D. (2006) Mol. Cancer Ther. 5, 1647–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]