Abstract
Morphine is a powerful analgesic natural product produced by the opium poppy Papaver somniferum. Although formal syntheses of this alkaloid have been reported, the morphine molecule contains five stereocenters and a C-C phenol linkage that to date render a total synthesis of morphine commercially unfeasible. The C-C phenol-coupling reaction along the biosynthetic pathway to morphine in opium poppy is catalyzed by the cytochrome P450-dependent oxygenase salutaridine synthase. We report herein on the identification of salutaridine synthase as a member of the CYP719 family of cytochromes P450 during a screen of recombinant cytochromes P450 of opium poppy functionally expressed in Spodoptera frugiperda Sf9 cells. Recombinant CYP719B1 is a highly stereo- and regioselective enzyme; of forty-one compounds tested as potential substrates, only (R)-reticuline and (R)-norreticuline resulted in formation of a product (salutaridine and norsalutaridine, respectively). To date, CYP719s have been characterized catalyzing only the formation of a methylenedioxy bridge in berberine biosynthesis (canadine synthase, CYP719A1) and in benzo[c]phenanthridine biosynthesis (stylopine synthase, CYP719A14). Previously identified phenol-coupling enzymes of plant alkaloid biosynthesis belong only to the CYP80 family of cytochromes. CYP719B1 therefore is the prototype for a new family of plant cytochromes P450 that catalyze formation of a phenol-couple.
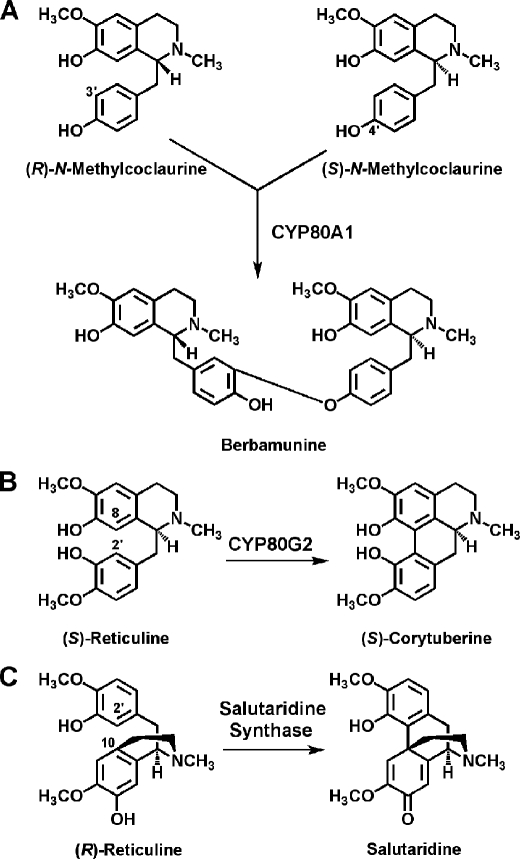
The C-O or C-C phenol-couple is widely present in the plant kingdom in natural product biosynthetic processes such as alkaloid (1), lignan (2), lignin (3), and gallotannin (4) formation. Phenol-coupling reactions in nature were thought to be catalyzed by a variety of oxidative enzymes with broad substrate specificity such as peroxidases, polyphenol oxidases, and laccases. More recently, several enzymes discovered to be responsible for the formation of intermolecular C-O phenol and intramolecular C-C phenol-couples were found to be highly regio- and/or stereoselective catalysts. The first intermolecular C-O phenol-coupling enzyme identified was the cytochrome P450-dependent oxidase berbamunine synthase (CYP80A1) of bisbenzylisoquinoline alkaloid biosynthesis in Berberis cell cultures (5, 6) (Fig. 1). This enzyme is regiospecific, but will accept either (R)- and (S)-N-methylcoclaurine to form R-R and R-S phenol-coupled products. Absolute regio- and stereospecificity is demonstrated in the formation of the lignan (+)-pinoresinol from two molecules of coniferyl alcohol, a reaction guided by dirigent proteins that can be catalyzed by a range of oxidases or oxidants (7). The aporphine alkaloid intramolecular C-C phenol-couple is catalyzed in Coptis japonica cell cultures by the cytochrome P450-dependent oxidase CYP80G2; this enzyme accepts six tetrahydrobenzylisoquinoline alkaloids as substrate (8).
FIGURE 1.
Selected phenol-coupling reactions of alkaloid biosynthesis. Berbamunine synthase (CYP80A1) catalyzes the C-O intermolecular phenol-coupling reaction of bisbenzyisoquinoline alkaloid biosynthesis. (S)-Corytuberine synthase (CYP80G2) catalyzes formation of the intramolecular C-C phenol-couple in magnoflorine biosynthesis. Salutaridine synthase forms the C-C intramolecular phenol-couple of salutaridine in morphine biosynthesis.
Morphine has often been described as the king of alkaloids. Although formal syntheses of this powerful analgesic have been reported, yields are low (Ref. 9 and references therein); attempts in organic chemistry to mimic the biosynthetic formation of the C-C phenol-couple of salutaridine (Fig. 1) have been either unsuccessful, yielding rather isoboldine or pallidine (10), or have resulted in very low yield of salutaridine (11) or in a mixture of isoboldine and salutaridine, with the reaction favoring formation of isoboldine by a factor of ∼5 (12). Along with the five stereocenters present in this molecule, the C-C phenol-couple renders a chemical synthesis of morphine commercially unfeasible. The enzyme catalyzing this reaction in planta was sought unsuccessfully for many years and was discovered finally in the opium poppy Papaver somniferum to be a cytochrome P450-dependent oxidase that stereo- and regiospecifically produces salutaridine by C-C phenol-coupling of (R)-reticuline (Fig. 1) (1, 13). The native enzyme salutaridine synthase was unstable, which precluded protein purification for further characterization.
Herein, we describe the identification and functional expression of opium poppy salutaridine synthase, a member of the cytochrome P450 family, in Spodoptera frugiperda Sf9 cells. The recombinant enzyme was sufficiently stable in insect cell culture to be characterized with respect to substrate specificity and steady state kinetic values. Recombinant salutaridine synthase converted (R)-reticuline exclusively to salutaridine and (R)-norreticuline exclusively to norsalutaridine (N-demethylsalutaridine).
EXPERIMENTAL PROCEDURES
Plant Material and Natural Products
P. somniferum L. plants were grown outdoors in summer in either Munich or Halle, Germany. Stem material (2 cm immediately below capsule) was excised within 1–3 days after petal fall, immediately frozen in liquid nitrogen, and stored at −80 °C until extracted. Total RNA was extracted as previously described (14). The natural products used in this study were from the collection of Prof. Meinhart H. Zenk (Donald Danforth Plant Science Center).
Generation of Cytochrome P450 Gene Fragments
Four degenerate oligodeoxynucleotide primers corresponding to highly conserved regions of cytochromes P450 (CYP75A3 (15), CYP80A1 (6), CYP80B1 (16), CYP51 (17), CYP71A1 (18), and CYP71A4 (19)) were used for reverse transcriptase-polymerase chain reaction (RT-PCR)6 to amplify cDNA fragments from P. somniferum RNA. FCS1, which covers the motif YGP(K)Y(L)W, is located near the N terminus of the cytochrome P450, whereas FCS2, FCS3, and FCAS, which cover the motifs EWVMSLL, EEFRPE, and PFGAGRIICP, respectively, are close to the C terminus of the protein. The oligodeoxynucleotide primer sequences were as follows: FCS1 (forward) 5′-TAC/T GGIA/C A/C ITAC/T TGG-3′, FCS2 forward (5′-GAA/G TGGGTIATGT/A C/G IT/C TIT/C TI-3′), FCS3 forward (5′-GAA/G GAA/G TTC/T A/C GICCIGAA/G A/C G-3′), FCAS reverse (5′-CCIGGA/G CAIATIA/C T/G C/T C/T TICCIGCICCA/G AAIGG-3′). First strand cDNA was synthesized from 10 μg of total RNA isolated from stems of P. somniferum. Either FCS1, FCS2, or FCS3 as forward primers and FCAS as reverse primer were used to amplify cDNA fragments of ∼1000, 400, and 120 bp, respectively. The temperature program used was 2 min 94 °C, 1 cycle; 45 s 94 °C, 45 s 50 °C, 1 min 72 °C, 30 cycles; final extension at 72 °C for 10 min. The polymerase chain reaction (PCR) products were ligated into pGEM-T or pGEM-T Easy vector (Promega). 119 randomly selected clones (78 clones of 120 bp, 15 clones of 400 bp, and 26 clones of 1000 bp) were sequenced and a homology search was performed using TFASTA (HUSAR 4.0) to ascertain their identity as putative cytochromes P450. 25 of 28 unigenes were found homologous to cytochrome P450 sequences. Each of these 25 cDNA fragments were obtained as either full-length or near full-length clones by screening a primary cDNA library prepared from P. somniferum stem RNA in lambda ZAPII according to the manufacturer's instructions (Stratagene). The cDNAs were amplified by PCR and were ultimately introduced into the vector pCR2.1 (Invitrogen) for further study.
Transcript Accumulation Profile
P. somniferum suspension cell culture elicitation were performed as described by Ref. 20. Papaver interspecies comparison and macroarray preparation, hybridization, and evaluation were carried out exactly according to Refs. 21, 22.
Construction of Expression Vectors for CYP719B1 and CPR cDNAs
Full-length P. somniferum CYP719B1 cDNA was generated by amplification out of bacterial vector pCR2.1 by PCR and inserted into baculovirus transfer vector pVL1392 using primers that introduced XbaI and BamHI recognition sites (underlined) into the 5′- and 3′-ends, respectively, and are as follows: forward (5′-GGGTCTAGAATGGCTCCGATTAATATAGAGGGG-3′) and reverse (5′-GGGGGATCCCTACTGTCGAAAAGGTTTTGTACG-3′). Pfu Hotstart polymerase (Stratagene) was used for PCR amplification with the following conditions: 3 min at 94 °C, succeeded by 30 cycles of amplification (30 s at 94 °C, 30 s at 52 °C, 2 min 30 s at 72 °C), and final extension of 5 min at 72 °C with a total reaction volume of 50 μl. The PCR products along with pVL1392 were subjected to digestion with XbaI and BamHI, followed by gel purification and ligation with T4 DNA ligase (Promega). Transformation and recombinant vector amplification were performed with BP5α Competent Cells (BioPioneer). Full-length Eschscholzia californica cytochrome P450 reductase (CPR) cDNA was developed with PstI and XbaI sites on the 5-′ and 3′-ends, respectively, by amplification from the vector pZL1 and insertion into pVL1392 using the following primers: forward (5′-GGGCTGCAGATGGAACAAACTGCGGTTAAA-3′) and reverse (5′GGGTCTAGATCACCACACATCACGTAGATA-3′). Primers used to amplify the petunia CPR cDNA from pFastBac were: forward (5′-GGGTCTAGAATGGAGTCGAGTTCGTCGGAG-3′) and reverse (5′-GGGGGATCCTCACCACACATCCCTGAGATATC-3′) containing XbaI and BamHI restriction sites, respectively. Primers used to amplify the Arabidopsis thaliana CPR cDNA from pFastBac were designed with the incorporation of BglII and EcoRI restriction sites and were as follows: forward (5′-GGGAGATCTATGACTTCTGCTTTGTATGC-3′) and reverse (5′-GGGGAATTCTCACCAGACATCTCTGAGG-3′). Subsequent procedures were identical to that of CYP719B1.
Transfection and Virus Amplification
Both CYP719B1 and CPR independently underwent homologous recombination with Baculogold Linearized Baculovirus DNA using a BaculoGold Transfection Kit (BD Biosciences) and S. frugiperda Sf9 cells according to the manufacturer's instructions. Sf9 cells were grown in TC-100 medium (Sigma) supplemented with 10% (v/v) fetal bovine serum (FBS, Hyclone) and were regularly maintained as previously described (23). Viral amplification was achieved by adding 100 μl of Sf9 cell spent medium containing recombinant virus to 1 ml of uninfected cells (three rounds of amplification, 27 °C, 4–5 days of incubation each) in a 24-well plate and then adding 100 μl of Sf9 cell spent medium containing virus into 5 ml of uninfected cells grown in a T-25 flask. Final amplification followed by growing 50 ml of Sf9 cells in a 100 ml flask (3–4 days, 27 °C at 140 rpm) until a density of 2 × 106 cells/ml was reached. Cells were centrifuged (900 × g, 10 min, room temperature), resuspended in 7.5 ml of suspension medium (TC-100/10% (v/v) fetal bovine serum, 0.1% (w/v) pluronic (Sigma)), and combined with 2.5 ml of supernatant from the fourth amplification. Cells were allowed to incubate for 1 h (27 °C, 80 rpm), after which an additional 40 ml of suspension medium was added. The flask was incubated for 3 days (27 °C, 140 rpm), and cells were collected by centrifugation (3,000 × g, 10 min, 4 °C). The supernatant (virus stock) was stored at 4 °C and subsequently subjected to verification by DNA isolation and PCR analysis for the presence of the transgene.
Infection for Protein Production
Sf9 cells were grown, collected, and resuspended in the same manner as the final amplification step (described above) and transferred to a new 50-ml flask to which 2.5 ml of virus containing either the CYP719B1 or CPR cDNA (single infection), or 1.25 ml of virus containing CYP719B1 cDNA and 1.25 ml of virus containing CPR cDNA (double infection) were added. Incubation was allowed to proceed for 1 h (27 °C, 80 rpm), followed by the addition of 40 ml of suspension medium supplemented with 50 μl of hemin (2 mg/ml). Infected cells were incubated for 3 days (27 °C, 140 rpm) and collected by centrifugation (3,000 × g, 10 min, 4 °C). The pellet was washed twice with 10 ml of PBS buffer (130 mm NaCl, 7 mm Na2HPO4, 3 mm NaH2PO4, pH 7.4), resuspended in 3.5 ml of suspension buffer (100 mm Tricine/NaOH buffer (pH 7.4) containing 5 mm thioglycolic acid) and frozen in 1-ml aliquots at −80 °C.
Quantitation of CYP719B1 Protein
Insect cells (750 ml) expressing CYP719B1 and CPR were collected by centrifugation (1,000 × g for 5 min at room temperature) and washed twice with 40 ml of phosphate-buffered saline buffer. Cells were then resuspended in 30 ml of ice-cold suspension buffer. Half were frozen in liquid nitrogen, and the remaining 15 ml were sonicated four times on ice (10× pulsed, 50 watts) while being protected from light. Cellular debris was removed by centrifugation (8,000 × g for 20 min at 4 °C), and microsomes were collected by subjecting the supernatant to ultracentrifugation (107,000 × g for 65 min at 4 °C). Microsomes were carefully homogenized in 1.5 ml of ice-cold suspension buffer until the solution was uniform. The reduced CO difference spectrum of P450 heme protein was measured with a Cary 300 UV-visible spectrophotometer on whole microsomes after a 3-fold dilution with suspension buffer. The resulting spectrum was used to determine microsomal P450 protein concentration using an extinction coefficient of 91 cm−1 mm−1.
Recombinant CYP719B1 protein quantitation was also performed by running a series of bovine serum albumin standards (200, 400, 600, 800, 1000, and 1,200 ng) against 10 μl of cell suspension on a 10% SDS-PAGE gel. Staining was allowed to proceed for 30 min in Coomassie Brilliant Blue G-250. Destaining solution (25% (v/v) methanol, 7.5% (v/v) acetic acid) was applied to the gel for 1 h, then fresh solution was added, and the gel was destained overnight. A Gelpix scanner (Genetix) coupled to Phoretix 1D software was used to photograph the gel and quantify the bands.
Enzyme Assay for Analysis of Substrate Specificity
Because of the poor aqueous solubility of a number of the substrates tested, the enzyme assays performed on various natural products (Table 1) as potential substrates were carried out under mildly acidic conditions and contained 30 mm potassium phosphate buffer (pH 6.5), 1.25 mm NADPH, 0.25 mm EDTA, 70 μl of hypotonically lysed Sf9 cell suspension (expressing both CYP719B1 (∼3 μg) and CPR) and 5 μm substrate in a final volume of 200 μl. Control assays without enzyme, without NADPH, with only CYP719B1 or only CPR were also analyzed. Each reaction was allowed to proceed for 2 h at 37 °C and was terminated by chloroform extraction. Chloroform extraction was performed in basic conditions with the addition of 400 μl of 1 m sodium carbonate buffer (pH 9.5) and 400 μl of chloroform, followed by rapid mixing for 1 min, and centrifugation (2 min at 13,000 rpm). The organic layer was collected and extraction was repeated once. The combined organic phase was dried under N2, and the sample resuspended in 200 μl of 80% methanol.
TABLE 1.
Substrate specificity of recombinant CYP719B1
Enzyme assays were analyzed by LC-MS/MS. In the mass spectrum, products were sought that were substrate molecular ion minus 2 (phenol-coupling or methylenedioxy bridge formation); minus 14 (demethylation) and plus 16 (hydroxylation). The chemical structures are provided in supplemental Fig. S1.
| Substrate | Conversion kcat |
|---|---|
| % | |
| Isoquinoline alkaloids | |
| Cheilanthifoline | nda |
| (S)-Coclaurine | nd |
| Codeine | nd |
| (S)-Coreximine | nd |
| (R,S)-3'-Hydroxy-N-methylcoclaurine | nd |
| (R,S)–Isococlaurine | nd |
| (R,S)–Isoorientaline | nd |
| (R,S)-Laudanine | nd |
| (R,S)-Laudanosoline | nd |
| (R,S)-N-Methylcoclaurine | nd |
| (R,S)-4'-O-Methylcoclaurine | nd |
| (R,S)-4'-O-Methyllaudanosoline | nd |
| (S)-4'-O-Methyllaudanosoline | nd |
| (R,S)–6-O-Methyllaudanosoline | nd |
| Morphine | nd |
| (R,S)–Norcoclaurine | nd |
| (R,S)-Nororientaline | nd |
| (R)-Norprotosinomenine | nd |
| (S)-Norprotosinomenine | nd |
| (R)-Norreticuline | 8 |
| (S)-Norreticuline | nd |
| (R,S)-Orientaline | nd |
| Oripavine | nd |
| Papaverine | nd |
| (R,S)–Protosinomenine | nd |
| (R)-Reticuline | 100b |
| (S)-Reticuline | nd |
| Salutaridine | nd |
| (S)-Scoulerine | nd |
| (S)-Tetrahydrocolumbamine | nd |
| Thebaine | nd |
| Phenylpropanoids/flavonoids/coumarins | |
| Ferulic acid | nd |
| Isoeugenol | nd |
| 3-O-Methylquercitin | nd |
| Naringenin | nd |
| Scopoletin | nd |
| Sinapic acid | nd |
| Simple phenols | |
| Guaiacol | nd |
| 3-Methoxycatechol | nd |
| 3-Methoxytyramine | nd |
| Vanillic acid | nd |
a nd, not detected.
b100% activity is 1.64 min−1.
Enzyme Assay for Kinetic Analysis
Enzyme assays for kinetic analysis contained 30 mm potassium phosphate buffer (pH 8.0), 1.25 mm NADPH, increasing concentrations of (R)-reticuline (0, 0.5, 1, 5, 10, 15, 20, 30, 40, and 50 μm), and 70 μl of hypotonically lysed Sf9 cell suspension (expressing both CYP719B1 (∼3 μg) and CPR) brought to a final volume of 200 μl. Each reaction was allowed to proceed linearly for 15 min at 30 °C and was terminated by freezing in liquid nitrogen. Chloroform extraction was performed in the same manner as described above. The enzymatic product was quantified by LC-MS/MS analysis and the kinetic parameters (Km and kcat) were estimated by non-linear regression with GraphPad Prism in three independent experiments.
LC-MS/MS Analysis
Detection of product and subsequent identification and quantification was accomplished with a 4000 QTrap (AB Sciex Instruments) for mass spectroscopic analysis connected to an LC-20AD (Shimadzu) for chromatographic separation. Program parameters included a TurboIonSpray ionization source temperature of 500 °C and low resolution for Q1 and Q3 done with a multiple reaction monitoring (MRM) scan in the positive ion mode. Specific parameters in the (R)-reticuline program used for kinetic analysis are shown in supplemental Table S1. Fragmentation patterns for (R)-reticuline and salutaridine were identified with enhanced product ion (EPI) scans for m/z 330 and m/z 328 ions, respectively.
Samples containing (R)-reticuline were diluted 50-fold prior to injection of 10 μl into a Phenomenex Gemini C18 column (150 mm × 2 mm 5 micron) and subjected to resolution in the following solvent system: solvent A (10 mm ammonium acetate in 90% (v/v) methanol) and solvent B (0.1% (v/v) acetic acid in acetonitrile). A flow rate of 0.2 ml/min was used with the following gradient: 0–3 min, 0–100% solvent B; 3–5.5 min, 100% solvent B; 5.5–6.5 min, 100–0% solvent B; and 0% solvent B was held for 3.5 min. Analyst 1.4.2 was used in data analysis and quantification.
Retention times for each compound included: (R)-reticuline: 2.37 min; m/z 330 [M + H]+ and salutaridine: 2.48 min; m/z 328 [M + H]+. Monitored mass transitions for the quantification were from m/z 330 to m/z 192 for (R)-reticuline and m/z 328 to m/z 237 for salutaridine. Standard curves with pmol of authentic alkaloid versus peak area were created for the quantification.
Samples containing (R)-norreticuline were concentrated 4-fold by dissolving the N2-dried chloroform extract in 50 μl of 80% (v/v) methanol for product detection. Conditions used for (R)-norreticuline included solvent A (0.2% (v/v) acetic acid in water) and solvent B (0.2% (v/v) acetic acid in acetonitrile) with a flow rate of 0.4 ml/min and the following gradient: 0–4 min, 0–100% solvent B; 4–6 min, 100% solvent B; 6–7 min, 100–0% solvent B; and 0% solvent B was held for 3 min. EPI scans were run using m/z 316 [M + H]+ ((R)-norreticuine) and m/z 314 [M + H]+ (norsalutaridine) as parent ion for identification of mass spectra. Specific method parameters are shown in supplemental Table S2. All other parameters were identical to that of (R)-reticuline. Retention times for (R)-norreticuline was 3.18 min and norsalutaridine was 3.13 min. Monitored mass transitions for the quantification were from m/z 316 to m/z 178 for (R)-norreticuline and from m/z 314 to m/z 237 for norsalutaridine. Fragmentation of (R)-reticuline at 35 V of collision energy: m/z 330 ([M+H]+, 3), 299 (5), 267 (10), 239 (8), 227 (6), 207 (7), 192 (100), 177 (10), 175 (14), 143 (8), 137 (16). Fragmentation of salutaridine at 40 V of collision energy: m/z 328 ([M+H]+, 11), 297 (5), 282 (19), 270 (23), 267 (21), 265 (23), 255 (27), 253 (11), 250 (10), 239 (20), 237 (100), 233 (12), 222 (12), 211 (28), 209 (9), 207 (30), 205 (19), 183 (9). Fragmentation of (R)-norreticuline at 40 V of collision energy: m/z 316 ([M+H]+, 0.2), 284 (5), 267 (13), 239 (9), 235 (6), 207 (9), 178 (100), 175 (9), 163 (10), 143 (8), 137 (21). Fragmentation of norsalutaridine at 40 V of collision energy: m/z 314 ([M+H]+, 13), 299 (12), 298 (6), 282 (16), 270 (10), 267 (32), 265 (16), 254 (6), 250 (16), 237 (100), 233 (23), 222 (23), 211 (13), 207 (35), 205 (29), 183 (10), 178 (10), 137 (10).
RESULTS
Generation and Characterization of Opium Poppy Cytochrome P450
Native salutaridine synthase had been characterized as a cytochrome P450-dependent oxidase, but the protein could not be purified because of its instability (1, 13). An approach to identifying a cDNA encoding the enzyme was, therefore, chosen that entailed generating a collection of putative cytochrome P450 cDNAs by PCR using degenerate oligodeoxynucleotide primers based on conserved amino acid sequences. An amino acid sequence comparison of conserved regions of CYP75A3 (15), CYP80A1 (6), CYP80B1 (16), CYP51 (17), CYP71A1 (18), and CYP71A4 (19) yielded the sequence motifs YGP(K)Y(L)W located near the N terminus and EWVMSLL, EEFRPE, and PFGAGRIICP located near the C terminus of the cytochromes P450. The details of the PCR reactions are given under “Experimental Procedures.” 25 of the 28 unique sequences were found homologous to cytochrome P450 sequences in the public databases. To obtain these cDNAs as full-length clones, ∼2 × 106 plaques of a P. somniferum stem primary cDNA library (in lambda ZAPII) were screened using each PCR fragment as hybridization probe. After sequencing 154 clones resulting from the primary screen, 18 unigenes were identified that corresponded to 17 PCR fragments. 9 clones were full-length, 9 clones were partial cDNAs.
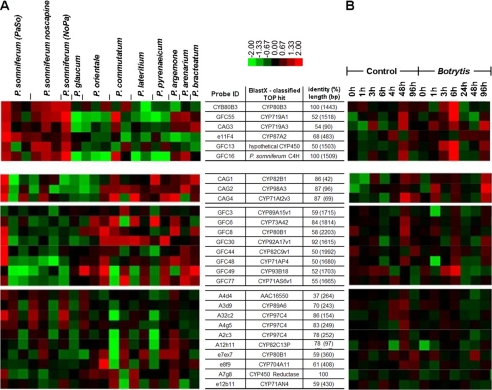
These 18 cytochrome P450-encoding unigenes from P. somniferum stem were added to a collection of ∼2000 ESTs that were used to prepare macroarrays that were hybridized to radioactively labeled cDNA synthesized from RNA from 16 Papaver species, only one of which, P. somniferum, produces morphine (21, 22). In these studies, 69 unigenes showed increased expression in morphinan alkaloid producing Papaver species. 6 of these 69 sequences were members of the family of cytochrome P450 enzymes (22). Two of these, CAG3 and GFC55, were nearly identical and belonged to the collection of 18 unigenes generated by PCR as described above. GFC55 contained a full-length open reading frame, and could be classified as a member of the CYP719 family. Expression analysis in 9 Papaver species showed elevated GFC55 expression in morphinan-producing P. somniferum varieties, similar to that of the other identified cytochrome P450 of morphine biosynthesis, CYP80B3 (Fig. 2A). The amino acid sequence of GFC55 (assigned CYP719B1) was similar (50.4% identity) to CYP719A1 encoding canadine synthase from C. japonica (24) and two methylenedioxy bridge-forming cytochromes involved in benzo[c]phenanthridine biosynthesis in the Mexican prickly poppy Argemone mexicana (CYP719A13, CYP719A14 (49.4 and 63.6% identity, respectively)).7 Canadine synthase is an enzyme that catalyzes the formation of the methylenedioxy bridge of (S)-canadine from (S)-tetrahydrocolumbamine in the biosynthesis of berberine alkaloids (25).
FIGURE 2.
Clustered display of P. somniferum P450 gene expression analysis. A, six P450 encoding cDNAs demonstrated higher expression in morphine-producing P. somniferum varieties when compared with eight other Papaver species that do not synthesize morphine. B, five of the same six genes were induced between 3–6 h in P. somniferum cell suspension culture after addition of a B. cinerea elicitor preparation to the culture medium.
The CYP719B1 transcript profile was consistent with the induction of transcript accumulation of known benzylisoquinoline biosynthetic genes in P. somniferum cell cultures by Botrytis cinerea and the increased accumulation of known benzylisoquinoline biosynthetic transcripts in the morphine-containing opium poppy compared with morphine free Papaver species. In detail, the CYP719B1 transcript accumulation was induced 3–6 h after addition of a B. cinerea preparation (2 ml/100 ml suspension culture (20)) to the medium of P. somniferum cell suspension cultures (Fig. 2B). The increase of alkaloid accumulation in the elicited cell culture is limited to those from benzo[c]phenanthridine (sanguinarine), protopine (protopine), protoberberine (N-methylstylopine), aporphine (N-methylcorytuberine), and berberine types (14).8 Although morphine is not accumulated in culture, the elicitation of P. somniferum cell suspension cultures by Botrytis cinerea induces the accumulation of transcript that encoded salutaridinol 7-O-acetyltransferase of morphine biosynthesis (26, 27). In a comparison of different Papaver species, the CYP719B1 transcript accumulates to the highest level in P. somniferum. Increased transcript levels could also be detected in the morphinan alkaloid-containing plants P. bracteatum and P. arenarium (Fig. 2A) (22). Because of the amino acid homology to CYP719A1 and the expression profile, and since a number of cytochromes P450 remain unidentified in morphine and in sanguinarine biosynthesis (28), CYP719B1 was selected for functional expression.
Expression of CYP719B1
The CYP719B1 cDNA clone was obtained as a full-length clone of 1869 bp encoding a protein of 505 amino acids and containing 67 bp of the 5′-flanking region and 284 bp of the 3′-flanking region. The reading frame was generated by PCR amplification out of pCR2.1 and was ligated into the baculovirus transfer vector pVL1392. This particular heterologous expression system was chosen because several alkaloid biosynthetic cytochromes P450 have been functionally expressed with this system (CYP80A1, (6); CYP80B1, (16); CYP80B3, (14)). Because selected cytochromes P450 can require the presence of a plant cytochrome P450 reductase (CPR) for either optimal enzyme activity, such as the (S)-reticuline biosynthetic enzyme (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1) from the California poppy Eschscholzia californica (16), or for activity per se, such as the same enzyme CYP80B3 from P. somniferum (14), the CPR cDNA from E. californica ((29); accession no. O24425), A. thaliana ((30); accession no. X66016), or petunia (accession DQ099545) was also introduced into pVL1392. Both CYP719B1 and CPR in pVL1392 were introduced into the linearized baculovirus by homologous recombination and the S. frugiperda Sf9 cells were then transfected with the individual viruses. After several rounds of virus amplification (23), recombinant CYP719B1 was tested for enzyme activity. Sf9 cells were infected with baculovirus containing either CYP719B1 alone, CPR alone or a 1:1 mixture of CYP719B1 and CPR as was performed for CYP80B1 from E. californica (16).
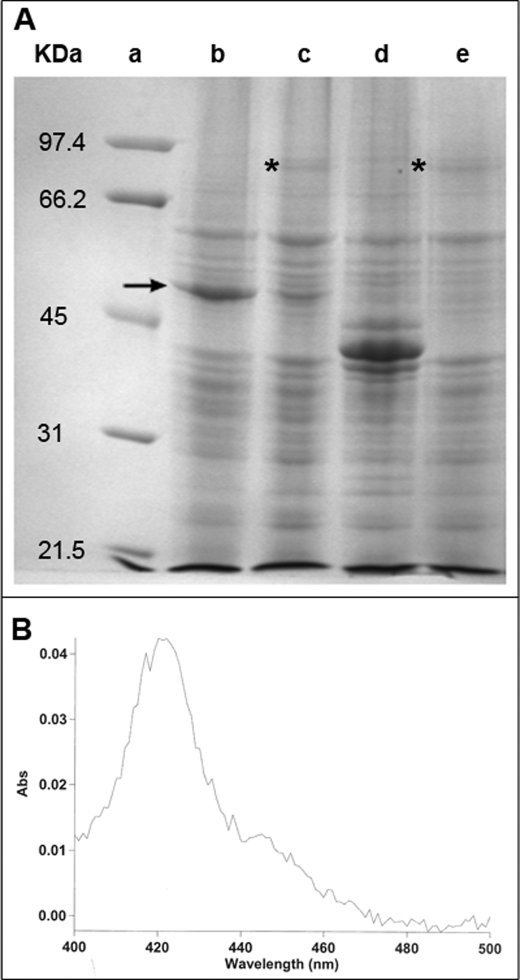
Aliquots of Sf9 cells from each of the three infection experiments using CYP719B1 and the E. californica CPR were subjected to SDS-PAGE to analyze for the presence of recombinant protein. As can be seen in Fig. 3A, CYP719B1 was readily visualized with Coomassie Brilliant Blue G-250 staining of the gel (Fig. 3A, lane b), the E. californica CPR protein was more difficult to identify (Fig. 3A, lane e) and the co-infection with baculoviruses containing CYP719B1 and E. californica CPR lead to a reduction in the quantity of CYP719B1 accumulated in the insect cells (Fig. 3A, lane c). Quantitation of recombinant CYP719B1 indicated that when expressed alone in Sf9 cells, the protein accumulated to 4440 nmol/750 ml Sf9 culture; when co-expressed with E. californica CPR, CYP719B1 accumulated to 597 nmol/750 ml Sf9 culture. Because protein that is detected by SDS-PAGE can also represent denatured cytochrome P450, microsomes were isolated from Sf9 cells co-infected with baculoviruses containing CYP719B1 and E. californica CPR, and the reduced CO difference spectrum of whole microsomes was measured (Fig. 3B). Quantitation of recombinant CYP719B1 as determined by reduced CO difference spectrum was 320 nmol/750 ml Sf9 culture. The reduction (46%) in the amount of P450 measured by the reduced CO difference spectrum compared with SDS-PAGE of whole cells was even greater with respect to salutaridine synthase enzyme activity. Hypotonically lysed Sf9 cells yielded six times more enzyme activity than isolated microsomes when normalized to 750 ml of cell culture (17 and 2.9 nmol salutaridine/min, respectively). Because hypotonically lysed cells were used as enzyme source for further characterization, the CYP719B1 quantity was determined by SDS-PAGE of whole cells.
FIGURE 3.
Detection of recombinant CYP719B1 by SDS-PAGE and reduced CO difference spectrum. A, SDS-PAGE of recombinant CYP719B1 and CPR. Recombinant CYP719B1 and CPR produced in Sf9 insect cell cultures were resolved by SDS-PAGE and visualized with Coomassie Brilliant Blue G-250. Lane a, molecular mass markers: 97.4 kDa, phosphorylase b; 66.2, bovine serum albumin; 45, ovalbumin; 31, carbonic anhydrase; 21.5, soybean trypsin inhibitor; lane b, CYP719B1 expressed in Sf9 cells; lane c, CYP719B1 and CPR co-expressed in Sf9 cells; lane d, STR of Psychotria ipecacuanha expressed in Sf9 cells (as control, Nomura et al., unpublished); lane e, CPR expressed in Sf9 cells. Arrow indicates position of CYP719B1 protein; asterisk indicates position of CPR protein. B, reduced CO difference spectrum obtained with microsomes isolated from Sf9 cells co-infected with baculoviruses containing CYP719B1 and E. californica CPR (38).
Characterization of Recombinant CYP719B1
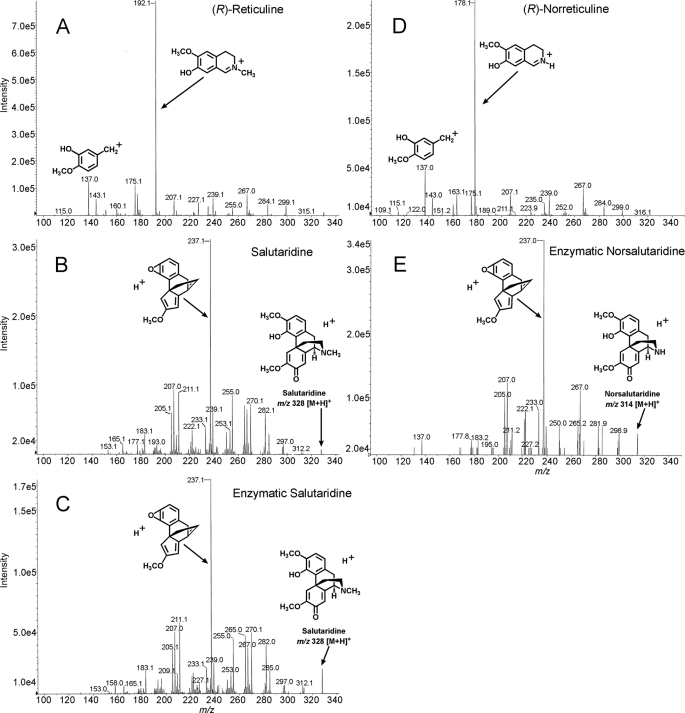
Recombinant cytochrome P450 was sufficiently expressed in the Sf9 cells such that resuspended cells could be used directly for enzyme assays. Aliquots of the cells were taken through multiple freeze-thaw cycles without loss of enzyme activity. 31 alkaloids were tested as substrate (Table 1). In addition, six phenylpropanoids/flavonoids and four simple phenols were tested (Table 1). The chemical structures are given in supplemental Fig. S1. Of the 41 compounds tested, only (R)-reticuline and (R)-norreticuline were converted into a product, salutaridine and norsalutaridine, respectively. All enzyme assays were analyzed by LC-MS/ MS and the enzyme assay results obtained with (R)-reticuline and (R)-norreticuline as substrate are shown in Fig. 4. In the mass spectrum, products were sought that were substrate molecular ion minus 2 (phenol-coupling or methylenedioxy bridge formation); minus 14 (demethylation) and plus 16 (hydroxylation).
FIGURE 4.
Mass spectrometric analyses of reaction products produced by CYP719B1 from (R)-reticuline and (R)-norreticuline. Selected ion monitoring (m/z 330 for (R)-reticuline, m/z 316 for (R)-norreticuline, m/z 328 for salutaridine and m/z 314 for norsalutaridine is shown. A, (R)-reticuline standard; B, salutaridine standard; C, product of the enzyme assay containing (R)-reticuline and recombinant CYP719B1 and CPR; D, (R)-norreticuline standard; E, product of the enzyme assay containing (R)-norreticuline and recombinant CYP719B1 and CPR. Assay and MS conditions are given under “Experimental Procedures.”
Only those Sf9 cells co-expressing both P. somniferum CYP719B1 and a plant CPR were able to convert (R)-reticuline and (R)-norreticuline into product. Neither cells expressing CYP719B1 alone nor a plant CPR alone resulted in product formation when assayed with substrate indicating that this cytochrome P450 requires a plant cytochrome P450 reductase for activity. The enzyme activity obtained when co-expressed with either the E. californica, Arabidopsis, or petunia did not substantially vary, so the remainder of the characterization of recombinant CYP719B1 with (R)-reticuline and (R)-norreticuline as substrate was carried out with enzyme produced by co-infection of Sf9 cells with CYP719B1 and E. californica CPR. Formation of the intramolecular C-C phenol-couple was sufficiently high in the conversion of (R)-reticuline to salutaridine to allow for quantification and the determination of kinetic parameters. The reaction has a temperature optimum at 30 °C and a pH optimum at 8.5. This varied somewhat from the native enzyme, which showed temperature and pH optima at 20–25 °C and 7.5, respectively (13). CYP719B1 followed Michaelis Menten-type reaction kinetics when the (R)-reticuline concentration was varied; the kinetic parameters Km and kcat were estimated by non-linear regression with GraphPad Prism to be 6.20 ± 0.93 μm and 1.64 ± 0.06 min−1, respectively. The sequence of CYP719B1 reported herein was deposited in the GenBankTM database under the accession number EF451150.
DISCUSSION
The C-C and C-O phenol-couple are important features in the structures of a number of pharmacologically active plant natural products. To date, members of the CYP80 family of cytochromes P450 have been shown to catalyze the formation of intermolecular C-O and intramolecular C-C phenol-couples in the isoquinoline class of alkaloids. It is demonstrated herein that a member of the CYP719 family of cytochromes P450, CYP719B1 (salutaridine synthase), catalyzes formation of the intramolecular C-C phenol-couple of salutaridine, a biosynthetic intermediate of the narcotic analgesic morphine.
CYP719B1 from P. somniferum required a plant CPR for enzyme activity when heterologously expressed in Sf9 cells. The ability to catalyze the formation of salutaridine from (R)-reticuline did not substantially vary when CYP719B1 was co-expressed with either E. californica, A. thaliana, or petunia CPR. The results obtained thus far in our hands with recombinant alkaloid biosynthetic P450s with respect to the type of reductase that is required for sufficient transfer of electrons has varied. Recombinant berbamunine synthase (CYP80A1, intermolecular C-O phenol-coupling) from Berberis stolonifera (Berberidaceae) was functionally expressed in Sf9 cells in the absence of a plant cytochrome P450 reductase (6). The hydroxylating CYP80B1 from E. californica was functionally expressed in Sf9 cells without a plant cytochrome P450 reductase, but the enzyme activity was increased when the cells were co-infected with recombinant E. californica CPR (16). In contrast, the hydroxylating CYP80B3 from P. somniferum required a plant cytochrome P450 reductase for functional expression (14). To date, it appears that morphine biosynthetic cytochromes P450 from P. somniferum are not active in Sf9 cells in the absence of a plant CPR.
A recombinant cytochrome P450 CYP80G2 has recently been reported from C. japonica cell culture that catalyzes the formation of an intramolecular C-C phenol-couple with (R,S)-reticuline to form the aporphine alkaloid (S)-corytuberine (8). The enzyme accepted (R,S)-norreticuline, orientaline, (S)-N-methylcoclaurine, (S)-coclaurine and codamine as substrate to form multiple products. CYP80G2 O-demethylated (R,S)-codamine at the 4′ position to form (R,S)-orientaline. In comparison, CYP719B1 was specific for catalyzing the formation of an intramolecular C-C phenol-couple with only (R)-reticuline and (R)-norreticuline. In each case, only one product (salutaridine or norsalutaridine) could be detected. Because CYP719B1 was highly specific for catalyzing the formation of the morphine biosynthetic intermediate salutaridine from (R)-reticuline ((R)-norreticuline and norsalutaridine are not known to be natural products in P. somniferum), it is concluded that it is the cytochrome P450-dependent oxidase salutaridine synthase. Salutaridine synthase is now the fourth enzyme of the morphine-specific portion of the pathway for which a cDNA has been characterized (Fig. 5). The cDNAs encoding the two subsequent enzymes, salutaridine reductase and salutaridinol acetyltransferase as well as the penultimate gene in the pathway encoding codeinone reductase have been characterized (22, 26, 31). Salutaridine synthase is also the first CYP719 cytochrome P450 family member to be shown to catalyze a phenol-coupling reaction. C-C phenol-coupling of reticuline has also been found to be catalyzed by the human cytochromes P450 2D6 and 3A4. In the presence of (R)-reticuline, not a single product, but rather (−)-isoboldine, (−)-corytuberine, (+)-pallidine, and the morphine precursor salutaridine were formed. (S)-Reticuline, a substrate of both 2D6 and 3A4, yielded the phenol-coupled alkaloids (+)-isoboldine, (+)-corytuberine, (−)-pallidine, and sinoacutine (40). CYP719B1, however, demonstrated a strict stereo- and regiospecificity. Even though each enzyme catalyzes the formation of a C-C phenol-couple with (R)-reticuline as substrate, a comparison of the amino acid sequences of CYP719B1 to the human cytochromes P450 revealed low homology (identity: 2D6 21.9% and 3A4 20.0%).
FIGURE 5.
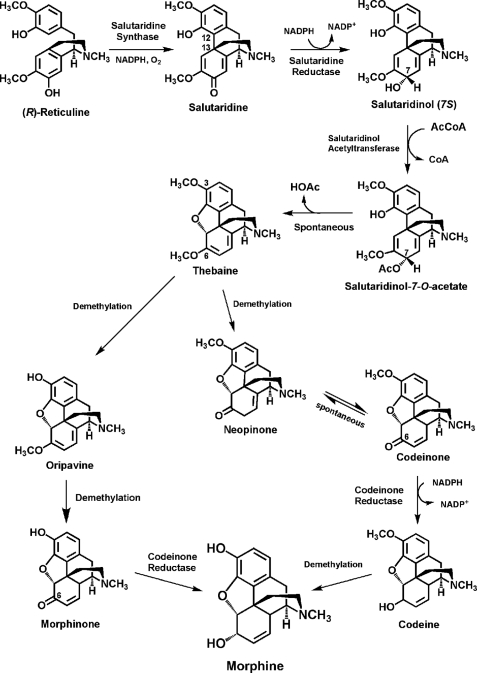
Schematic presentation of the biosynthetic pathway leading from (R)-reticuline to morphine in the opium poppy. Along the biosynthetic pathway from (R)-reticuline to morphine, salutaridine synthase catalyzes the first step, the C-C phenol-coupling of (R)-reticuline to salutaridine. The genes encoding the two subsequent enzymes, salutaridine reductase and salutaridinol acetyltransferase as well as the penultimate gene in the pathway encoding codeinone reductase have been characterized (22, 26, 31).
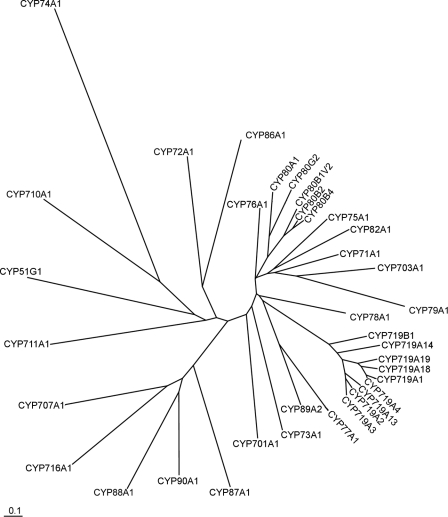
The most commonly observed cytochrome P450 catalyzed reaction is the monooxygenation of any of a plethora of organic molecules. Salutaridine synthase differs from most other cytochromes P450 in that oxygen is not incorporated into the enzymatic product. It was not to be readily predicted that salutaridine synthase would cluster closely with the CYP719 family of cytochromes (Fig. 6). To date, CYP719s have been characterized catalyzing only the formation of a methylenedioxy bridge in berberine biosynthesis (canadine synthase, CYP719A1, (24)) and in benzo[c]phenanthridine biosynthesis (stylopine synthase, CYP719A14, (32)). The distance between the CYP80 and the CYP719 clusters on the phylogenetic tree (Fig. 6) suggest that the ability to catalyze a C-C phenol-couple has arisen at least twice in the plant kingdom.
FIGURE 6.
Phylogenetic comparison of several plant cytochrome P450 enzymes. Amino acid sequences used for the tree were obtained from GenBankTM or SwissProt with the following accession numbers: AB014459, CYP51G1 (obtusifoliol 14-demethylase), A. thaliana; AF212990, CYP701A1 (ent-kaurene oxidase), Cucurbita maxima; AB006790, CYP703A1 (lauric acid monooxygenase), petunia; NM_202845, CYP707A1 (Abscisic acid 8′-hydroxylase), A. thaliana; M32885, CYP71A1, Persea americana; NM_129002, CYP710A1 (C22-sterol desaturase), A. thaliana; NP_850074, CYP711A1, A. thaliana; NM_123002, CYP716A1, A. thaliana; AB026122, CYP719A1 (canadine synthase), C. japonica; AB126257, CYP719A2, and AB126256, CYP719A3 (stylopine synthase), E. californica; AY610513, CYP719A4 (canadine synthase), Thalictrum flavum; EF451151, CYP719A13 (stylopine synthase), A. mexicana; EF451152, CYP719A14 (cheilanthifoline synthase), A. mexicana; AB374407, CYP719A18, C. japonica; AB374408, CYP719A19, C. japonica; EF451150, CYP719B1 (salutaridine synthase), P. somniferum; L10081, CYP72A1 (secologanin synthase), Catharanthus roseus; Z17369, CYP73A1 (cinnamate 4-hydroxylase), H. tuberosus; U00428, CYP74A1 (allene oxide synthase), Linum usitatissimum; Z22545, CYP75A1 (flavonoid 3′,5′-hydroxylase), petunia; X71658, CYP76A1, Solanum melongena; X71656, CYP77A1 (hydroxylase), Solanum melongena; P48420, CYP78A1, Zea mays; U32624, CYP79A1 (tyrosine N-hydroxylase), Sorghum bicolor; U09610, CYP80A1 (berbamunine synthase), B. stolonifera; AF014801, CYP80B1V2 ((S)-N-methylcoclaurine 3′-hydroxylase), E. californica; AB025030, CYP80B2 ((S)-N-methylcoclaurine 3′-hydroxylase), C. japonica; AY610509, CYP80B4 ((S)-N-methylcoclaurine 3′-hydroxylase), T. lavum; AB288053, CYP80G2 ((S)-corytuberine synthase), C. japonica; Q43068, CYP82A1, Pisum sativum; P48422, CYP86A1 (fatty acid omega-hydroxylase), A. thaliana; AF216313, CYP87A1, Helianthus annuus; U32579, CYP88A1 (ent-kaurenoic acid oxidase), Z. mays; U61231, CYP89A2, A. thaliana; Q42569, CYP90A1 (6-oxo-cathasterone 23a-hydroxylase), A. thaliana. The alignment was obtained using ClustalW (version 1.83) based on the neighbor-joining method (39); TreeView (version 1.6.6; Yves van de Peer, University of Konstanz, Germany) was used for visualization of the phylogenetic tree. A 10% change is indicated by the scale bar.
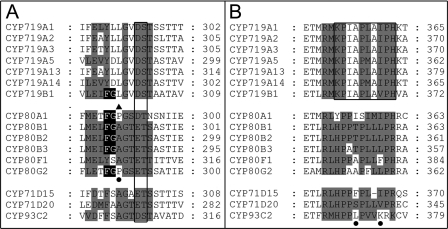
A comparison of the substrate recognition sites (SRS) according to (33) revealed a phenylalanine-glycine sequence within the SRS4 that is conserved in all tetrahydrobenzylisquinoline-binding CYP80s and in CYP719B1, distinguishable from the protoberberine-binding members of CYP719A subfamily and the unrelated CYP80 members (Fig. 7A). The following amino acids in the CYP80 enzymes distinguish the hydroxylases of CYP80B subfamily (alanine) and the phenol-coupling CYP80A1 (proline) and CYP80G2 (proline); however, mutation of the corresponding proline residue in CYP80G2 did not interfere with C-C phenol-coupling to form aporphines (8) (Fig. 7A).
FIGURE 7.
Substrate recognition site comparison. A, in the SRS4 substrate recognition site comparison, the highly conserved cytochrome P450 motif (D/E)T is boxed. The black background in SRS4 is the FG motif shared by the tetrahydrobenzylisoquinoline binding cytochromes P450. The filled triangle indicates the amino acid residue in SRS4 that distinguishes the CYP80 hydroxylases from the phenol-coupling enzymes. The filled circle indicates the position of the available mutation experiments: CYP80G2 P290A/P290G. The gray shading emphasizes the conservation of amino acids of CYP719s compared with CYP80Bs and other non-alkaloid transforming cytochromes P450 (CYP71s and CYP93). B, in the SRS5 substrate recognition site comparison, the KPIAPXXXPH motif thus far unique to the CYP719 family is boxed. The filled circles indicate the position of the available mutation experiments: CYP71D20 S368V; CYP71D15 F363I; CYP93C2 K375T. The gray shading in SRS5 emphasizes the differences in amino acids of CYP719s compared with CYP80Bs and other non-alkaloid transforming cytochromes P450 (CYP71s and CYP93). Accession numbers are provided in the legend to Fig. 6.
In contrast to the SRS4, the SRS5 residues are clearly distinguishable between the CYP719s and CYP80s. The CYP80 family is thereby related to hydroxylases of phenylpropanoid metabolism whereas the SRS5 KPIAPXXXPH motif is thus far unique to the CYP719 family (Fig. 7B). Mutations in the SRS5 region of terpenoid (CYP71D15, F363I; CYP71D20, S368V) and isoflavonoid (CYP93C2, K375T) hydroxylases are interfering with product formation as summarized in Ref. 34. The sequence similiarities in CYP719 SRS5 support a common mechanism for methylenedioxy bridge formation and the C-C phenol-coupling reaction that is catalyzed by CYP719B1.
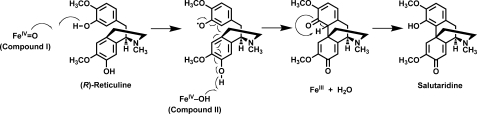
Mechanistically, stereochemistry of methylenedioxy bridge formation in berberine biosynthesis has been investigated using chiral methyl-labeled (14RS,methyl-R-)-[3-O-methyl-2H1,3H]tetrahydrocolumbamine administered to Thalictrum tuberosum cell suspension cultures (35). The reaction proceeded with 20% enantiomeric excess of configuration resulting from retention of configuration during cyclization to form the methylenedioxy bridge. However, a partial racemization was observed that was attributed to the low barrier of rotation around the C-O bond resulting from the assumed formation of an oxymethylene radical intermediate. The amino acid sequence of salutaridine synthase is most similar to cheilanthifoline synthase and formation of the C-C phenol-couple of salutaridine has long been presumed to be a radical reaction (13). Binding of the guaiacol moiety of (R)-reticuline to the active site of CYP719B1 is likely similar to that of canadine synthase (CYP719A1) (25, 24, 35), such that we favor that the formation of salutaridine from (R)-reticuline as catalyzed by CYP719B1 results from the initial formation of a phenoxy radical and does not involve a diradical intermediate (Fig. 8). We currently do not have insights into the structure and flexibility of the CYP719B1 substrate binding pocket and look to future enzyme structure studies that will directly test the mechanism proposed herein.
FIGURE 8.
Proposed mechanism of the reaction catalyzed by CYP719B1. The phenol-coupling of (R)-reticuline to salutaridine catalyzed by CYP719B1 passes through a single cycle of iron oxidation (Modified from Ref. 40).
CYP719B1 is the fourth cDNA specific to morphine biosynthesis that has been isolated and characterized. Attempts in organic chemistry to mimic the biosynthetic formation of the C-C phenol-couple of morphine biosynthesis have been either unsuccessful, yielding incorrectly coupled products, or have resulted in very low yield of salutaridine. (R,S)-reticuline has been produced in both Escherichia coli and Saccharomyces cerevisiae (36, 37). A biotechnological production of morphine from (R)-reticuline could now utilize the recombinant plant enzymes salutaridine synthase, salutaridine reductase, and salutaridinol acetyltransferase to form an advanced intermediate en route to morphine.
Acknowledgments
We thank Dr. Leslie Hicks and Dr. Baichen Zhang of the Proteomics & Mass Spectrometry Facility of the Donald Danforth Plant Science Center and Dr. Jürgen Schmidt of the Institut für Pflanzenbiochemie, Halle (Saale), Germany for help with the mass spectrometric analyses, Dr. Thomas Vogt of the Institut für Pflanzenbiochemie, Halle (Saale), Germany for the generous gift of phenylpropanoids and flavonoids and Dr. Jan Jaworski of the Donald Danforth Plant Science Center for Arabidopsis and petunia CPR cDNAs. We thank Dr. Taiji Nomura (Donald Danforth Plant Science Center) for critical reading of the manuscript. We especially thank Dr. F. Peter Guengerich of Vanderbilt University for helpful discussions on the mechanism of CYP719B1.
This work was supported in part by the Deutsche Forschungsgemeinschaft, Bonn, Germany, Fonds der Chemischen Industrie, Frankfurt, Germany, and the Mallinckrodt Foundation, St. Louis, MO.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) EF451150.
M. L. Diaz Chávez, M. Rolf, and T. M. Kutchan, in preparation.
T. M. Kutchan, unpublished results.
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- CPR
- cytochrome P450 reductase
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- SRS
- substrate recognition sites
- EPI
- enhanced product ion
- Tricine
- N-[2-hydroxy-1,1- bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Zenk M. H., Gerardy R., Stadler R. (1989) J. Chem. Soc., Chem. Commun. 1725–1727 [Google Scholar]
- 2.Davin L. B., Lewis N. G. (2005) Curr. Opin. Biotechnol. 16, 398–406 [DOI] [PubMed] [Google Scholar]
- 3.Lewis N. G. (1999) Curr. Opin. Plant. Biol. 2, 153–162 [DOI] [PubMed] [Google Scholar]
- 4.Niemetz R., Gross G. G. (2005) Phytochemistry 66, 2001–2011 [DOI] [PubMed] [Google Scholar]
- 5.Stadler R., Zenk M. H. (1993) J. Biol. Chem. 268, 823–831 [PubMed] [Google Scholar]
- 6.Kraus P. F., Kutchan T. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2071–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davin L. B., Wang H. B., Crowell A. L., Bedgar D. L., Martin D. M., Sarkanen S., Lewis N. G. (1997) Science 275, 362–366 [DOI] [PubMed] [Google Scholar]
- 8.Ikezawa N., Iwasa K., Sato F. (2008) J. Biol. Chem. 283, 8810–8821 [DOI] [PubMed] [Google Scholar]
- 9.Novak B. H., Hudlicky T., Reed J. W., Mulzer J., Trauner D. (2000) Curr. Org. Chem. 4, 343–362 [Google Scholar]
- 10.Kametani T., Kozuka A., Fukumoto K. (1971) J. Chem. Soc. 1021–1024 [Google Scholar]
- 11.Barton D. H. R., Bhakuni D. S., James R., Kirby G. W. (1967) J. Chem. Soc. 128–132 [DOI] [PubMed] [Google Scholar]
- 12.Szántay C., Bárczai-Beke M., Péchy P., Blaskó G., Dörnyei G. (1982) J. Org. Chem. 47, 594–596 [DOI] [PubMed] [Google Scholar]
- 13.Gerardy R., Zenk M. H. (1993) Phytochemistry 32, 79–86 [Google Scholar]
- 14.Huang F. C., Kutchan T. M. (2000) Phytochemistry 53, 555–564 [DOI] [PubMed] [Google Scholar]
- 15.Holton T. A., Brugliera F., Lester D. R., Tanaka Y., Hyland C. D., Menting J. G., Lu C. Y., Farcy E., Stevenson T. W., Cornish E. C. (1993) Nature 366, 276–279 [DOI] [PubMed] [Google Scholar]
- 16.Pauli H. H., Kutchan T. M. (1998) Plant J. 13, 793–801 [DOI] [PubMed] [Google Scholar]
- 17.Bak S., Kahn R. A., Olsen C. E., Halkier B. A. (1997) Plant J. 11, 191–201 [DOI] [PubMed] [Google Scholar]
- 18.O'keefe D. P., Leto K. J. (1989) Plant Physiol. 89, 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umemoto N., Kobayashi O., Ishizaki-Nishizawa O., Toguri T. (1993) FEBS Lett. 330, 169–173 [DOI] [PubMed] [Google Scholar]
- 20.Eilert U., Kurz W. G., Constabel F. (1985) J. Plant Physiol. 199, 65–76 [Google Scholar]
- 21.Ziegler J., Diaz-Chávez M. L., Kramell R., Ammer C., Kutchan T. M. (2005) Planta 222, 458–471 [DOI] [PubMed] [Google Scholar]
- 22.Ziegler J., Voigtländer S., Schmidt J., Kramell R., Miersch O., Ammer C., Gesell A., Kutchan T. M. (2006) Plant J. 48, 177–192 [DOI] [PubMed] [Google Scholar]
- 23.Kutchan T. M., Bock A., Dittrich H. (1994) Phytochemistry 35, 353–360 [DOI] [PubMed] [Google Scholar]
- 24.Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inouye K., Sato F. (2003) J. Biol. Chem. 278, 38557–38565 [DOI] [PubMed] [Google Scholar]
- 25.Rueffer M., Zenk M. H. (1994) Phytochemistry 36, 1219–1223 [Google Scholar]
- 26.Grothe T., Lenz R., Kutchan T. M. (2001) J. Biol. Chem. 276, 30717–30723 [DOI] [PubMed] [Google Scholar]
- 27.Zulak K. G., Cornish A., Daskalchuk T. E., Deyholos M. K., Goodenowe D. B., Gordon P. M., Klassen D., Pelcher L. E., Sensen C. W., Facchini P. J. (2007) Planta 225, 1085–1106 [DOI] [PubMed] [Google Scholar]
- 28.Kutchan T. M. (1998) in The Alkaloids ( Cordell G. ed), Vol. 50, pp. 257–316, Academic Press, San Diego, CA [Google Scholar]
- 29.Rosco A., Pauli H. H., Priesner W., Kutchan T. M. (1997) Arch. Biochem. Biophys. 348, 369–377 [DOI] [PubMed] [Google Scholar]
- 30.Urban P., Mignotte C., Kazmaier M., Delorme F., Pompon D. (1997) J. Biol. Chem. 272, 19176–19186 [DOI] [PubMed] [Google Scholar]
- 31.Unterlinner B., Lenz R., Kutchan T. M. (1999) Plant J. 18, 465–475 [DOI] [PubMed] [Google Scholar]
- 32.Ikezawa N., Iwasa K., Sato F. (2007) FEBS J. 274, 1019–1035 [DOI] [PubMed] [Google Scholar]
- 33.Gotoh O. (1992) J. Biol. Chem. 267, 83–90 [PubMed] [Google Scholar]
- 34.Rupasinghe S., Schuler M. A. (2006) Phytochem. Rev. 5, 473–505 [Google Scholar]
- 35.Bjorklund J. A., Frenzel T., Rueffer M., Kobayashi M., Mocek U., Fox C., Beale J. M., Gröger S., Zenk M. H., Floss H. G. (1995) J. Am. Chem. Soc. 117, 1533–1545 [Google Scholar]
- 36.Minami H., Kim J. S., Ikezawa N., Takemura T., Katayama T., Kumagai H., Sato F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawkins K. M., Smolke C. D. (2008) Nat. Chem. Biol. 4, 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 39.Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 40.Grobe N., Zhang B., Fisinger U., Kutchan T. M., Zenk M. H., Guengerich F. P. (2009) J. Biol. Chem. 284, 24425–24431 [DOI] [PMC free article] [PubMed] [Google Scholar]