Abstract
SIRT3 is a major mitochondrial NAD+-dependent protein deacetylase playing important roles in regulating mitochondrial metabolism and energy production and has been linked to the beneficial effects of exercise and caloric restriction. SIRT3 is emerging as a potential therapeutic target to treat metabolic and neurological diseases. We report the first sets of crystal structures of human SIRT3, an apo-structure with no substrate, a structure with a peptide containing acetyl lysine of its natural substrate acetyl-CoA synthetase 2, a reaction intermediate structure trapped by a thioacetyl peptide, and a structure with the dethioacetylated peptide bound. These structures provide insights into the conformational changes induced by the two substrates required for the reaction, the acetylated substrate peptide and NAD+. In addition, the binding study by isothermal titration calorimetry suggests that the acetylated peptide is the first substrate to bind to SIRT3, before NAD+. These structures and biophysical studies provide key insight into the structural and functional relationship of the SIRT3 deacetylation activity.
Sirtuins are class III histone deacetylases that couple lysine deacetylation with NAD+ hydrolysis and are highly conserved in prokaryotes and eukaryotes (1). Mammals possess seven sirtuins, SIRT1–7, that occupy different subcellular compartments such as the nucleus (SIRT1, -6, -7), cytoplasm (SIRT2), and the mitochondria (SIRT3, -4, and -5) (2). They deacetylate lysines not only on histone substrates (3, 4) but also on non-histone substrates such as p53 tumor suppressor protein (5), Foxo transcription factors (6, 7), PGC-1α (8), α-tubulin (9), acetyl-CoA synthetases (10–12), and glutamate dehydrogenase (13). SIRT4 and SIRT6 have been shown to have ADP-ribosyltransferase activity (14–16). Sirtuins have been reported to play important roles in gene silencing (17), cell cycle regulation (18, 19), metabolism (8, 10–12, 14, 20–22), apoptosis (5, 23, 24), the lifespan-extension effects of calorie restriction (25, 26), and circadian rhythms (27–30). Sirtuins have emerged as therapeutic targets for diseases (31) such as type 2 diabetes (32), neurodegenerative diseases (33, 34), inflammation (35), and cancers (36, 37).
Several crystal structures of sirtuins have been reported from Thermotoga maritima (Tm) (38–40), Archaeoglobus fulgidus (Af1 and Af2) (38, 41–44), Escherichia coli (45), yeast (Hst2) (46–49), human SIRT2 (50), and SIRT5 (51). Sirtuins contain a conserved enzymatic core with two domains; that is, a large Rossmann fold domain that binds NAD+ and a small domain formed by two insertions of the large domain that binds to a zinc atom. The acetylated peptide substrate binds to the cleft between the two domains. Some of the known structures are apo structures with sirtuin protein alone, whereas others are bound to acetylated peptide substrate and/or NAD+ and its analogs. These structures revealed the mechanism of action for the deacetylation activity and substrate specificity.
SIRT3 localizes in mitochondria (13, 52–54) and is a major mitochondrial deacetylase. Hyperacetylation of mitochondrial proteins have been observed in SIRT3 knock-out mice (13, 55). Several key enzymes involved in energy production in the mitochondria have been identified as SIRT3 substrates. Acetyl-CoA synthetase 2 (AceCS2)2 converts acetate into acetyl-CoA in the mitochondria. Deacetylation of AceCS2 at lysine 642 by SIRT3 activates acetyl-CoA synthetase activity, providing increased acetyl-CoA to feed into the tricarboxylic acid cycle (10, 11). Glutamate dehydrogenase converts glutamate to α-ketoglutarate in mitochondria, promoting the metabolism of glutamate and glutamine to be used as fuels for the tricarboxylic acid cycle for ATP production (56). Glutamate dehydrogenase also induces amino acid-stimulated insulin secretion in insulinoma cells (14). Deacetylation of glutamate dehydrogenase by SIRT3 activates its enzyme activity (13, 57). SIRT3 is reported to deacetylate isocitrate dehydrogenase 2 in turn to activate the enzyme (57). Deacetylate isocitrate dehydrogenase 2 oxidatively decarboxylates isocitrate to α-ketoglutarate while converting NAD+ to NADH that promotes regeneration of antioxidants. Recently, Ahn et al. (55) reported that SIRT3−/− mouse embryonic fibroblasts have decreased ATP levels and abnormal activity of Complex 1 of the electron transport chain. Mice lacking SIRT3 showed reduced basal levels of ATP in the heart, kidney, and liver. Therefore, SIRT3 could play an important role in cell metabolism and energy balance. Caloric restriction (CR) extends the lifespan in Caenorhabditis elegans (58), Drosophila (59), yeast (60), and rodents (61) and showed beneficial effects in primates (62, 63) and human subjects (reported by CALORIE Pennington Team). SIRT3 activity is known to be increased by caloric restriction (64). Deacetylate isocitrate dehydrogenase 2 has been shown to be acetylated in the mitochondria of the fed mouse liver and deacetylated in mitochondria of the fasted mouse liver (65). The activity of glutamate dehydrogenase in the liver was also reported to increase by CR (66). Therefore, SIRT3 could be a key player in CR response. Lanza et al. (67) reported that exercise can increase SIRT3 expression level and prevent the decreasing of SIRT3 levels with increasing age. In addition, variability in the SIRT3 gene, which up-regulates SIRT3 expression, is enriched in long-lived individuals (68, 69). SIRT3 mRNA expression level also increased in leptin-treated ob/ob mice that links SIRT3 with the beneficial effects of leptin in regulating body weight and lipid metabolism (70). SIRT3 has also been implicated in selective apoptotic pathways and cell growth control (71) as well as the NAD+ salvage pathway that regulates the NAD+ level, which is crucial to cell survival (72).
Sirtuins have emerged as therapeutic targets to treat many diseases (31). The potential, important roles of SIRT3 in cell metabolism and CR suggest that SIRT3 could be a promising therapeutic target. Our human SIRT3 crystal structures reported here provide molecular information on the conformational changes induced by substrate binding. This is the essential first step for using structural based ligand design for developing SIRT3 modulators.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
Human SIRT3-(118–399) was cloned into a modified pET21b vector (Novagen) between BamHI and XhoI, which places expression under the control of the T7-lacO promoter. The protein was expressed in E. coli BL21-Gold(DE3) cells (Stratagene) as an N-terminal fusion to a hexahistidine affinity tag with integrated TEV protease site. A single colony was inoculated in LB media containing 100 μg/ml ampicillin at 37 °C, 250 rpm until the A600 reached 0.3. The culture was then transferred to 18 °C, 250 rpm until the A600 reached 0.6–0.8. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.3 mm, and expression was continued at 18 °C, 160 rpm overnight. Cells were collected by centrifugation, and the pellet was resuspended in lysis buffer (200 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol, and 25 mm HEPES-NaOH, pH 7.5) and sonicated to open the cells. Supernatant was separated from cell debris by centrifugation at 10,000 × g for 40 min at 4 °C and loaded onto a Ni-NTA column (Qiagen) that equilibrated with the buffer containing 200 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol, 20 mm imidazole, and 25 mm HEPES-NaOH, pH 7.5. The column was washed with 5 column volumes of the buffer containing 200 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol, 50 mm imidazole, and 25 mm HEPES-NaOH, pH 7.5, and eluted with the buffer containing 200 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol, 250 mm imidazole, and 25 mm HEPES-NaOH, pH 7.5. The eluted protein was dialyzed in lysis buffer and digested with TEV protease (Invitrogen) to remove the N-terminal His tag at 4 °C overnight. The protein was loaded on a second Ni-NTA column equilibrated with lysis buffer. The untagged protein was eluted by the buffer containing 200 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol, 5 mm imidazole, and 25 mm HEPES-NaOH, pH 7.5. The purified protein was dialyzed against the dialyzing buffer containing 200 mm NaCl, 5 mm 2-mercaptoethanol, and 20 mm Tris-HCl, pH 8.0, and concentrated. The protein was further purified by a S200 column (GE Healthcare) to >95% purity as assessed by SDS-PAGE analysis stained by Coomassie Brilliant Blue R-250 and concentrated to 10–15 mg/ml in the dialyzing buffer. SeMet protein was expressed in M9 media (in-house) and purified by the same method described as above.
Human SIRT3-(102–399) was cloned into the same vector as SIRT3-(118–399) and expressed in BL21(DE3) cells (Novagen) with co-expression of chaperone pG-KJE8 (Takara). A single colony was inoculated in LB media containing 20 μg/ml chloramphenicol and 100 μg/ml ampicillin for plasmid selection and 5 ng/ml tetracycline and 0.5 mg/ml l-arabinose for chaperone induction at 37 °C, 250 rpm until the A600 reached 0.3. The culture was then transferred to 16 °C, 250 rpm until the A600 reached 0.6. isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.3 mm, and expression was continued at 16 °C, 180 rpm overnight. Cells were collected by centrifugation, and the pellet was resuspended in lysis buffer (10 mm imidazole, 300 mm NaCl, 10% glycerol, 50 mm Tris-HCl, pH 7.8, 10 ml/gm of cell pellet) and sonicated to open the cells. Supernatant was separated from cell debris by centrifugation at 11,000 × g for 40 min at 4 °C and loaded onto a Ni-NTA column (Qiagen). The column was washed with 30 column volumes of the lysis buffer and 20 column volumes of the buffer containing 20 mm imidazole, 300 mm NaCl, 10% glycerol, 50 mm Tris-HCl, pH 7.8, and eluted with the buffer containing 100 mm imidazole, 300 mm NaCl, 10% glycerol, 50 mm Tris-HCl, pH 7.8. The eluted protein was dialyzed against the TEV buffer (300 mm NaCl, 10% glycerol, 50 mm Tris-HCl, pH 7.8) and then digested with TEV protease (Invitrogen) to remove the N-terminal His tag. The tag-free enzyme was isolated by a second Ni-NTA column using the TEV buffer. The pooled protein was concentrated by Amicon Ultra-15 Centrifugal Filter device (Millipore) and loaded onto a S200 column (GE Healthcare) with the buffer containing 200 mm NaCl, 20 mm Tris-HCl, pH 7.8. The protein was further loaded on a Q-Sepharose column (GE Healthcare) and eluted by the buffer containing 500 mm NaCl, 20 mm Tris-HCl, pH 7.8. The purity of the final protein was >98%, judged by SDS-PAGE analysis stained by Coomassie Brilliant Blue R-250.
Enzyme Assay
The purified SIRT3-(102–399) and SIRT3-(118–399) were tested for deacetylation activity with the mass spectrometry based assay reported previously (32). Km values for peptide substrate and NAD+ were determined by fixing one substrate at saturated concentration and varying the concentration of the other substrate. For the enzyme titration, the two enzymes were tested side-by-side with excess amounts of peptide substrate (80 μm) and NAD+ (6 mm). The enzyme concentration was started at 62.5 nm and serial-diluted in a 1:2 ratio to 0.24 nm. Reactions were incubated at 25 °C and stopped at 0-, 15-, 30-, 60-, 90-, 120-, 150-, and 180-min time points with the stopping solution containing 10% formic acid and 50 mm nicotinamide. The conversion of substrate to product was determined by mass spectrometry in conjunction with a RapidFire system (BioTrove). The percentage of conversions from peptide substrate to product was calculated.
Protein Crystallization
AceCS2-Kac peptide is a 12-mer containing residues 638–649 (638TRSGKacVMRRLLR, the numbering is for human AceCS2 sequence number, and Kac is acetylated lysine). The peptide was dissolved in the buffer containing 200 mm NaCl and 20 mm Tris-HCl, pH 7.0, and then adjusted to pH 7.0, achieving a final concentration of 40 mm. Purified native or SeMet containing SIRT3 and peptide mixture was prepared before crystallization and reached a final concentration of 6 mg/ml protein and 0.965 mm peptide. The SIRT3-AceCS2-Kac crystals were obtained by the hanging drop method at 18 °C. The drop was composed of 1 μl of the protein-peptide mixture and 1 μl of the crystallization solution containing 0.2 m lithium sulfate monohydrate, 15% (w/v) polyethylene glycol (PEG) 12000, and 0.1 m Bis-Tris, pH 5.5.
For the SIRT3-NAD+ co-crystals, NAD+ was dissolved in 10 mm HEPES-NaOH, pH 7.4, and then the pH was re-adjusted to 7.4 to a final concentration of 100 mm. Native SIRT3-(118–399) was mixed with NAD+ to a final protein concentration of 6 mg/ml and NAD+ concentration of 10 mm. Crystals were grown by the hanging drop method with the crystallization buffer containing 0.2 m lithium sulfate monohydrate, 25% (w/v) PEG 3350, and 0.1 m Bis-Tris, pH 5.5 at 18 °C.
The AceCS2-Ks-ac peptide has the same sequence as the AceCS2-Kac peptide with the acetyl lysine replaced with thioacetyl lysine using Fmoc-Lys(CSCH3)-OH synthesized according to a published procedure (87). The preparation of the peptide stock solution, SIRT3-peptide complex, and crystallization setup are the same. The crystallization solution contained 0.2 m lithium sulfate monohydrate, 17% w/v PEG 12000, and 0.1 m Bis-Tris, pH 5.5. The SIRT3-AceCS2-Ks-ac crystals were then soaked for 1–16 h in 0.125 mm NAD+ in the crystallization buffer.
Data Collection and Structure Determination
The SIRT3-AceCS2-Kac crystals with and without SeMet was cryo-protected in mother liquor containing 20% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at beamline 24-ID-E at Advanced Photon Source at Argonne National Laboratory and processed using the HKL2000 program (73). The SIRT3-AceCS2-Kac crystal belongs to the space group C2221 with one SIRT3-AceCS2-Kac peptide complex in the asymmetric unit. The structure was solved by the single wavelength anomalous dispersion method using data collected from a SeMet SIRT3-AceCS2-Kac crystal by CNX (Accelrys). With the four SeMet residues in the protein, the figure-of-merit for the single wavelength anomalous dispersion phasing was 0.4. Auto chain tracing was performed by Arp/Warp (74) with 262 of 295 residues built. Further structure refinement was carried out with the native data set to 1.8 Å using CNX, and model building was performed in Quanta (Accelrys). Detailed information of the diffraction data, refinement, and structure statistics are listed in Table 1. In Table 1, all the parameters for each diffraction data set are from that reprocessed by Mosflm (75) and Scala (76), and refinement statistics are from Refmac5 (77) in the CCP4 suite (78) for consistency.
TABLE 1.
Data collection and refinement statistics
B-factor, temperature factor.
| SIRT3-AceCS2-Kac |
SIRT3 apo | SIRT3-AceCS2-Ks-ac-ADPR NAD+-soak (1 h) | SIRT3-AceCS2 NAD+-soak (16 h) | ||
|---|---|---|---|---|---|
| SeMet | Native | ||||
| Diffraction data | |||||
| Resolution (Å)a | 30.0-2.1 (2.18-2.10) | 65.0-1.8 (1.90-1.80) | 65.0-2.7 (2.85-2.70) | 65.0-2.1 (2.21-2.10) | 65.0-2.5 (2.64-2.50) |
| Space group | C2221 | C2221 | P21 | C2221 | C2221 |
| Unit cell parameters | |||||
| a (Å) | 77.74 | 78.19 | 85.37 | 80.72 | 77.50 |
| b (Å) | 129.26 | 129.06 | 143.93 | 127.28 | 129.46 |
| c (Å) | 77.95 | 77.9 | 91.66 | 76.83 | 78.60 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| β (°) | 90.00 | 90.00 | 117.10 | 90.00 | 90.00 |
| γ (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Completeness (%)a | 99.5 (96.1) | 99.9 (99.9) | 85.0 (87.6) | 99.9 (99.6) | 95.7 (96.9) |
| Redundancy | 3.9 (3.1) | 6.6 (4.4) | 3.9 (3.8) | 4.0 (4.0) | 4.1 (4.2) |
| Average I/σIa | 22.8 (3.2) | 17.7 (3.1) | 9.9 (4.5) | 12.6 (6.5) | 10.3 (3.3) |
| Rmerge (%)a | 6.7 (31.7) | 6.2 (41.2) | 12.1 (41.7) | 9.0 (28.9) | 8.7 (44.7) |
| Refinement statistics | |||||
| Data (Å) | 65.0-1.8 (1.85-1.80) | 65.0-2.7 (2.77-2.70) | 65.0-2.1 (2.15-2.10) | 65.0-2.5 (2.57-2.50) | |
| Rworking (%)/No. of reflections | 20.3/33143 (36.8/2550) | 18.2/41169 (26.4/3311) | 19.5/21021 (22.3/1615) | 20.5/11989 (27.8/664) | |
| Rfree (%)/No. of reflections | 22.6/1838 (35.7/119) | 24.4/2313 (35.6/152) | 22.8/1210 (29.8/106) | 24.8/664 (38.5/40) | |
| r.m.s.d. in bond lengths (Å) | 0.014 | 0.016 | 0.015 | 0.012 | |
| r.m.s.d. in bond angles (°) | 1.44 | 1.45 | 1.52 | 1.45 | |
| Mean B factors (Å2) | 29.43 | 27.32 | 21.11 | 39.22 | |
| Ramachandran plot | |||||
| Favored (%) | 93.5 | 89.8 | 92.2 | 91.8 | |
| Additionally allowed (%) | 6.5 | 10 | 7.8 | 8.2 | |
| Generously allowed (%) | 0 | 0.2 | 0 | 0 | |
| Disallowed (%) | 0 | 0 | 0 | 0 | |
a Values in parentheses are for the highest resolution shell.
The diffraction data for the SIRT3 apo crystal were collected and processed in the same way as previously described. The crystal is in the space group of P21 with six molecules in the asymmetric unit. The structure was solved by molecular replacement using Molrep (79) with the coordinates of the SIRT3-AceCS2-Kac structure as the search model. Refinement was carried out by Refmac5, and the model building was carried out by COOT (80). Detailed information of the diffraction data, refinement, and structure statistics are listed in Table 1.
The diffraction data for the SIRT3-AceCS2-Ks-ac crystals after NAD+ soaking were collected and processed in the same way as above. The crystals were in the space group of C2221. The structures were solved by molecular replacement using Molrep with the coordinates of the SIRT3-AceCS2-Kac structure as the search model. Refinement was carried out by Refmac5, and model building was carried out by COOT. Detailed information of the diffraction data, refinement, and structure statistics are listed in Table 1.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a VP-ITC system (MicroCal) at 26 °C in a buffer composed of 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 2 mm Tris(2-carboxyethyl)phosphine, 5% (v/v) glycerol, and 50 mm HEPES-NaOH, pH 7.3. The SIRT3-(118–399) protein was purified as above and extensively dialyzed against the buffer, centrifuged, and degassed before the experiment. The AceCS2-Kac peptide and NAD+ were dissolved in the final dialysis buffer, and their pH values were adjusted to match that of the protein solution. Typically, 1 mm AceCS2-Kac peptide or 5 mm NAD+ was injected in 35 aliquots of 8 μl (except the first injection, which was 2 μl) into a 1.4699-ml sample cell containing 100 μm SIRT3. Data were corrected for the heat of dilution and fitted using a nonlinear least-squares routine using a single-site binding model with Origin for ITC v7.0383 (MicroCal) with the stoichiometry (n), the enthalpy of the reaction (ΔH), and the association constant (Ka) calculated.
RESULTS AND DISCUSSION
SIRT3 Protein for Crystallization
Human SIRT3 contains 399 residues with an N-terminal mitochondrial targeting sequence and locates in the mitochondria. The full-length protein is enzymatically inactive. Mitochondrial matrix processing peptidase has been proposed to cleave the N-terminal 101 residues in turn to activate the enzyme (53). Recombinant SIRT3-(102–399) did not crystallize readily. During the course of our studies, we designed several truncations of SIRT3. SIRT3-(118–399) expressed abundant soluble protein in E. coli and could be purified to high purity. In addition, SIRT3-(118–399) and SIRT3-(102–399) had similar deacetylation activity (see below) and responded similarly to a panel of SIRT3 modulators (data not shown). Therefore, we selected SIRT3-(118–399) for crystallization work.
Overall Structure of SIRT3 with the AceCS2 Substrate Peptide (SIRT3-AceCS2-Kac)
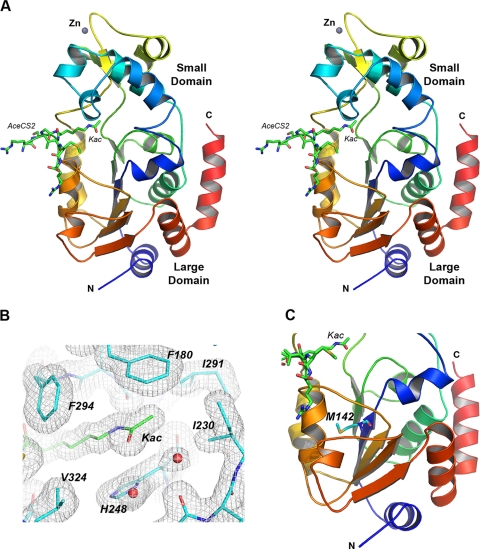
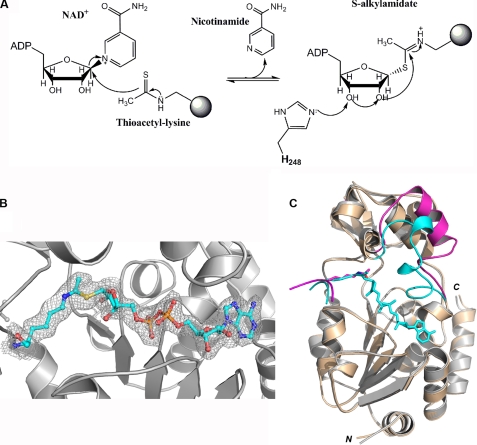
AceCS2 is the natural substrate of SIRT3 in mitochondria (10, 11). We cocrystallized SIRT3 with a 12-mer AceCS2 peptide (638TRSGKacVMRRLLR) containing acetylated lysine 642 that has been identified to be deacetylated by SIRT3. The crystal is in the C2221 space group, with one molecule in the asymmetric unit. Residues 121–394 of SIRT3-(118–399) are visible in the structure, and several residues on the N terminus (residues 118–120) and C terminus (residues 395–399) are disordered. Similar to other sirtuin structures, SIRT3 has a two-domain structure; that is, a large Rossmann fold domain for NAD+ binding and a smaller domain comprising a helical bundle and a zinc binding motif formed by two extending loops from the large domain (Fig. 1A).
FIGURE 1.
Structure of human SIRT3 in complex with the AceCS2-Kac peptide. A, cross-eye stereo view of the SIRT3-AceCS2-Kac structure. SIRT3 is presented in a ribbon representation and colored by a color range from blue of the N terminus to red of the C terminus. The top half of the molecule is the small domain binding to a zinc atom, and the bottom half is the large domain. The acetyl lysine of the AceCS2 peptide inserts in the cleft between the two domains and is in stick representation with carbon atoms in green, oxygen atoms in red, nitrogen atoms in blue, and the sulfur atom in yellow. The N and C termini of SIRT3 are labeled. B, residues surrounding the acetyl lysine binding site are in stick representation with the water molecules presented in red balls. The 2Fo − Fo map is shown in gray wires and contoured at 1 σ. The residues involved in the binding of the acetyl lysine are labeled and discussed under “Results and Discussion.” C, the location of Met-142 in the large domain is indicated.
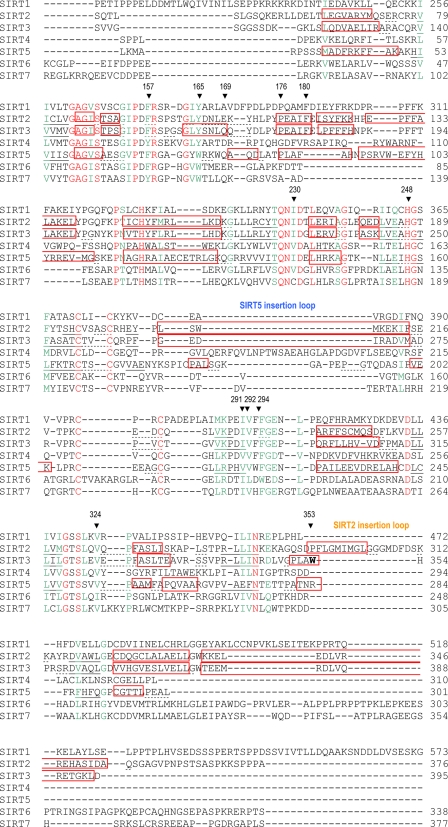
The acetylated lysine of the AceCS2 peptide is in an extended conformation inserting into the cleft between the two domains (Fig. 1A). Three residues on either side of the acetyl lysine (residues 639–645) are visible in the 12-mer AceCS2 peptide, and the residues on the two ends of the peptide (residues 638 and 646–649) are disordered. The main-chain atoms of the AceCS2 peptide form hydrogen bonds with the main-chain atoms of one loop from the small domain (residue 295, 296, and 298) and another loop from the large domain (residue 323 and 325) of the protein, forming an enzyme-substrate anti-parallel β sheet. As shown in Fig. 1B, the aliphatic portion of the acetyl lysine side chain is packed by hydrophobic residues (Phe-294 and Val-324) that are functionally conserved in SIRT1–7 (Fig. 2). Nζ of the acetyl lysine forms a hydrogen bond with the carbonyl oxygen of Val-292, and the acetyl group is sandwiched between His-248 and Phe-180. His-248 is critical for the deacetylation activity of sirtuins (42, 81–83) and absolutely conserved in SIRT1–7, whereas Phe-180 is not conserved. The methyl group on the acetyl lysine is in van der Waals contact with Ile-291 and Ile-230 (both are functionally conserved in SIRT1–7), and the oxygen atom faces the NAD+ binding groove. In addition to the zinc molecule, there are two sulfate, one carbonate, and one glycerol molecules in the structure trapped from the crystallization solution along with 298 water molecules.
FIGURE 2.
Sequence alignment of the core deacetylase domains of human SIRT1–7. The included sequences are SIRT1-207-573 (full-length, 747 a.a.), SIRT2-46-376 (389 a.a.), SIRT3-110-395 (399 a.a.), SIRT4-36-310 (314 a.a.), SIRT5-31-301 (310 a.a.), SIRT6-17-338 (355 a.a.), and SIRT7-67-377 (400 a.a.). The conserved residues are colored in red, and the functionally conserved residues are colored in green. Based on the crystal structures of SIRT2, -3, and -5, the α-helical regions are in red boxes, the β-sheets are underlined with solid lines, and the turn regions are underlined by dashed lines. The residues discussed under “Results and Discussion” are pointed by ▾ above the residues and labeled with the SIRT3 sequence number. The SIRT2 and SIRT5 insertion loops discussed under “Results and Discussion” are labeled. The alignment was done by T-Coffee (90).
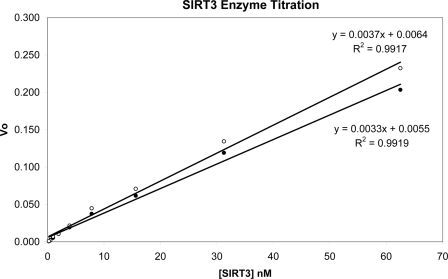
We have determined the deacetylation activities of our recombinant SIRT3-(102–399) and SIRT3-(118–399) proteins (Fig. 3). The two enzymes have similar Km values for the peptide substrate (33.0 μm for SIRT3-(102–399) and 28.7 μm for SIRT3-(118–399)) and for NAD+ (600 μm for SIRT3-(102–399) and 598 μm for SIRT3-(118–399)). When the two proteins were tested side-by-side with excess amounts of peptide substrate and NAD+, they also displayed very similar activity for their deacetylase function (0.0037 for SIRT3-(102–399) and 0.0033 for SIRT3-(118–399) in the unit of percentage-of-conversion of acetylated peptide substrate to deacetylated product over time and enzyme concentration). This is in disagreement with a recent report that SIRT3-(114–399) has a ∼50-fold-higher specific activity than SIRT3-(102–399) (57). Based on their SIRT3 model generated with SIRT2 structure (PDB code 1J8F), they proposed the activity difference was caused by removing the N-terminal short helix that interacts with the C-terminal helix. We found it difficult to obtain crystals for SIRT3-(102–399). Secondary structure prediction from the sequence using several programs suggests that residues 102–117 are in extended conformation, not helical. The cause of the discrepancy of activity between our SIRT3-(118–399) and the published SIRT3-(114–399) is not clear.
FIGURE 3.
Deacetylation activities of SIRT3-(102–399) and SIRT3-(118–399) determined by the mass spectrometry-based assay. The percentage of conversions from peptide substrate to product was determined with excess amounts of peptide substrate and NAD+. Vo is the initial velocity determined at each enzyme concentration in the unit of percentage of conversion of peptide substrate to product over time in minutes. The values of Vo and enzyme concentration were fit to a linear equation. The fitting result and statistics are included. The slope of the curve reflects the specific activity of the enzyme. The closed circle is for SIRT3-(118–399), and the open circle is for SIRT3-(102–399).
Previously, we reported that human SIRT3-142-399 and the equivalent short mouse SIRT3 (containing 257 residues) had poor solubility and no detectable deacetylation activity (84) in vitro. The shorter SIRT3 of human and mouse start with a methionine residue and have been suggested to be the active forms of SIRT3 in the mitochondria (53, 64). This methionine (Met-142 in human SIRT3) is located in the middle of the β-sheet in the large domain (Fig. 1C). Truncation of the residues before Met-142 would disrupt the core β-sheet in the large domain, expose a hydrophobic patch to the solvent that is shielded by the N-terminal helix, and therefore, affect the folding and solubility of the protein. This structure provides insight into why human SIRT3-142-399 and the short mouse SIRT3 are not likely to be the active forms of SIRT3 in mitochondria.
Structural Comparison of the SIRT3 Apo Structure with the SIRT3-AceCS2-Kac Structure
Borra et al. (85) reported that the NAD+-dependent deacetylation activity of Sir2-like enzymes (human SIRT2 and yeast HST2) follows a sequential mechanism with the acetylated substrate binding first followed by NAD+ binding. Although NAD+ has been cocrystallized with several sirtuin enzymes such as Sir2-Af1 and Sir2-Af2 from A. fulgidus (41–43), the presence of acetyl lysine peptide promotes NAD+ to adopt a strained conformation required for the deacetylation activity to proceed (43, 86). The order of binding of substrates for SIRT3 is previously unknown. We obtained the crystal structure of SIRT3 with acetylated AceCS2 peptide showing that SIRT3 can form a stable complex with the substrate peptide in the absence of NAD+. We then tried to cocrystallize SIRT3 in the presence of 10 mm NAD+. The space group of the crystal changed to P21 with six SIRT3 molecules in the asymmetric unit named from chain A to F. When the six molecules were examined, no NAD+ could be observed in the structure. Two of the six molecules have a PEG molecule from the crystallization solution occupying the acetyl lysine binding site (chain C and F). PEG molecules were also observed in the Sir2-AF2 structures previously (38, 43). This result suggests that NAD+ cannot bind to SIRT3 efficiently in the absence of the substrate peptide. NAD+ has been observed in nonproductive binding conformations in the structures of Sir2-Af1 (41) and Sir2-Af2 (42, 43) and productive binding conformation in the structures of Sir2-Af2 (42, 43) and Sir2-Tm (86). The binding of substrate peptide or PEG in the acetyl lysine binding site is required for NAD+ to adopt the productive binding conformation. Some of the NAD+-bound structures were obtained by cocrystallization of the Sir2-NAD+ complex (41, 43) and the others by soaking NAD+ into preformed Sir2-substrate peptide crystals (86). The different modes of NAD+ binding were observed in the same crystal, as well as ADP-ribose-bound structure with no density for nicotinamide (NAM). We attempted to obtain NAD+-bound structure by soaking NAD+ into SIRT3-AceCS2-Kac crystals and SIRT3-H248Y-AceCS2-Kac crystals and were not successful. SIRT3 behaves differently from Sir2-Af1, Sir2-Af2, and Sir2-Tm. The cause of the different responses to NAD+ among different Sir2 proteins needs further investigation.
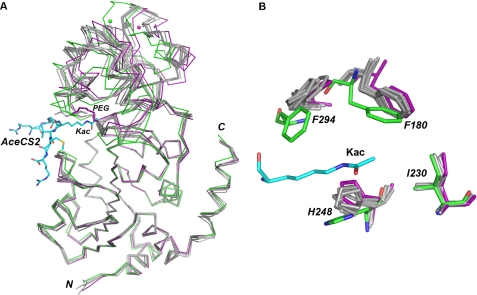
The overall structures of the six molecules are similar to the SIRT3-AceCS2-Kac structure (Fig. 4A). When the Cα atoms of each molecule were aligned with the Cα atoms of SIRT3-AceCS2-Kac, the root mean square deviations (r.m.s.d.) were 1.25, 1.33, 1.41, 1.35, 1.47, and 1.87 Å for chain A to F, respectively. When the Cα atoms of the large domains of the six molecules aligned with the large domain of SIRT3-AceCS2-Kac (residues 138–149, 207–250, and 300–319) individually, the large domain superimposed nicely with an r.m.s.d. of 0.29–0.43 Å. However, the relative position of the small domain was shifted. In the SIRT3-AceCS2-Kac structure, the AceCS2 peptide pulls the two adjacent loops from the large and small domain closer, therefore pulling the two domains closer to each other. In the SIRT3 apo structure, the distance between the two loops that would be involved in the peptide binding increases, and the two domains open up, with molecule F adopting the most open conformation. Although the PEG molecule occupies the acetyl lysine binding site, it cannot form a β sheet with the peptide binding loops to pull the two domains together. This conformational change of SIRT3 induced by substrate peptide binding was observed in Sir2Tm structures (39), which is important for proper NAD+ binding and deacetylation activity.
FIGURE 4.
Structural comparison of the human SIRT3 structures with and without the AceCS2-Kac peptide. A, the superimposition of the Cα traces of the SIRT3-AceCS2-Kac structure and the six molecules (chains A–F) of the SIRT3 apo structure. The SIRT3-AceCS2-Kac structure is colored in green with the AceCS2-Kac peptide in the stick representation with carbon atoms colored in blue. For the SIRT3 apo structure, chain F is colored in purple, chain A-E are in gray, and the PEG molecules are in the stick representation with one in gray binding to chain C and one in purple binding to chain F. The large domains of the SIRT3 molecules are aligned well, and the relative positions of the small domains are shifted. Chain F (purple) of the apo structure is in the most open conformation compared with the SIRT3-AceCS2-Kac structure (green). B, a close up view of the acetyl lysine binding site. The color code is the same as in A. The orientations of His-248 and Phe-180 are shifted in the absence of the acetyl lysine at the binding site.
The residues directly interacting with acetyl lysine were also shifted because of the lack of substrate peptide (Fig. 4B). The ring of His-248 was parallel to the acetyl group of the acetyl lysine in the SIRT3-AceCS2-Kac structure. In the apo structure, the imidazole ring of His-248 rotated to occupy the acetyl group binding site. The ring of Phe-180 was also shifted. The relative orientations of Phe-294 and Ile-230 did not change. In summary, the binding of the substrate peptide to SIRT3 pulls the two domains of SIRT3 structure together and also organizes the residues in the binding site for the deacetylation reaction.
SIRT3 Structure with the Reaction Intermediate Trapped Using an AceCS2 Peptide Containing the Thioacetyl Lysine (Ks-ac)
Thioacetyl lysine is a structural mimic of the acetyl lysine and has been found to inhibit SIRT1, -2, -3, and Hst2 with exceptional potency (87, 88). Because of the presence of the thioacetyl lysine, the reaction stalls at an intermediate step after NAM formation and an ADP-ribose-peptidyl intermediate (S-alkylamidate) was detected by mass spectral analysis (Fig. 5A). Smith and Denu (88) have also shown that Hst2 displayed rapid NAM formation but ∼80 fold slower in overall turnover with a histone H3 peptide containing thioacetyl lysine compared with that containing acetyl lysine. However the thioacetyl lysine peptide is still a substrate of Hst2, allowing the dethioacetylation reaction to proceed. The S-alkylamidate intermediate has been trapped in the crystal structure of Sir2Tm (PDB code 3d81) (40). We crystallized SIRT3 with the same AceCS2 peptide containing a thioacetyl lysine instead of the acetyl lysine. The crystal structure of SIRT3-AceCS2-Ks-ac is the same as that of SIRT3-AceCS2-Kac (data not shown). We soaked the SIRT3-AceCS2-Ks-ac crystals in 0.125 mm NAD+ from 1 to 16 h. Diffraction data were obtained for the crystals with 1- and 16-h soaking times.
FIGURE 5.
Structure of the SIRT3-AceCS2-Ks-ac crystal with a 1-h NAD+ soak (SIRT3-AceCS2-Ks-ac-ADPR). A, the initial step of dethioacetylation reaction by SIRT3. The NAM moiety of NAD+ is released first followed by the ADPR moiety of NAD+ transferred to the thioacetyl lysine, generating the S-alkylamidate. B, the Fo − Fo omit electron density map (1 σ) for the S-alkylamidate intermediate (in stick representation) is presented by gray wires. C, superimposition of the SIRT3-AceCS2-Kac and SIRT3-AceCS2-Ks-ac-ADPR structures. The regions that have similar conformations are colored in gray for the SIRT3-AceCS2-Kac structure and in tinted yellow for the SIRT3-AceCS2-Ks-ac-ADPR structure. The flexible-loop region that has significant conformational difference is highlighted in magenta for the SIRT3-AceCS2-Kac structure and in blue for the SIRT3-AceCS2-Ks-ac-ADPR structure.
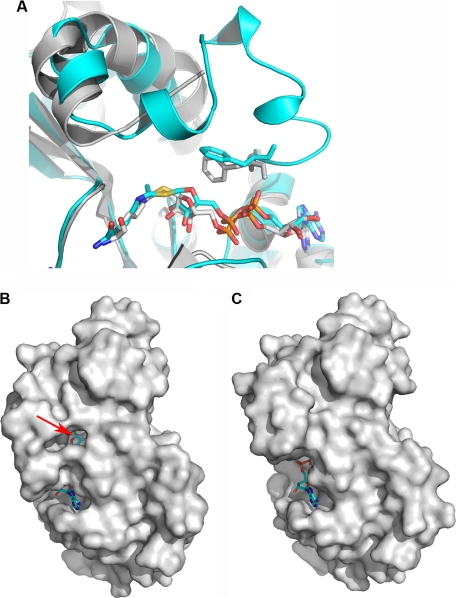
In the structure derived from the 1-h NAD+ soak, the S-alkylamidate intermediate is trapped. The sulfur atom on the thioacetyl lysine of the AceCS2 peptide is covalently bound to the C1 atom of the ribosyl ring on the ADP-ribose (ADPR) from the α face as reported by Sauve and Schramm (89) for the deacetylation reaction (Fig. 5, A and B). There is no electron density for NAM. This is consistent with the result of kinetic studies that show NAM is the first product released (88). We named the structure with the S-alkylamidate intermediate SIRT3-AceCS2-Ks-ac-ADPR. As shown in Fig. 5C, when the large domains of the SIRT3-AceCS2-Ks-ac-ADPR and SIRT3-AceCS2-Kac are superimposed, the r.m.s.d. for the same chosen Cα atoms as above is 0.15 Å, indicating the large domains of the two structures are very similar. In the small domain, a large portion of the structure does not change significantly except for the flexible loop in response to the ADPR binding. The helix in the flexible loop of the SIRT3-AceCS2-Kac structure unwinds itself and folds down to interact with ADPR to shield the S-alkylamidate intermediate from solvent exposure. Most of the other residues that interact with ADPR in the large domain do not change upon ADPR binding, except the side chain of Val-366 rotates to pack against the adenine ring of ADPR. This suggests that the ADPR binding pocket in the large domain is pre-existing, and the structural change of the flexible loop is induced after ADPR binding.
When comparing the SIRT3-AceCS2-Ks-ac-ADPR structure with the Sir2Tm structure (PDB code 3d81), the S-alkylamidate intermediate adopts a similar conformation (Fig. 6A). The flexible loop in the Sir2Tm was disordered. An interesting difference is the orientation of the gate keeper residue, Phe-157 in SIRT3 and the equivalent Phe-33 in Sir2Tm. The side chain of Phe-33 in Sir2Tm is stacked against the ribosyl ring of ADPR face-to-face, which blocks the base-exchange reaction with NAM (40), whereas the side chain of Phe-157 in SIRT3 is oriented with the ribosyl ring edge-to-face, similar to the Phe-33 in the Sir2Tm ternary complex structure with acetylated p53 peptide and NAD+ bound (PDB code 2H4F) (86). Therefore, there is a small opening exposing the β face of the ribosyl ring of ADPR to the solvent in the SIRT3-AceCS2-Ks-ac-ADPR structure (Fig. 6B). The opening does not exist in the SIRT3-AceCS2-Kac structure (Fig. 6C). The β face is the side where NAM could react for the base-exchange reaction (89). This could be the opening for NAM to diffuse away for the deacetylation reaction.
FIGURE 6.
Structural comparison for the S-alkylamidate structures and surface representation of the SIRT3 structures. A, structural comparison of SIRT3-S-alkylamidate intermediate (SIRT3-AceCS2-Ks-ac-ADPR, in blue) with Sir2-Tm-S-alkylamidate intermediate (PDB code 3d81, in gray). The carbon atoms of the gatekeeper phenylalanine residues and the S-alkylamidate are in stick representation and colored in the same color as the respective structures. The phenyl ring of Phe-157 in SIRT3 is perpendicular relative to the ribosyl ring of the ADPR, whereas that of Phe-33 in Sir2Tm is in parallel. B, the SIRT3-AceCS2-Ks-ac-ADPR structure is presented in a surface representation, showing the opening to the β face of the ribosyl ring of ADPR that is in stick representation. The arrow points to the C1 atom of the ribosyl ring of ADPR where NAM connects to in NAD+. C, the SIRT3-AceCS2-Kac structure is presented in a surface representation in the same orientation as in B. ADPR is left in the structure to serve as a reference for the comparison between C and B.
Thioacetyl AceCS2 Peptide Is a Substrate for SIRT3
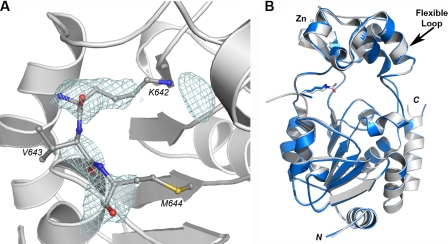
Although we trapped the S-alkylamidate intermediate in the SIRT3-AceCS2-Ks-ac crystal with a 1 h NAD+ soak, another crystal that soaked in NAD+ overnight yielded a structure with weak density for the AceCS2 peptide with the thioacetyl group removed (Fig. 7A). Nζ of the dethioacetylated lysine is disordered. There is a globular electron density near the tip of the lysine. However, the density is disconnected from the density for the dethioacetylated lysine, and a water molecule fits well during refinement. This is structural evidence that the AceCS2-Ks-ac peptide is still a substrate for SIRT3, consistent with the report that histone H3 peptide with thioacetyl lysine could be dethioacetylated by Hst2 (88). It is interesting that the reaction is carried out in the crystal with the flexible loop changed back to the original conformation that is observed in the SIRT3-AceCS2-Kac structure (Fig. 7B) after the dethioacetyl reaction. The deacetylated peptide has been captured previously in the structure of the Sir2Tm-acetylated p53 peptide crystal soaked in NAD+ for 16 h (86). However, this is the first time that the conformational changes of the flexible loop have been captured by a set of crystal structures representing the different stages of the reaction.
FIGURE 7.
Structure of the SIRT3-AceCS2-Ks-ac crystal with 16 h NAD+ soak. A, the Fo − Fo omit electron density map (2 σ) for the dethioacetylated AceCS2 (in stick representation) is presented by the blue wires. B, the structure of the SIRT3-AceCS2-Ks-ac crystal with 16 h NAD+ soak (in blue) is superimposed with the structure of SIRT3-AceCS2-Kac (in gray). The arrow points to the flexible loops that are superimposed well between the two structures.
Substrate Binding Study by Isothermal Titration Calorimetry
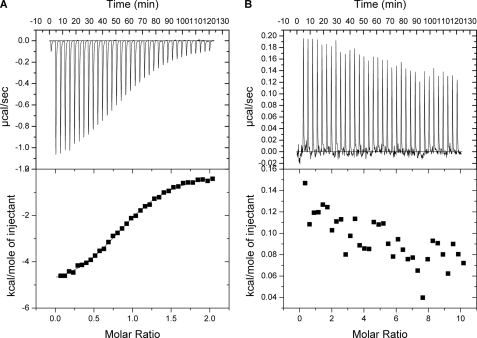
To determine the binding order for the substrate peptide and NAD+ toward SIRT3, ITC experiments were performed by titrating the AceCS2-Kac peptide or NAD+ to SIRT3. Fig. 8A shows that the AceCS2-Kac peptide binds to SIRT3. The data were fit to a one-site binding model with a binding constant of 15 μm, and binding stoichiometry was determined to be one. NAD+ did not bind to SIRT3 directly (Fig. 8B). Taken together the data suggest that the substrate peptide binds first to SIRT3 followed by NAD+ binding. The ITC result is in agreement with the structural information we obtained for SIRT3.
FIGURE 8.
Isothermal titration calorimetry study of the substrate binding to SIRT3. A, the sequential heat pulses and the integrated heat data for the AceCS2-Kac peptide titrating to SIRT3 show a direct binding fitted to a single-site binding model. B, the sequential heat pulses and the integrated heat data for the NAD+ titrating to SIRT3 show no direct binding.
Structural Comparison for Human SIRT3-AceCS2-Kac, SIRT2 (PDB Code 1J8F), and SIRT5 (PDB Code 2B4Y)
The core structure for deacetylase activity is conserved in SIRT2, SIRT3, and SIRT5 (Fig. 9 and Fig. 2). However, the conformation of the flexible loop is different in each structure. The flexible loop contains NAD+ binding residues. A small region of the flexible loop (residues 68–74) is disordered in the ADPR-bound SIRT5 structure (PDB code 2B4Y) (51). However, the flexible loop is ordered and interacts directly with suramin in the SIRT5 structure with the inhibitor suramin bound (PDB code 2NYR) (51). The changes of the flexible loop are also observed in the set of SIRT3 structures we present here. Therefore, the conformation of the flexible loop is determined by what binds in the substrate binding pocket.
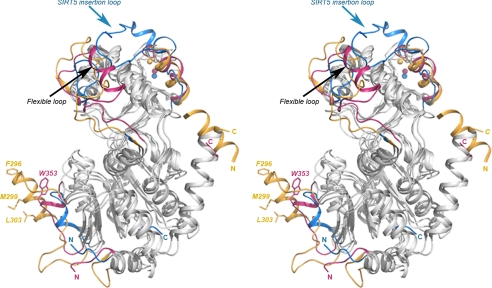
FIGURE 9.
Structure comparison of SIRT3-AceCS2-Kac with SIRT2 (PDB code 1J8F) and SIRT5 (PDB code 2B4Y). The structurally conserved regions are colored in gray, and the structurally divergent regions are highlighted in different colors with SIRT3 in pink, SIRT2 in gold, and SIRT5 in blue. The black arrow points to the flexible loop region. Part of the flexible loop in SIRT5 is disordered. The blue arrow points to the insertion loop in SIRT5 that covers the top of the helix bundle region of the small domain. SIRT2 (in gold) has an insertion loop in the large domain that contains a helix with a hydrophobic surface formed by residues Phe-296, Met-299, and Leu-303 (in stick representation) and a unique N-terminal helix (there is a disordered loop after the N-terminal helix). Trp-353 of SIRT3 is presented in stick representation colored in pink. The N and C termini of all the structures are colored and labeled with letters in the respective colors.
In addition, there are structural divergences on the surface of the proteins away from the deacetylation site. SIRT5 has an insertion in the zinc binding motif (Fig. 2) extending over to cover the surface of the helical bundle region of the small domain (Fig. 9). This insertion loop of SIRT5 blocks a groove between the zinc binding motif and the helical bundle region that exists in the small domain of the SIRT2 and SIRT3 structures. In the SIRT3 apo structure, this groove is involved in crystal packing; therefore, it could be a potential protein-protein interaction site. SIRT2 has a unique N-terminal helix that is absent in the SIRT3 and SIRT5 structures and essential for the crystal packing in SIRT2 structure (50). SIRT2 also has an insertion in the large domain near the position that Trp-353 locates in SIRT3 (labeled in Fig. 9). The α-helix in this region of SIRT2 has a hydrophobic surface formed by Phe-296, Met-299, Ile-300, and Leu-303. This hydrophobic surface of SIRT2 inserts into the binding site of the acetyl lysine in a symmetry-related molecule with Phe-296 occupying the space where the acetyl group binds. Trp-353 of SIRT3 in this region protrudes on the surface in the SIRT3-AceCS2-Kac and is involved in crystal packing in SIRT3 apo-structure. However, the SIRT5 structure shows a hydrophilic surface at this region. Although the protein-protein interaction observed in the SIRT2 and SIRT3 apo structures could be a crystal packing artifact, the unique features on the surface of each protein could be important for its substrate specificity and/or protein-protein interaction.
Previously we have determined that residues 118–225 of SIRT1 are important for the function of the SIRT1 activators (32). SIRT3 does not contain the corresponding region. However, the SIRT3-(118–399) protein for this crystallization work has deacetylation activity and can be activated and inhibited by SIRT3 modulators. This suggests that SIRT3 modulation could be different from SIRT1. We are in the process of obtaining the SIRT3 structures with a SIRT3 modulator bound to identify the modulator binding site(s). The SIRT3 structures we reported here shed light on the structure-function relationship of SIRT3 deacetylation activity. It is an important first step toward structure-based ligand design of SIRT3 modulators for drug discovery.
Acknowledgment
We thank Yu Xia for the support of coordinating cloning and protein purification effort.
The atomic coordinates and structure factors (codes 3GLR, 3GLS, 3GLT, and 3GLU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AceCS2
- acetyl-CoA synthetase 2
- ADPR
- ADP ribose
- CR
- caloric restriction
- ITC
- isothermal titration calorimetry
- Kac
- acetyl lysine
- Ks-ac
- thioacetyl lysine
- NAM
- nicotinamide
- PEG
- polyethylene glycol
- SIRT3-AceCS2-Kac
- SIRT3 crystal structure with AceCS2 peptide containing acetyl lysine
- SIRT3-AceCS2-Ks-ac
- SIRT3 crystal structure with AceCS2 peptide containing thioacetyl lysine
- SIRT3-AceCS2-Ks-ac-ADPR
- SIRT3 crystal structure with AceCS2 peptide containing thioacetyl lysine that binds to ADPR
- Ni-NTA
- nickel-nitrilotriacetic acid
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- r.m.s.d.
- root mean square deviations
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Dutnall R. N., Pillus L. (2001) Cell 105, 161–164 [DOI] [PubMed] [Google Scholar]
- 2.North B. J., Verdin E. (2004) Genome Biology 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., Sternglanz R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004) Mol. Cell 16, 93–105 [DOI] [PubMed] [Google Scholar]
- 5.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 6.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 7.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 8.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 9.North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003) Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 10.Hallows W. C., Lee S., Denu J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 13.Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 15.Liszt G., Ford E., Kurtev M., Guarente L. (2005) J. Biol. Chem. 280, 21313–21320 [DOI] [PubMed] [Google Scholar]
- 16.Sauve A. A., Schramm V. L. (2004) Curr. Med. Chem. 11, 807–826 [DOI] [PubMed] [Google Scholar]
- 17.Rusche L. N., Kirchmaier A. L., Rine J. (2003) Annu. Rev. Biochem. 72, 481–516 [DOI] [PubMed] [Google Scholar]
- 18.Dryden S. C., Nahhas F. A., Nowak J. E., Goustin A. S., Tainsky M. A. (2003) Mol. Cell. Biol. 23, 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straight A. F., Shou W., Dowd G. J., Turck C. W., Deshaies R. J., Johnson A. D., Moazed D. (1999) Cell 97, 245–256 [DOI] [PubMed] [Google Scholar]
- 20.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 21.Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007) Mol. Cell 28, 91–106 [DOI] [PubMed] [Google Scholar]
- 22.Schwer B., Verdin E. (2008) Cell Metab. 7, 104–112 [DOI] [PubMed] [Google Scholar]
- 23.Langley E., Pearson M., Faretta M., Bauer U. M., Frye R. A., Minucci S., Pelicci P. G., Kouzarides T. (2002) EMBO J. 21, 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 25.Blander G., Guarente L. (2004) Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 26.Haigis M. C., Guarente L. P. (2006) Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 27.Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., Schibler U. (2008) Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- 28.Belden W. J., Dunlap J. C. (2008) Cell 134, 212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimaldi B., Nakahata Y., Kaluzova M., Masubuchi S., Sassone-Corsi P. (2009) Int. J. Biochem. Cell Biol. 41, 81–86 [DOI] [PubMed] [Google Scholar]
- 30.Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. (2008) Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milne J. C., Denu J. M. (2008) Curr. Opin. Chem. Biol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 32.Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garske A. L., Smith B. C., Denu J. M. (2007) ACS Chem. Biol. 2, 529–532 [DOI] [PubMed] [Google Scholar]
- 34.Qin W., Yang T., Ho L., Zhao Z., Wang J., Chen L., Zhao W., Thiyagarajan M., MacGrogan D., Rodgers J. T., Puigserver P., Sadoshima J., Deng H., Pedrini S., Gandy S., Sauve A. A., Pasinetti G. M. (2006) J. Biol. Chem. 281, 21745–21754 [DOI] [PubMed] [Google Scholar]
- 35.Rajendrasozhan S., Yang S. R., Kinnula V. L., Rahman I. (2008) Am. J. Respir. Crit. Care Med. 177, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firestein R., Blander G., Michan S., Oberdoerffer P., Ogino S., Campbell J., Bhimavarapu A., Luikenhuis S., de Cabo R., Fuchs C., Hahn W. C., Guarente L. P., Sinclair D. A. (2008) PLoS ONE 3, e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders L. R., Verdin E. (2007) Oncogene 26, 5489–5504 [DOI] [PubMed] [Google Scholar]
- 38.Avalos J. L., Bever K. M., Wolberger C. (2005) Mol. Cell 17, 855–868 [DOI] [PubMed] [Google Scholar]
- 39.Cosgrove M. S., Bever K., Avalos J. L., Muhammad S., Zhang X., Wolberger C. (2006) Biochemistry 45, 7511–7521 [DOI] [PubMed] [Google Scholar]
- 40.Hawse W. F., Hoff K. G., Fatkins D. G., Daines A., Zubkova O. V., Schramm V. L., Zheng W., Wolberger C. (2008) Structure 16, 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Min J., Landry J., Sternglanz R., Xu R. M. (2001) Cell 105, 269–279 [DOI] [PubMed] [Google Scholar]
- 42.Chang J. H., Kim H. C., Hwang K. Y., Lee J. W., Jackson S. P., Bell S. D., Cho Y. (2002) J. Biol. Chem. 277, 34489–34498 [DOI] [PubMed] [Google Scholar]
- 43.Avalos J. L., Boeke J. D., Wolberger C. (2004) Mol. Cell 13, 639–648 [DOI] [PubMed] [Google Scholar]
- 44.Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., Wolberger C. (2002) Mol. Cell 10, 523–535 [DOI] [PubMed] [Google Scholar]
- 45.Zhao K., Chai X., Marmorstein R. (2004) J. Mol. Biol. 337, 731–741 [DOI] [PubMed] [Google Scholar]
- 46.Sanders B. D., Zhao K., Slama J. T., Marmorstein R. (2007) Mol. Cell 25, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao K., Chai X., Clements A., Marmorstein R. (2003) Nat. Struct. Biol. 10, 864–871 [DOI] [PubMed] [Google Scholar]
- 48.Zhao K., Chai X., Marmorstein R. (2003) Structure 11, 1403–1411 [DOI] [PubMed] [Google Scholar]
- 49.Zhao K., Harshaw R., Chai X., Marmorstein R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8563–8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finnin M. S., Donigian J. R., Pavletich N. P. (2001) Nat. Struct. Biol. 8, 621–625 [DOI] [PubMed] [Google Scholar]
- 51.Schuetz A., Min J., Antoshenko T., Wang C. L., Allali-Hassani A., Dong A., Loppnau P., Vedadi M., Bochkarev A., Sternglanz R., Plotnikov A. N. (2007) Structure 15, 377–389 [DOI] [PubMed] [Google Scholar]
- 52.Onyango P., Celic I., McCaffery J. M., Boeke J. D., Feinberg A. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13653–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwer B., North B. J., Frye R. A., Ott M., Verdin E. (2002) J. Cell Biol. 158, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper H. M., Spelbrink J. N. (2008) Biochem. J. 411, 279–285 [DOI] [PubMed] [Google Scholar]
- 55.Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brosnan J. T. (2000) J. Nutr. 130, 988S–990S [DOI] [PubMed] [Google Scholar]
- 57.Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 58.Lakowski B., Hekimi S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mair W., Goymer P., Pletcher S. D., Partridge L. (2003) Science 301, 1731–1733 [DOI] [PubMed] [Google Scholar]
- 60.Jiang J. C., Jaruga E., Repnevskaya M. V., Jazwinski S. M. (2000) FASEB J. 14, 2135–2137 [DOI] [PubMed] [Google Scholar]
- 61.Weindruch R., Walford R. L. (1982) Science 215, 1415–1418 [DOI] [PubMed] [Google Scholar]
- 62.Ramsey J. J., Colman R. J., Binkley N. C., Christensen J. D., Gresl T. A., Kemnitz J. W., Weindruch R. (2000) Exp. Gerontol. 35, 1131–1149 [DOI] [PubMed] [Google Scholar]
- 63.Qin W., Chachich M., Lane M., Roth G., Bryant M., de Cabo R., Ottinger M. A., Mattison J., Ingram D., Gandy S., Pasinetti G. M. (2006) J. Alzheimers Dis. 10, 417–422 [DOI] [PubMed] [Google Scholar]
- 64.Shi T., Wang F., Stieren E., Tong Q. (2005) J. Biol. Chem. 280, 13560–13567 [DOI] [PubMed] [Google Scholar]
- 65.Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 66.Hagopian K., Ramsey J. J., Weindruch R. (2003) Exp. Gerontol. 38, 267–278 [DOI] [PubMed] [Google Scholar]
- 67.Lanza I. R., Short D. K., Short K. R., Raghavakaimal S., Basu R., Joyner M. J., McConnell J. P., Nair K. S. (2008) Diabetes 57, 2933–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bellizzi D., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Franceschi C., Passarino G., De Benedictis G. (2005) Genomics 85, 258–263 [DOI] [PubMed] [Google Scholar]
- 69.Rose G., Dato S., Altomare K., Bellizzi D., Garasto S., Greco V., Passarino G., Feraco E., Mari V., Barbi C., BonaFe M., Franceschi C., Tan Q., Boiko S., Yashin A. I., De Benedictis G. (2003) Exp. Gerontol. 38, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 70.Zhang W., Della-Fera M. A., Hartzell D. L., Hausman D., Baile C. A. (2008) Life Sci. 83, 35–42 [DOI] [PubMed] [Google Scholar]
- 71.Allison S. J., Milner J. (2007) Cell Cycle 6, 2669–2677 [DOI] [PubMed] [Google Scholar]
- 72.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 74.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 75.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 BBSRC Collaborative Computational Project Number 4 on Protein Crystallography, Daresbury Laboratory, Daresbury, Warrington, UK [Google Scholar]
- 76.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 77.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 78.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 79.Vagin A. A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 80.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 81.Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003) J. Biol. Chem. 278, 50985–50998 [DOI] [PubMed] [Google Scholar]
- 82.Smith B. C., Denu J. M. (2006) Biochemistry 45, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., Schramm V. L. (2001) Biochemistry 40, 15456–15463 [DOI] [PubMed] [Google Scholar]
- 84.Jin L., Galonek H., Israelian K., Choy W., Morrison M., Xia Y., Wang X., Xu Y., Yang Y., Smith J. J., Hoffmann E., Carney D. P., Perni R. B., Jirousek M. R., Bemis J. E., Milne J. C., Sinclair D. A., Westphal C. H. (2009) Protein Sci. 18, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 86.Hoff K. G., Avalos J. L., Sens K., Wolberger C. (2006) Structure 14, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 87.Fatkins D. G., Monnot A. D., Zheng W. (2006) Bioorg. Med. Chem. Lett. 16, 3651–3656 [DOI] [PubMed] [Google Scholar]
- 88.Smith B. C., Denu J. M. (2007) Biochemistry 46, 14478–14486 [DOI] [PubMed] [Google Scholar]
- 89.Sauve A. A., Schramm V. L. (2003) Biochemistry 42, 9249–9256 [DOI] [PubMed] [Google Scholar]
- 90.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]