Abstract
Co-translational import into the endoplasmic reticulum (ER) is primarily controlled by N-terminal signal sequences that mediate targeting of the ribosome-nascent chain complex to the Sec61/translocon and initiate the translocation process. Here we show that after targeting to the translocon the secondary structure of the nascent polypeptide chain can significantly modulate translocation efficiency. ER-targeted polypeptides dominated by unstructured domains failed to efficiently translocate into the ER lumen and were subjected to proteasomal degradation via a co-translocational/preemptive pathway. Productive ER import could be reinstated by increasing the amount of α-helical domains, whereas more effective ER signal sequences had only a minor effect on ER import efficiency of unstructured polypeptides. ER stress and overexpression of p58IPK promoted the co-translocational degradation pathway. Moreover polypeptides with unstructured domains at their N terminus were specifically targeted to proteasomal degradation under these conditions. Our study indicates that extended unstructured domains are signals to dispose ER-targeted proteins via a co-translocational, preemptive quality control pathway.
To ensure cellular homeostasis and to preclude toxic effects of aberrant protein conformers quality control mechanisms have evolved to recognize and degrade non-functional and misfolded proteins. In the cytosol the ubiquitin-proteasome system is the main pathway for regulated protein turnover (for reviews, see Refs. 1–3). Moreover the proteasome is part of a quality control system designated endoplasmic reticulum (ER)5-associated degradation (ERAD), which mediates post-translational degradation of non-native proteins generated in the ER. ERAD is the primary response to eliminate non-native ER proteins. It involves recognition of non-native polypeptides by ER-resident proteins and retrograde transport to the cytosol where proteasomal degradation occurs (for reviews, see Refs. 4–6). In case ERAD substrates accumulate in the ER lumen intracellular signaling pathways are induced that are collectively called the unfolded protein response (for reviews, see Refs. 7 and 8). Recently a preemptive, co-translocational quality control pathway was described that operates before translocation into the ER is completed (9, 10). Regulated translocation could act as an early quality control step to prevent an overload of the ER with non-native proteins. This regulation relies on an interplay between intrinsic features of the polypeptides and accessory factors able to modulate the translocation efficiency (for a review, see Ref. 11). Although numerous factors are known to be involved in ERAD less is known about mediators of the preemptive, co-translocational quality control pathway. In one study p58IPK was identified as a key regulator (9), whereas in another it was shown that ER translocation during conditions of acute ER stress is controlled by the signal peptide (10). Indeed the signal peptide is an important intrinsic determinant. Although an exceptionally diverse set of sequences can target proteins for ER import it has been demonstrated that the translocation efficiency is modulated in a signal peptide sequence-specific manner (for reviews, see Refs. 12–14). Another attractive candidate for an intrinsic factor to regulate translocation is the folding state of the nascent polypeptide chain. Formation of secondary structure occurs already in the ribosome exit tunnel (15–18). Moreover it was shown that the polypeptide structure within the ribosomal exit tunnel can modulate translocation of distal parts of the nascent chain (19).
The mammalian prion protein (PrP) is a suitable model protein to study whether formation of secondary structure could modulate translocation efficiency because it has an intriguing modular composition: the N-terminal domain spanning 120 amino acids is flexibly disordered followed by a highly structured C-terminal domain of ∼110 amino acids. This autonomously folding domain contains three α-helical regions and a short, two-stranded β-sheet (20–22). Notably folding of the C-terminal domain is one of the most rapid folding reactions measured in vitro (23). Interestingly previous studies indicated that the C-terminal folded domain of PrP is necessary and sufficient to promote ER import. In cultured cells and neurons of transgenic animals PrP-S230X (also known as PrPΔGPI or GPI−PrP), a mutant devoid of the C-terminal glycosylphosphatidylinositol (GPI) anchor signal peptide, is efficiently imported into the ER and secreted (24–27). However, by deleting parts of the α-helical domains located in the C-terminal domain a fraction is directed to proteasomal degradation in the cytosol (28).
We have now analyzed the underlying mechanisms of this impaired ER import and show that ER-targeted proteins require a certain amount of structured domains to be imported into the ER. In addition, our study indicates that extended unstructured domains are signals for a preemptive/co-translocational degradation pathway.
EXPERIMENTAL PROCEDURES
Generation of PrP and PrP Mutants
The coding region of mouse prion protein gene (GenBankTM accession number M18070) modified to express PrP-L108M/V111M was inserted into the mammalian expression vector pcDNA3.1/Zeo (Invitrogen), allowing detection by the monoclonal antibody (mAb) 3F4. PrP was used as a template to introduce the following deletions and mutations by standard PCR cloning techniques. The generation of the constructs S230X, Δ27–89/S230X, Δ27–156/S230X, and mouse Doppel (Dpl) was described earlier (27–29). In PrP/31CHO, Q159X/31CHO, W144X/31CHO, M133X/31CHO, A115X/31CHO, 115α2α3/31CHO, 115α2α3/31CHO/N196Q, 115α2α3/31CHO/N196Q/N180Q, 115/31CHOGFR, 115/31CHO+115, 115/31CHO+Tau, 115/31CHO+αsyn, and 31CHO+αsyn, the amino acids (aa) Trp and Asn at position 31/32 were substituted into Asn and Phe thereby generating an additional glycosylation acceptor site (NFT-motif). For the generation of cytosolic versions of PrP mutants the ER signal sequence (aa 2–22) was deleted. The Cre5p-SS and GH-SS mutants were generated by replacing aa 1–27 of PrP containing the endogenous signal sequence with the yeast Cre5p signal peptide (MRLLALVLLLLCAPLRA) or the rat growth hormone (GH) signal sequence (MAADSQTPWLLTFSLLCLLWPQEAGA). The following PrP mutants were cloned: 115α2α3 (aa 1–114 + 171–221), α2α3115 (aa 1–22 + 171–221 + 23–114), 115/31CHO+115 (aa 1–114 + 23–114), 115Dpl (PrP aa 1–114 + Dpl aa 98–148), Dpl115 (PrP aa 1–22 + Dpl aa 98–148 + PrP aa 23–114), 115/31CHOGFR (PrP aa 1–114 + GFR aa 40–89), 115/31CHO+αsyn (PrP aa 1–114 + αsyn aa 2–114), 31CHO+αsyn (PrP aa 1–34 + αsyn aa 2–114), and 115/31CHO+Tau (PrP aa 1–114 + Tau40/P301L aa 103–197). The plasmid for human Tau40/P301L was a kind gift from Eva-Maria Mandelkow (Max-Planck-Arbeitsgruppen für strukturelle Molekularbiologie, Hamburg, Germany). The construct 115/31CHO+Tau contains an amino acid change at position 108 of Tau (I108N). The expression plasmid for rat glial cell line-derived neurotrophic factor receptor α (GFR; GenBankTM accession number CAA05171) was a kind gift from Zhe-Yu Chen (Second Military Medical University, Shanghai, China).
The plasmids for the expression of p58IPK and p58IPKΔJ were kindly provided by David Ron (9) and were modified with a FLAG tag at the C terminus of the protein. For the generation of p58IPKΔSS the ER signal sequence (aa 2–31) was deleted. The plasmid for EYFP-Hsp70 was a kind gift from Richard I. Morimoto (30). The constructs for hamster BiP (kindly provided by Linda Hendershot) and the Hsp40 members Hdj1 and Hdj2 (a kind gift from Richard I. Morimoto) were subcloned into the pcDNA3.1/Zeo vector for mammalian expression. Hdj2 was modified to bear a C-terminal FLAG tag.
Antibodies and Reagents
All standard chemicals and reagents were purchased from Sigma if not otherwise noted. The following antibodies were used: mouse mAb 3F4 (31), mouse M2 anti-FLAG mAb (Sigma), mouse anti-α-synuclein mAb (mAb42, BD Biosciences), mouse mAb anti-Hsp70 (kindly provided by William J. Welch), mouse anti-KDEL mAb (Stressgen), and horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibody (Amersham Biosciences). Rabbit anti-Hsp40 polyclonal antibody was a kind gift from F. Ulrich Hartl (32).
Cell Culture and Secretion Analysis
Mouse N2a (ATCC number Ccl 131) cells were cultivated as described previously (33). Cells were transfected by a liposome-mediated method using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. To examine the secretion of proteins into the cell culture supernatant, cells were cultivated in cell culture medium without supplements for at least 3 h at 37 °C. The medium was collected, and proteins were precipitated with trichloroacetic acid and then analyzed by Western blotting.
Inhibitor Treatment
To transiently inhibit the proteasome, cells were treated with MG132 (Calbiochem; 30 μm in DMSO) for 3 h at 37 °C prior to cell lysis. To induce ER stress transfected cells were cultivated with thapsigargin (Sigma; 1 μm in DMSO) for the time indicated prior to Western blot analysis.
Cell Lysis, Endoglycosidase H (EndoH) Digestion, and Western Blot Analysis
As described earlier (34), cells were rinsed twice with ice-cold phosphate-buffered saline, scraped off the plate, pelleted by centrifugation, and lysed in cold detergent buffer (0.5% Triton X-100, 0.5% sodium deoxycholate in phosphate-buffered saline). For EndoH digestion, protein lysates were adjusted to 0.5% SDS, boiled, and then treated with EndoH (New England Biolabs) for 1 h at 37 °C as specified by the manufacturer. Laemmli sample buffer was added, and after boiling samples were examined by immunoblotting as described previously (35).
Metabolic Labeling and Immunoprecipitation
As described previously (36), cells were starved for 30 min in methionine-free modified Eagle's medium (Invitrogen) and subsequently labeled for the time indicated with 300 μCi/ml l-[35S]methionine (Hartmann Analytics; >37 TBq/mmol) in methionine-free modified Eagle's medium (Invitrogen). When indicated, the proteasomal inhibitor MG132 was present during the starving and the labeling periods (50 μm in DMSO). For the chase, the labeling medium was removed, and cells were washed twice and then incubated in complete medium for the time indicated. Immunoprecipitation of PrP was performed using the mAb 3F4 as described earlier (27). The immunopellet was analyzed by SDS-PAGE.
In Vitro Transcription, Translation, and Translocation
In vitro transcription/translation was performed using the TnT T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions in the presence or absence of canine pancreatic microsomal membranes (Promega). If indicated samples were subjected to EndoH digestion before SDS-PAGE.
Protein Hydrophobicity Plot
The hydrophobicity of the signal sequences was analyzed using the Kyte and Doolittle hydropathicity plot (37). Regions with values above 0 are hydrophobic in character.
Statistical Analysis
Quantifications were based on at least three independent experiments. Data were expressed as means ± S.E. Statistical analysis was performed using Student's t test.
RESULTS
Loss of α-Helical Domains Directs ER-targeted Prion Protein to Proteasomal Degradation in the Cytosol
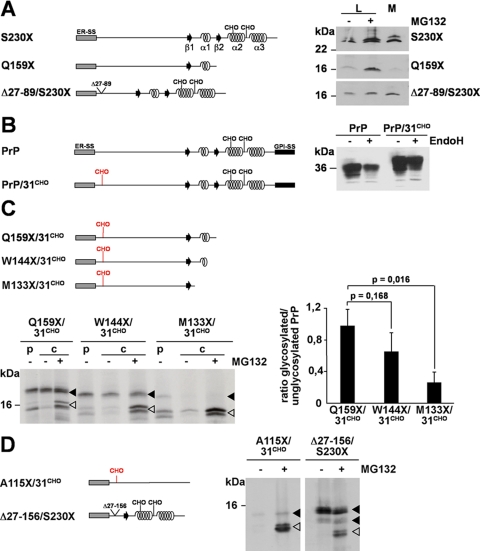
Wild type PrP is characterized by a set of co- and post-translational modifications (for a review, see Ref. 38) such as two N-linked glycans of complex structure (Asn180 and Asn196 in murine PrP) (39), a disulfide bond between Cys178 and Cys213, and a GPI anchor attached to Ser230 (40). Deletion of the C-terminal GPI anchor signal sequence prevents membrane attachment of PrP-S230X but does not interfere with import into the ER or further trafficking through the secretory pathway (24, 27, 41). PrP-S230X is secreted both by cultured cells (Fig. 1A, right panel, M) (27) and by neurons of transgenic mice (26). Please note that PrP-S230X is incompletely glycosylated. Thus, two bands corresponding to a glycosylated and an unglycosylated fraction are detectable on Western blots (27). Interestingly deletion of additional parts of the C terminus interferes with ER import. PrP-Q159X, a pathogenic mutant linked to Gerstmann-Sträussler-Scheinker syndrome in humans lacking helix 2 and 3 (α2α3) of the C-terminal domain, was barely found in the cell culture supernatant in contrast to PrP-S230X (Fig. 1A, M). Moreover upon transient proteasomal inhibition the relative amount of PrP-Q159X in the lysate increased indicating that a fraction was subjected to proteasomal degradation (Fig. 1A, L, MG132+ and Ref. 28). Two different mechanisms could explain the phenomenon that a significant fraction of PrP-Q159X is subjected to proteasomal degradation whereas PrP-S230X is not. Either the decreasing length of the polypeptide chain impairs ER import, or information present in the C-terminal globular domain is required. To test for the possibility that the α-helical domains are important for efficient ER import we generated a mutant with a deletion in the unstructured N-terminal domain. PrPΔ27–89/S230X and PrP-Q159X are of similar length; however, PrPΔ27–89/S230X was efficiently imported into the ER and secreted (Fig. 1A, M). In addition, this mutant was not subjected to proteasomal degradation. The relative amount of PrPΔ27–89/S230X did not significantly increase upon transient inhibition of the proteasome (Fig. 1A, L, +MG132).
FIGURE 1.
Loss of α-helical domains directs ER-targeted prion protein to proteasomal degradation in the cytosol. A, deletion of α-helical domains impairs ER import. Left panel, schematic presentation of the constructs. ER-SS, ER signal sequence; α, α-helical region; β, β-strand; CHO, N-linked glycosylation acceptor site; straight line, unstructured regions. Right panel, N2a cells were transiently transfected with the mutants depicted, and PrP present in the cell lysate (L) or in the cell culture medium (M) was analyzed by immunoblotting using the mAb 3F4. In addition, cells lysates were analyzed treated with or without the proteasomal inhibitor MG132 for 3 h prior to lysis (± MG132). B, an additional N-linked glycosylation acceptor site is functional. Left panel, schematic presentation of the constructs. GPI-SS, GPI signal sequence. The additional glycosylation site (CHO) at amino acid 31 in PrP/31CHO is marked in red. Right panel, N2a cells were transiently transfected with wild type PrP (PrP) or PrP/31CHO. Total cell lysates were either treated with the endoglycosidase H (+ EndoH) or left untreated (− EndoH), and PrP was detected by Western blotting. C and D, ER import efficiency correlates with the amount of α-helical domains. C, N2a cells transiently transfected with the constructs depicted in the upper panel were pulse-labeled (p) for 1 h with [35S]methionine and then chased (c) for 1 h in the presence (+ MG132) or absence (− MG132) of the proteasomal inhibitor MG132 (50 μm). PrP was immunoprecipitated using the mAb 3F4 and analyzed by SDS-PAGE. Quantification of three independent experiments is shown in the right panel. Data represent the ratio of glycosylated/unglycosylated PrP species present in the chase + MG132 (mean ± S.E.). p values were determined by Student's t test. D, α-helical domains are necessary and sufficient for ER import. N2a cells transiently transfected with the PrP mutants depicted in the left panel were metabolically labeled with [35S]methionine and then incubated in fresh medium for 1 h in the presence (+ MG132) or absence (− MG132) of MG132 (50 μm). PrP was immunoprecipitated using the mAb 3F4 and analyzed by SDS-PAGE. Open arrowheads represent unglycosylated PrP species; closed arrowheads represent glycosylated forms.
To facilitate our further analysis of monitoring productive ER import we introduced a new acceptor site for N-linked glycosylation shortly behind the ER signal sequence. In PrP/31CHO the amino acids Trp31 and Asn32 were changed to Asn and Phe, respectively, to create an additional NFT motif. The slightly decreased mobility on SDS-PAGE indicates that PrP/31CHO was modified with an additional N-linked glycan. Obviously the additional glycan was processed into complex structure, similarly to the two other glycans attached to Asn180 and Asn196, as treatment with endoglycosidase H did not cause a shift in electrophoretic mobility (Fig. 1B, right panel, +EndoH). PrP/31CHO is GPI-anchored at the outer leaflet of the plasma membrane, and after peptide-N-glycosidase F treatment wild type PrP and PrP/31CHO displayed a similar migration pattern on SDS-PAGE (data not shown). These data indicate that the new acceptor site for N-linked glycosylation located close to the N terminus of full-length PrP is functional and does not interfere with cellular trafficking.
Next we inserted the N-terminal acceptor site for N-linked glycosylation in PrP-Q159X. Analysis of PrP-Q159X/31CHO revealed two distinct species: a glycosylated fraction (Fig. 1C, filled arrowhead) and an unglycosylated fraction (Fig. 1C, open arrowhead). Interestingly two distinct species of unglycosylated molecules were detectable. As shown below, these two fractions are apparently due to polypeptides with or without the N-terminal ER signal peptide. The relative amount of the unglycosylated fraction was significantly increased in cells transiently incubated with MG132, indicating that a fraction of the mutant was subjected to proteasomal degradation (Fig. 1C, lower part, +MG132). To provide further support for a role of the C terminus in productive ER import we deleted additional parts of the C terminus to create PrP-W144X/31CHO and PrP-M133X/31CHO. Indeed the relative amount of the glycosylated fraction (Fig. 1C, filled arrowhead) successively decreased by increasing the C-terminal deletions (Fig. 1C, right panel). Of note, after transient proteasomal inhibition an unglycosylated fraction was stabilized, indicating that the loss of glycosylated proteins was not due to a decreased synthesis. To address the question of whether the length of the polypeptide chain is a limiting factor for a productive import we generated PrP-A115X/31CHO and PrPΔ27–156/S230X, two polypeptides of similar length. The major difference is that PrPΔ27–156/S230X is dominated by α-helical domains, whereas PrP-A115X/31CHO is composed solely of an unstructured domain (Fig. 1D). The analysis revealed that PrPΔ27–156/S230X was efficiently glycosylated, whereas PrP-A115X/31CHO was barely detectable (Fig. 1D, −MG132). Only after transient proteasomal inhibition could an unglycosylated fraction of PrP-A115X/31CHO be detected (Fig. 1D, +MG132). Metabolic labeling experiments further demonstrated that PrP-A115X/31CHO had obviously never been modified with N-linked glycans (supplemental Fig. 1A).
More Efficient ER Signal Sequences Do Not Restore Import of Intrinsically Disordered Proteins
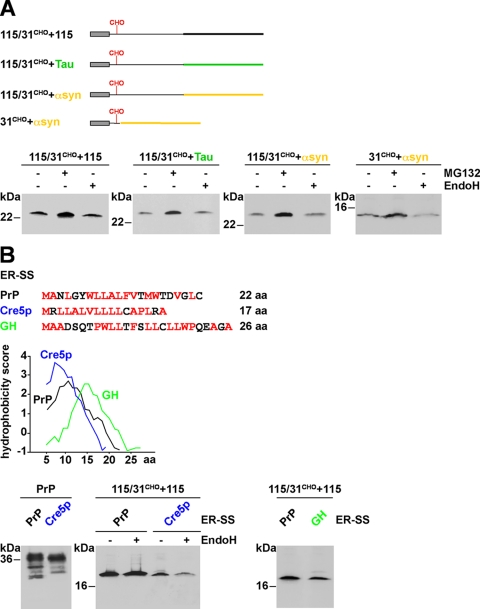
To test for the possibility that an increase in polypeptide chain length could promote translocation into the ER we fused an additional unstructured domain to PrP-115/31CHO, be it a second copy of PrP-A115X, an unstructured domain derived from the Tau protein, or an unstructured domain derived from α-synuclein (Fig. 2A) (42–44). However, ER import and glycosylation could not be restored (Fig. 2A, lower panel). To exclude the possibility that PrP-115/31CHO has a dominant negative effect on the import of a different intrinsically unstructured domain we constructed PrP-31CHO+αsyn. This construct consists of the ER signal sequence and the first 6 amino acids of PrP-115/31CHO to mediate ER targeting and N-linked glycosylation fused to the intrinsically disordered polypeptide derived from α-synuclein. Similarly to PrP-115/31CHO+αsyn, PrP-31CHO+αsyn was not glycosylated (Fig. 2A, lower panel). To address the possibility that overexpression of our PrP constructs leads to saturation of the translocation machinery we carried out titrations where cells were transfected with a broad range of expression plasmid (0.01–1.0 μg). However, even in cells transfected with low amounts of plasmid DNA we did not observe glycosylation of PrP-115/31CHO+115 or PrP-115/31CHO+Tau (supplemental Fig. 1B). In the same context we compared the kinetics of PrP maturation in cells transfected with 0.1 or 1 μg of DNA. The pulse/chase experiments indicated that formation of fully matured PrP, i.e. complex glycosylated, was relatively independent of the amount of the transfected PrP expression plasmid (supplemental Fig. 1C).
FIGURE 2.
More efficient ER signal sequences cannot restore import of intrinsically disordered proteins. A, increase in polypeptide chain length could not promote translocation into the ER. Upper panel, schematic presentation of the proteins analyzed. The following domains were fused to PrP-115/31CHO: 115/31CHO+115, the unstructured domain of PrP (black); 115/31CHO+Tau, an unstructured domain of the Tau protein (green); and 115/31CHO+αsyn, an unstructured domain of α-synuclein (yellow). In addition we generated 31CHO+αsyn, a construct consisting of the ER signal sequence and the first 6 amino acids of PrP-115/31CHO, which mediates ER targeting and N-linked glycosylation, fused to the intrinsically disordered polypeptide derived from α-synuclein. Lower panel, N2a cells were transfected with the constructs depicted and grown for 3 h in the presence or absence of MG132 (± MG132). If indicated cell lysates were treated with EndoH (± EndoH). PrP was analyzed by Western blotting using the mAb 3F4. 31CHO+αsyn was analyzed using the anti-α-synuclein mAb42. B, efficient ER signal sequences do not restore ER import. The ER signal sequences from the yeast protein Cre5p (blue) or the rat growth hormone (GH; green) were fused to PrP or PrP-115/31CHO+115. Amino acid sequences of the ER-SS are depicted. Hydrophobic amino acids are marked in red. The panel shows a Kyte and Doolittle hydrophobicity plot of the signal sequences. Regions with values above 0 are hydrophobic in character. Lower panel, N2a cells were transiently transfected with the constructs indicated and analyzed by Western blot using the mAb 3F4. If indicated, cell lysates were treated with EndoH (± EndoH) prior to Western blotting.
Previous studies revealed that signal sequences can vary in their efficiency to promote ER import (for a review, see Ref. 14). We have shown previously that expression of mammalian full-length PrP in yeast interferes with cellular viability because the endogenous ER signal sequence was insufficient to promote ER import. By using a more hydrophobic signal peptide derived from the yeast protein Cre5p ER import of PrP could be restored, and the adverse effects on cellular viability were prevented (45). To test whether the Cre5p signal sequence can increase ER import in mammalian cells we replaced the PrP signal sequence with that of Cre5p. In case of full-length PrP the Cre5p signal sequence appears to slightly increase ER import efficiency as the intensity of the unglycosylated fraction decreased. However, ER import and glycosylation of PrP-115/31CHO+115, a mutant consisting of unstructured domains, was not restored by the more hydrophobic signal sequence (Fig. 2B). In addition, we tested the ER signal sequence of rat growth hormone. This signal sequence was shown previously to enhance ER import of a secreted glycoprotein (46), yet it did not restore import of PrP-115/31CHO+115 (Fig. 2B) or of PrP-115 (28).
ER Import of Unstructured Domains Is Restored by Increasing the Content in α-Helical Domains
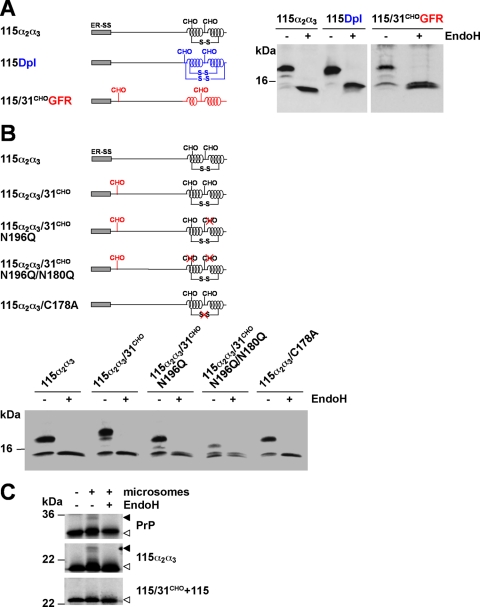
Assuming that the α-helical domains serve as a positive signal for ER import we fused the α-helical domains 2 and 3 (α2α3) of PrP to PrP-A115X. In this context it is important to note that in vitro experiments indicated that the isolated fragments comprising helix 2-helix 3 or only helix 3 show complete structural autonomy, i.e. adopt an α-helical conformation independently of the complete protein (47, 48). In addition, we constructed chimeric constructs (Fig. 3A) containing the unstructured domain of PrP-A115X fused to the α-helical domain of Dpl (49) or to the first two α-helices of the GFR (50). Importantly there is no significant sequence homology (<16%) between the α-helices of Dpl or GFR and PrP. The Western blot analysis of PrP-115α2α3, PrP-115Dpl, and PrP-115/31CHOGFR revealed that α-helical domains, be they from PrP, Dpl or GFR, efficiently restored import and glycosylation of an unstructured domain (Fig. 3A).
FIGURE 3.
ER import of unstructured domains is restored by increasing the content in α-helical domains. A and B, schematic presentation of the mutants analyzed. S-S, disulfide bond. The following domains were fused to PrP-115 or PrP-115/31CHO: 115α2α3, two α-helical domains of PrP (black); 115Dpl, two α-helical domains of Doppel (blue); and 115/31CHOGFR, two α-helical domains of GFR (red). A and B, N2a cells were transiently transfected with the mutants depicted. Cell lysates were either treated with EndoH (+ EndoH) or left untreated (− EndoH) and analyzed by Western blot using the mAb 3F4. C, α-helical but not unstructured domains restore in vitro translocation. PrP, 115α2α3, and 115/31CHO+115 were synthesized in vitro in the presence (+ microsomes) or absence (− microsomes) of ER-derived rough microsomes. If indicated, radioactively labeled products were treated with EndoH (± EndoH) before SDS-PAGE. Open arrowheads, unglycosylated PrP species; closed arrowheads, glycosylated PrP species.
Two N-linked glycans (Asn180 and Asn196) and a disulfide bond (Cys178-Cys213) are located in the α-helical domains of PrP. To address their possible role in the translocation process we created a series of new mutants (Fig. 3B). The analysis revealed that neither the C-terminal glycan acceptor sites (115α2α3/31CHO/N196Q/N180Q) nor the disulfide bond (115α2α3/C178A) were necessary to promote efficient import (Fig. 3B). Finally we used an in vitro ER import assay to verify the results obtained in transfected cells. In contrast to PrP-115α2α3, PrP-115/31CHO+115 was not glycosylated, indicating that this construct was not imported into microsomes (Fig. 3C).
The Polypeptide Fraction Subjected to Proteasomal Degradation Contains an Uncleaved Signal Peptide
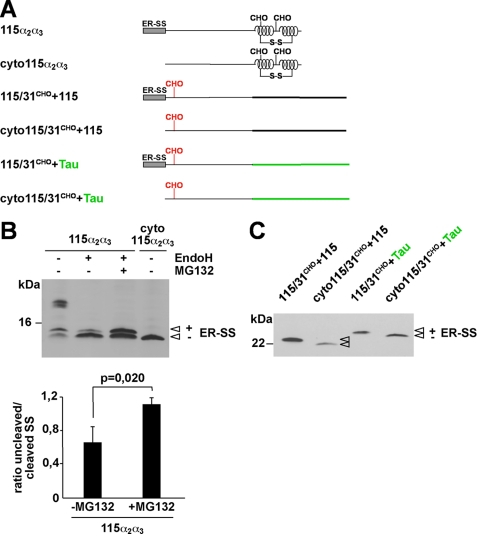
Cleavage of the N-terminal ER signal peptide occurs co-translocationally, whereas the degradation of misfolded ER proteins is mainly a post-translational event. However, a co-translocational or preemptive quality control pathway was described recently (9, 10) operating prior to productive import into the ER lumen. Notably polypeptides subjected to this quality control pathway retain their N-terminal signal peptide (9, 51). Two distinct unglycosylated species can be detected for PrP-115α2α3 (Fig. 4B). After deglycosylation with EndoH (Fig. 4B, +EndoH) the relative amount of the faster migrating band increased, indicating that this fraction consists of molecules devoid of the ER signal peptide (open arrowheads, −ER-SS). The slightly slower migrating band, apparently containing the ER signal peptide, was stabilized by MG132, i.e. was subjected to proteasomal degradation (Fig. 4B, +EndoH, +MG132). To provide more evidence that the slower migrating species of PrP-115α2α3 contained an uncleaved signal peptide we constructed PrP-cyto115α2α3 (Fig. 4A), a variant devoid of the N-terminal ER signal sequence comprising 23 aa. The migration pattern of PrP-cyto115α2α3 supports the notion that the glycosylated fraction of PrP-115α2α3 is devoid of the signal peptide whereas the unglycosylated species, which are subjected to proteasomal degradation, still contain the signal peptide. A quantitative analysis revealed that upon transient proteasomal inhibition there is a specific increase in the fraction of PrP-115α2α3 containing the signal peptide, whereas the relative amount of PrP-115α2α3 processed by the signal peptidase was not affected by proteasomal inhibition (Fig. 4B). Similarly both proteins PrP-115/31CHO+115 and PrP-115/31CHO+Tau that are entirely unstructured and were not imported into the ER displayed a slightly reduced migration pattern on SDS-PAGE compared with the corresponding constructs devoid of the ER signal peptide (Fig. 4C). In summary these data indicate that the polypeptide fractions subjected to proteasomal degradation were not co-translationally processed, i.e. contain their N-terminal signal peptide.
FIGURE 4.
Polypeptides subjected to proteasomal degradation contain an uncleaved signal sequence. A, schematic presentation of the mutants analyzed. Two versions of the mutants 115α2α3, 115/31CHO+115, and 115/31CHO+Tau were generated, one version with the original ER-SS and one lacking the ER-SS (cyto forms). B, N2a cells were transiently transfected and incubated in the presence or absence of MG132 (30 μm; 3 h). In addition, cell lysates were either treated with EndoH (+ EndoH) or left untreated (− EndoH) prior to Western blotting using the mAb 3F4. Unglycosylated PrP with (+ ER-SS) and without (− ER-SS) the ER signal peptide are marked. Lower panel, quantification of at least three independent experiments. Plotted is the ratio of the amount of 115α2α3 with an uncleaved versus cleaved SS in EndoH-treated samples with or without proteasomal inhibition (± MG132) (mean ± S.E.). The p value was determined by Student's t test. C, N2a cells transfected with the mutants depicted were lysed, and proteins analyzed by Western blot using the mAb 3F4. The protein fraction with (+ ER-SS) and without (− ER-SS) the ER signal peptide is marked.
p58IPK Promotes Proteasomal Degradation of ER-targeted Polypeptides with Extended Unstructured Domains at Their N Terminus
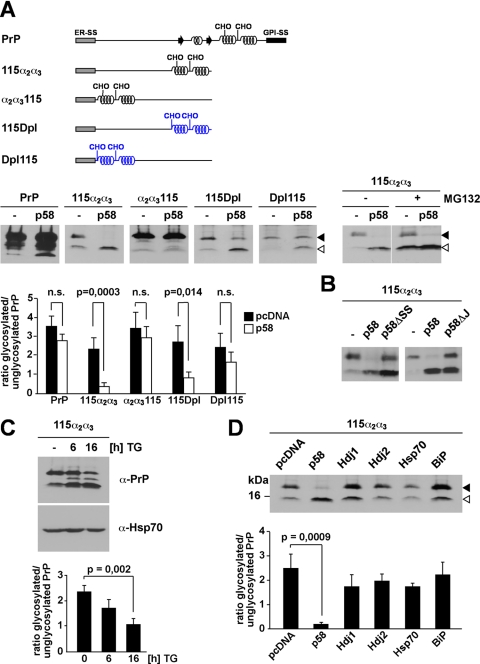
It was described previously that p58IPK is a mediator of co-translocational quality control (9). Consequently we tested a possible effect of p58IPK on ER import. Biogenesis of full-length PrP was not significantly affected by the co-expression of p58IPK. However, overexpression of p58IPK significantly interfered with ER import of PrP-115α2α3. As a consequence, the glycosylated fraction decreased whereas the unglycosylated fraction increased (Fig. 5A). As expected the unglycosylated fraction of PrP-115α2α3 in p58IPK-overexpressing cells was subjected to proteasomal degradation (Fig. 5A, right panel, ±MG132). Remarkably overexpression of p58IPK had little effect on the biogenesis of PrP-α2α3115. In cells co-expressing p58IPK the relative amount of glycosylated PrP-α2α3115 was not affected (Fig. 5A). The only difference of PrP-115α2α3 and PrP-α2α3115 is the arrangement of their structural elements: although in PrP-115α2α3 the α-helical domains are located C-terminally to an extended unstructured region, in PrP-α2α3115 the α-helical domains are N-terminal to the unstructured domain. To follow up on the idea that p58IPK specifically targets polypeptides with extended unstructured domains at their N terminus we included PrP-115Dpl and PrP-Dpl115 in our analysis. Both mutants have the same overall content in α-helical domains, but the arrangement is different: in PrP-115Dpl the unstructured domain is synthesized first followed by two α-helical domains; in PrP-Dpl115 the arrangement of structural elements is reversed. Remarkably overexpression of p58IPK significantly interfered with the ER import of PrP-115Dpl but not of PrP-Dpl115.
FIGURE 5.
p58IPK promotes proteasomal degradation of ER-targeted polypeptides with extended unstructured domains at their N terminus. A, p58IPK promotes a preemptive/co-translocational quality control pathway. N2a cells were transiently co-transfected with the constructs indicated and p58IPK (p58) or a vector control (−). Right panel, cells co-transfected with p58IPK and PrP-115α2α3 were incubated in the presence or absence of MG132 (30 μm; 3 h). The lanes MG132+ are positioned directly next to the lanes MG132− although all lanes originate from one gel. B, p58IPK requires the ER signal peptide and J-domain to interfere with ER import of PrP-115α2α3. N2a cells were transiently co-transfected with PrP-115α2α3 and either p58IPK (p58), p58IPKΔSS (p58ΔSS), p58IPKΔJ (p58ΔJ), or a vector control (−). A and B, expression of PrP was analyzed by immunoblotting using the mAb 3F4; an open arrowhead marks the unglycosylated protein species; a closed arrowhead marks the glycosylated forms. C, reduced import under conditions of ER stress. N2a cells transfected with PrP-115α2α3 were incubated with thapsigargin (TG; 1 μm) for the time indicated and then analyzed by Western blot using the mAb 3F4 for detection of PrP and anti-Hsp70 mAb for detection of endogenous Hsp70 as loading control. D, overexpression of cytosolic Hsp70 or BiP does not interfere with ER import. N2a cells were transiently co-transfected with PrP-115α2α3 and the constructs indicated. Expression of PrP-115α2α3 was then analyzed by immunoblotting using the mAb 3F4; an open arrowhead marks the unglycosylated PrP species; a closed arrowhead marks the glycosylated forms. A, B, and C, quantifications were based on at least three independent experiments. Data were expressed as the ratio of glycosylated versus unglycosylated PrP (mean ± S.E.). Statistical analysis was performed using Student's t test. n.s., not significant.
In one study it was suggested that p58IPK is a cytosolic co-chaperone and recruits Hsp70 to the cytosolic face of the translocation channel Sec61 (9). This concept was challenged later; instead it was proposed that p58IPK resides in the ER lumen where it interacts with BiP with its J-domain (51). Consequently we constructed mutants of p58IPK devoid of the N-terminal signal sequence (p58ΔSS) or the J-domain (p58ΔJ). In contrast to full-length p58IPK neither of the mutants impaired ER import of PrP-115α2α3, indicating that both ER targeting of p58IPK and an interaction with an Hsp70 molecule are required for the activity of p58IPK to impair ER translocation of PrP-115α2α3 (Fig. 5B).
It has been shown previously that p58IPK is up-regulated after ER stress (52). Under conditions of transient ER stress induced by thapsigargin treatment of PrP-115α2α3-expressing cells, we observed a significant decrease in the glycosylated fraction of PrP-115α2α3 along with an increase in the unglycosylated fraction (Fig. 5C).
To demonstrate that the observed effects were specific for p58IPK, we included the cytosolic chaperones and co-chaperones Hsp70, Hdj1, and Hdj2 as well as the ER chaperone BiP in our analysis. Cells were transiently co-transfected with the respective chaperones, and glycosylation of PrP-115α2α3 was analyzed by Western blotting. This comparative analysis revealed that only co-expression of p58IPK significantly interfered with productive ER import of PrP-115α2α3 (Fig. 5D and supplemental Fig. 2).
DISCUSSION
Our study presents evidence that the extent and localization of secondary structure in nascent polypeptides serve as a signal to regulate translocation into the ER: ER-targeted polypeptides dominated by unstructured domains failed to productively translocate into the ER lumen and are subjected to proteasomal degradation via a co-translocational/preemptive pathway. ER import could be restored by increasing the content in α-helical domains, whereas more effective ER signal sequences did not promote ER import of unstructured polypeptides. Our study also indicates that p58IPK has a specific role in controlling ER import of polypeptides with extended unstructured domains close to their N terminus.
To study a possible role of the folding state of ER-targeted polypeptides in the translocation process we made use of the prion protein as a model protein because of its particular modular structure. The N-terminal domain of about 120 amino acids is flexibly disordered followed by a highly structured C-terminal domain of ∼110 amino acids (20–22). A previous study already indicated that the folded C-terminal domain is essential and sufficient to promote ER import (28). In this study, we addressed the underlying mechanism and showed that ER-targeted polypeptides dominated by unstructured domains fail to be productively imported into the ER. This effect was relatively independent of length and primary sequence of the polypeptides. PrP-115+115, PrP-115+Tau, or PrP-115+αsyn, lacking α-helical domains, are around 230 aa in length yet are not efficiently imported. On the other hand, PrPΔ27–156/230X, characterized by α-helical domains, only comprises 120 aa but is efficiently imported into the ER. Importantly we could show a direct correlation between the amount of α-helical domains and ER import. A stepwise deletion of α-helical domains resulted in a gradual reduction in ER import efficiency (Fig. 1C). By using α-helical or unstructured domains derived from unrelated proteins we could demonstrate that the effect of α-helical domains on ER import is independent of the primary sequence. The α-helical domains of the Doppel protein as well as of the GFR efficiently promoted ER import, whereas polypeptides composed of the unstructured domains derived from the Tau protein or α-synuclein were not imported into the ER.
Previous studies clearly have demonstrated that the type of signal sequence can significantly modulate translocation efficiency (for a review, see Ref. 14). Interestingly more efficient ER signal sequences that improve import of full-length PrP (45) had little effect in promoting ER import of polypeptides characterized by unstructured domains such as PrP-115+115 or PrP-A115X. These findings could indicate a dominant effect of unstructured domains on the translocation efficiency.
The second important finding of our study was that the polypeptides with extended unstructured domains were disposed via a co-translocational/preemptive pathway. The major evidence was the finding that co-translational modifications, such as N-linked glycosylation and cleavage of the signal peptide, did not occur. We show that the protein fraction subjected to proteasomal degradation contained an uncleaved signal peptide, a characteristic feature of other substrates subjected to the co-translocational/preemptive pathway (9, 51). In addition, we could demonstrate an important role of p58IPK, a factor described recently to be involved in the co-translocational pathway (9). A mutational analysis indicated that both the ER signal peptide and the J-domain of p58IPK are required for this activity (Fig. 5B). Interestingly our study also revealed that p58IPK selectively targets polypeptides with unstructured domains close to their N terminus, such as PrP-115α2α3 or PrP-115Dpl, to proteasomal degradation. In contrast, when the structured α-helical domains were synthesized first (PrP-α2α3115 or Dpl115), import was not affected by overexpression of p58IPK (Fig. 5A).
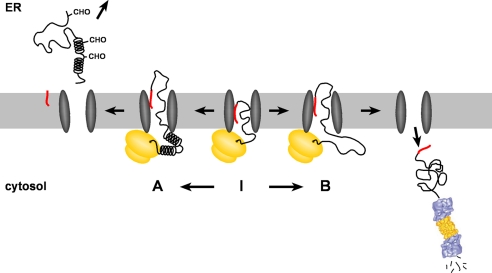
Based on our findings and studies described previously, the following model appears plausible (Fig. 6). After targeting of the ribosome-nascent chain complex to the translocon via the N-terminal signal peptide, the translation of an extended unstructured domain delays productive translocation into the ER lumen. Instead the growing polypeptide chain is kept in a translocationally competent state inside or at the cytosolic side of the translocon (Fig. 6I). Indeed previous studies indicated that the nascent chain can be exposed to the cytosol (53). Moreover it has been shown that the folding state of the growing polypeptide in the ribosome exit tunnel can be a signal to modulate translocation (19). Depending on the folding state of the growing polypeptide chain two alternative pathways are conceivable. In case α-helical domains are synthesized, the polypeptide chain is efficiently imported and modified by the signal peptidase and the oligosaccharyltransferase (Fig. 6A). However, if the remaining part of the polypeptide is still devoid of structured domains productive ER import is not pursued, and the protein is disposed via the proteasome (Fig. 6B).
FIGURE 6.
A model for the co-translocational quality control of ER-targeted polypeptides containing extended unstructured domains. After targeting of the ribosome-nascent chain complex to the translocon via the N-terminal signal peptide (red), the translation of an extended unstructured domain delays productive translocation into the ER lumen. Instead the growing polypeptide chain is kept in a translocationally competent state inside or at the cytosolic side of the translocon (I). Depending on the folding state of the growing polypeptide chain two alternative pathways are conceivable. In case α-helical domains are synthesized, the polypeptide chain is efficiently imported and modified by the signal peptidase and the oligosaccharyltransferase (A). If the remaining part of the polypeptide is still devoid of α-helical domains productive ER import is not pursued, and the protein is disposed via the proteasome (B). The proteasome picture was adopted with permission from Walz et al. (54).
What could be the physiological impact of such a system? Under physiological conditions only polypeptides were targeted to proteasomal degradation that are composed mainly or solely of unstructured domains. These proteins would be prime candidates for ERAD; thus the co-translocational control might provide an elegant mechanism to unburden the quality control machinery of the ER lumen. Strikingly the co-translocational quality control is more stringent under conditions of ER stress. Now ER import of polypeptides with extended unstructured domains was prevented. These polypeptides are able to fold in principle, but because of large unstructured domains they could be demanding clients for the ER chaperones. Preventing the import of such substrates could help to increase the folding capacity within the ER lumen.
Our study emphasizes the notion that the ribosome-nascent chain complex attached to the translocon complex is more dynamic than previously appreciated. It will now be interesting to see how the folding state of the emerging polypeptide chain and/or the sequence of the signal peptide is sensed to control ER translocation and to redirect ER-targeted polypeptides to the cytosol.
Supplementary Material
Acknowledgments
We are grateful to David Ron for providing the p58IPK constructs and antibody, to Rick Morimoto for the Hsp70-EYFP and Hsp40 constructs, to Zhe-Yu Chen for the rat GFR construct, to Eva-Maria Mandelkow for the human Tau40/P301L construct, to Linda Hendershot for the BiP construct, and to Ulrich Hartl for the Hsp40 antibody. We thank Richard Zimmermann for stimulating discussions, Viktoria Doll and Kerstin Lämmermann for technical help, and Christian Haass for continuous support.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 596), the Max Planck Society, and the Bundesministerium für Bildung und Forschung (BioDisc, DIP5.1).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- PrP
- prion protein
- GPI
- glycosylphosphatidylinositol
- mAb
- monoclonal antibody
- Dpl
- Doppel
- aa
- amino acids
- GFR
- glial cell line-derived neurotrophic factor receptor α
- αsyn
- α-synuclein
- SS
- signal sequence
- BiP
- immunoglobulin binding protein.
REFERENCES
- 1.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A., Orian A., Schwartz A. L. (2000) BioEssays 22, 442–451 [DOI] [PubMed] [Google Scholar]
- 4.Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 6.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 7.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 8.Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., Ron D. (2006) Cell 126, 727–739 [DOI] [PubMed] [Google Scholar]
- 10.Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. (2006) Cell 127, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hegde R. S., Kang S. W. (2008) J. Cell Biol. 182, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Heijne G. (1985) J. Mol. Biol. 184, 99–105 [DOI] [PubMed] [Google Scholar]
- 13.Martoglio B., Dobberstein B. (1998) Trends Cell Biol. 8, 410–415 [DOI] [PubMed] [Google Scholar]
- 14.Hegde R. S., Bernstein H. D. (2006) Trends Biochem. Sci. 31, 563–571 [DOI] [PubMed] [Google Scholar]
- 15.Whitley P., Nilsson I. M., von Heijne G. (1996) J. Biol. Chem. 271, 6241–6244 [DOI] [PubMed] [Google Scholar]
- 16.Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 17.Lu J., Deutsch C. (2005) Biochemistry 44, 8230–8243 [DOI] [PubMed] [Google Scholar]
- 18.Mingarro I., Nilsson I., Whitley P., von Heijne G. (2000) BMC Cell Biol. 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel C. J., Conti B., Johnson A. E., Skach W. R. (2008) J. Biol. Chem. 283, 20864–20873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., Dyson H. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. (1996) Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 22.Riek R., Hornemann S., Wider G., Glockshuber R., Wüthrich K. (1997) FEBS Lett. 413, 282–288 [DOI] [PubMed] [Google Scholar]
- 23.Wildegger G., Liemann S., Glockshuber R. (1999) Nat. Struct. Biol. 6, 550–553 [DOI] [PubMed] [Google Scholar]
- 24.Rogers M., Yehiely F., Scott M., Prusiner S. B. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3182–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocisko D. A., Come J. H., Priola S. A., Chesebro B., Raymond G. J., Lansbury P. T., Caughey B. (1994) Nature 370, 471–474 [DOI] [PubMed] [Google Scholar]
- 26.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M. (2005) Science 308, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 27.Winklhofer K. F., Heske J., Heller U., Reintjes A., Muranyi W., Moarefi I., Tatzelt J. (2003) J. Biol. Chem. 278, 14961–14970 [DOI] [PubMed] [Google Scholar]
- 28.Heske J., Heller U., Winklhofer K. F., Tatzelt J. (2004) J. Biol. Chem. 279, 5435–5443 [DOI] [PubMed] [Google Scholar]
- 29.Uelhoff A., Tatzelt J., Aguzzi A., Winklhofer K. F., Haass C. (2005) J. Biol. Chem. 280, 5137–5140 [DOI] [PubMed] [Google Scholar]
- 30.Kim S., Nollen E. A., Kitagawa K., Bindokas V. P., Morimoto R. I. (2002) Nat. Cell Biol. 4, 826–831 [DOI] [PubMed] [Google Scholar]
- 31.Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winklhofer K. F., Heller U., Reintjes A., Tatzelt J. (2003) Traffic 4, 313–322 [DOI] [PubMed] [Google Scholar]
- 34.Tatzelt J., Prusiner S. B., Welch W. J. (1996) EMBO J. 15, 6363–6373 [PMC free article] [PubMed] [Google Scholar]
- 35.Winklhofer K. F., Tatzelt J. (2000) Biol. Chem. 381, 463–469 [DOI] [PubMed] [Google Scholar]
- 36.Kiachopoulos S., Bracher A., Winklhofer K. F., Tatzelt J. (2005) J. Biol. Chem. 280, 9320–9329 [DOI] [PubMed] [Google Scholar]
- 37.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 38.Tatzelt J., Winklhofer K. F. (2004) Amyloid 11, 162–172 [DOI] [PubMed] [Google Scholar]
- 39.Haraguchi T., Fisher S., Olofsson S., Endo T., Groth D., Tarentino A., Borchelt D. R., Teplow D., Hood L., Burlingame A., Lycke E., Kobata A., Prusiner S. B. (1989) Arch. Biochem. Biophys. 274, 1–13 [DOI] [PubMed] [Google Scholar]
- 40.Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. (1987) Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 41.Blochberger T. C., Cooper C., Peretz D., Tatzelt J., Griffith O. H., Baldwin M. A., Prusiner S. B. (1997) Protein Eng. 10, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 42.Cleveland D. W., Hwo S. Y., Kirschner M. W. (1977) J. Mol. Biol. 116, 227–247 [DOI] [PubMed] [Google Scholar]
- 43.von Bergen M., Barghorn S., Jeganathan S., Mandelkow E. M., Mandelkow E. (2006) Neurodegener. Dis. 3, 197–206 [DOI] [PubMed] [Google Scholar]
- 44.Weinreb P. H., Zhen W., Poon A. W., Conway K. A., Lansbury P. T., Jr. (1996) Biochemistry 35, 13709–13715 [DOI] [PubMed] [Google Scholar]
- 45.Heller U., Winklhofer K. F., Heske J., Reintjes A., Tatzelt J. (2003) J. Biol. Chem. 278, 36139–36147 [DOI] [PubMed] [Google Scholar]
- 46.Rutkowski D. T., Ott C. M., Polansky J. R., Lingappa V. R. (2003) J. Biol. Chem. 278, 30365–30372 [DOI] [PubMed] [Google Scholar]
- 47.Eberl H., Glockshuber R. (2002) Biophys. Chem. 96, 293–303 [DOI] [PubMed] [Google Scholar]
- 48.Gallo M., Paludi D., Cicero D. O., Chiovitti K., Millo E., Salis A., Damonte G., Corsaro A., Thellung S., Schettini G., Melino S., Florio T., Paci M., Aceto A. (2005) Int. J. Immunopathol. Pharmacol. 18, 95–112 [DOI] [PubMed] [Google Scholar]
- 49.Mo H., Moore R. C., Cohen F. E., Westaway D., Prusiner S. B., Wright P. E., Dyson H. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2352–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leppänen V. M., Bespalov M. M., Runeberg-Roos P., Puurand U., Merits A., Saarma M., Goldman A. (2004) EMBO J. 23, 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutkowski D. T., Kang S. W., Goodman A. G., Garrison J. L., Taunton J., Katze M. G., Kaufman R. J., Hegde R. S. (2007) Mol. Biol. Cell 18, 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao S., Lin J., Do H., Johnson A. E. (1997) Cell 90, 31–41 [DOI] [PubMed] [Google Scholar]
- 54.Walz J., Erdmann A., Kania M., Typke D., Koster A. J., Baumeister W. (1998) J. Struct. Biol. 121, 19–29 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.