Abstract
In this study, we report that the purified wild-type FANCI (Fanconi anemia complementation group I) protein directly binds to a variety of DNA substrates. The DNA binding domain roughly encompasses residues 200–1000, as suggested by the truncation study. When co-expressed in insect cells, a small fraction of FANCI forms a stable complex with FANCD2 (Fanconi anemia complementation group D2). Intriguingly, the purified FANCI-FANCD2 complex preferentially binds to the branched DNA structures when compared with either FANCI or FANCD2 alone. Co-immunoprecipitation with purified proteins indicates that FANCI interacts with FANCD2 through its C-terminal amino acid 1001–1328 fragment. Although the C terminus of FANCI is dispensable for direct DNA binding, it seems to be involved in the regulation of DNA binding activity. This notion is further enhanced by two C-terminal point mutations, R1285Q and D1301A, which showed differentiated DNA binding activity. We also demonstrate that FANCI forms discrete nuclear foci in HeLa cells in the absence or presence of exogenous DNA damage. The FANCI foci are colocalized perfectly with FANCD2 and partially with proliferating cell nuclear antigen irrespective of mitomycin C treatment. An increased number of FANCI foci form and become resistant to Triton X extraction in response to mitomycin C treatment. Our data suggest that the FANCI-FANCD2 complex may participate in repair of damaged replication forks through its preferential recognition of branched structures.
Fanconi anemia (FA)3 is a genetic disorder characterized by chromosome instability, predisposition to cancer, hypersensitivity to DNA cross-linking agents, developmental abnormalities, and bone marrow failure (1–9). There are at least 13 distinct FA complementation groups, each of which is associated with an identified gene (2, 9, 10). Eight of them are components of the FA core complex (FANC A, B, C, E, F, G, L, and M) that is epistatic to the monoubiquitination of both FANCI and FANCD2, a key event to initiate interstrand cross-link (ICL) repair (2, 9, 11). Downstream of or parallel to the FANCI and FANCD2 monoubiquitination are the proteins involved in double strand break repair and breast cancer susceptibility (i.e. FANCD1/BRCA2, FANCJ/BRIP1, and FANCN/PALB2) (2, 9).
FANCI is the most recently identified FA gene (11–13). FANCI protein is believed to form a FANCI-FANCD2 (ID) complex with FANCD2, because they co-immunoprecipitate with each other from cell lysates and their stabilities are interdependent of each other (9, 11, 13). FANCI and FANCD2 are paralogs to each other, since they share sequence homology and co-evolve in the same species (11). Both FANCI and FANCD2 can be phosphorylated by ATR/ATM (ataxia telangiectasia and Rad3-related/ataxia telangiectasia-mutated) kinases under genotoxic stress (11, 14, 15). The phosphorylation of FANCI seems to function as a molecular switch to turn on the FA repair pathway (16). The monoubiquitination of FANCD2 at lysine 561 plays a critical role in cellular resistance to DNA cross-linking agents and is required for FANCD2 to form damage-induced foci with BRCA1, BRCA2, RAD51, FANCJ, FANCN, and γ-H2AX on chromatin during S phase of the cell cycle (17–25). In response to DNA damage or replication stress, FANCI is also monoubiquitinated at lysine 523 and recruited to the DNA repair nuclear foci (11, 13). The monoubiquitination of both FANCI and FANCD2 depends on the FA core complex (11, 13, 26), and the ubiquitination of FANCI relies on the FANCD2 monoubiquitination (2, 11). In an in vitro minimally reconstituted system, FANCI enhances FANCD2 monoubiquitination and increases its specificity toward the in vivo ubiquitination site (27).
FANCI is a leucine-rich peptide (14.8% of leucine residues) with limited sequence information to indicate which processes it might be involved in. Besides the monoubiquitination site Lys523 and the putative nuclear localization signals (Fig. 1A), FANCI contains both ARM (armadillo) repeats and a conserved C-terminal EDGE motif as FANCD2 does (11, 28). The EDGE sequence in FANCD2 is not required for monoubiquitination but is required for mitomycin C (MMC) sensitivity (28). The ARM repeats form α-α superhelix folds and are involved in mediating protein-protein interactions (11, 29). In addition, FANCI, at its N terminus, contains a leucine zipper domain (aa 130–151) that could be involved in mediating protein-protein or protein-DNA interactions (Fig. 1A) (30–33). FANCD2, the paralog of FANCI, was reported to bind to double strand DNA ends and Holliday junctions (34).
FIGURE 1.
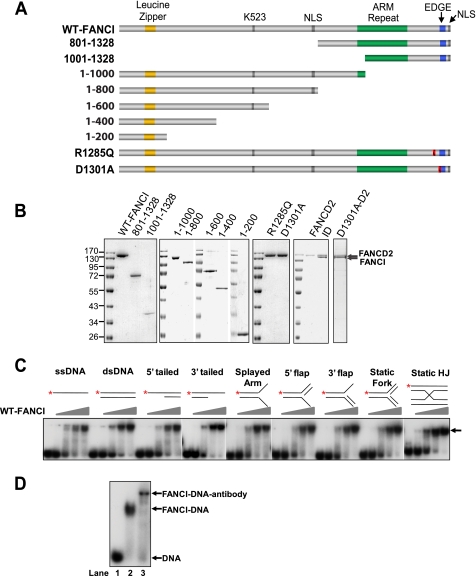
Purified human FANCI binds to DNA promiscuously. A, schematic diagram of predicted FANCI motifs and mutagenesis strategy to define the DNA binding domain. The ranges of numbers indicate how FANCI was truncated (e.g. 801–1328 represents FANCI-(801–1328)). NLS, predicted nuclear localization signal (aa 779–795 and 1323–1328); K523, lysine 523, the monoubiquitination site. The leucine zipper (orange bars, aa 130–151), ARM repeats (green bars), and EDGE motif (blue bars) are indicated. Red bars with a slash indicate the point mutations shown on the left. B, SDS-PAGE of the purified proteins stained with Coomassie Brilliant Blue R-250. R1285Q and D1301A are two point mutants of FANCI. All FANCI variants are tagged by hexahistidine. FANCD2 is in its native form. Protein markers in kilodaltons are indicated. C, titration of WT-FANCI for the DNA binding activity. Diagrams of the DNA substrates are shown at the top of each set of reactions. *, 32P-labeled 5′-end. HJ, Holliday junction. Concentrations of FANCI were 0, 20, 40, 60, and 80 nm (ascending triangles). The substrate concentration was 1 nm. Protein-DNA complex is indicated by an arrow. D, supershift assay. 1 nm of ssDNA was incubated with PBS (lane 1), 80 nm FANCI alone (lane 2), and 80 nm FANCI preincubated with a specific FANCI antibody (lane 3) in the condition described under “Experimental Procedures.”
In order to delineate the function of FANCI protein, we purified the recombinant FANCI from the baculovirus expression system. In this study, we report the DNA binding activity of FANCI. Unlike FANCD2, FANCI binds to different DNA structures, including single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), 5′-tailed, 3′-tailed, splayed arm, 5′-flap, 3′-flap, static fork, and Holliday junction with preference toward branched structures in the presence of FANCD2. Our data suggest that the dynamic DNA binding activity of FANCI and the preferential recognition of branched structures by the ID complex are likely to be the mechanisms to initiate downstream repair events.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant FA Proteins
cDNAs for human FANCI and FANCD2 were obtained by PCR amplification from a universal cDNA pool (BioChain Institute, Inc.). The FANCI cDNA matches the NCBI Reference Sequence NM_001113378, and the FANCD2 cDNA matches NM_001018115. The full-length open reading frames for both of them were sequenced before expression. Both FANCI and FANCD2 were expressed in insect High Five cells using the Bac-to-Bac expression system (Invitrogen). The site-directed mutations were introduced into the FANCI gene through a PCR-based method (35). The expression of these proteins was confirmed by Western blot analysis using the Pierce ECL kit. Antibodies against FANCI and FANCD2 were kindly provided by Weidong Wang (NIA, National Institutes of Health) or by the Fanconi Anemia Research Fund. A monoclonal antibody against the hexahistidine tag (Calbiochem) was also used to confirm expression and subsequent purification. Upon expression of the recombinant proteins in insect cells, the cells were homogenized using a Dounce homogenizer to prepare extracts. Hexahistidine-tagged wild-type and mutant FANCI proteins and FANCD2 were purified by using a HiTrap chelating column charged with nickel, Mono Q, Mono S, and/or Superdex 200 gel filtration column (GE Healthcare). Protein concentration was determined by the Coomassie (Bradford) protein assay reagent (Pierce). The purified proteins were stored at −80 °C in aliquots.
DNA Binding Assay and Supershift Assay
Oligonucleotides used to create ssDNA (61-mer), dsDNA (61 bp), 5′-tailed (30-mer for single-stranded part and 31-bp for double-stranded part), 3′-tailed (30 bp for double-stranded part and 31-mer for single-stranded part), splayed arm (30 for double-stranded part and 31-mer for single-stranded part), 5′-flap (with 31-mer flap), 3′-flap (with 31-mer flap), static fork (all three arms are 30 bp), and static Holliday junction (all four arms are symmetrically 30 bp) were adopted from the excellent design by Gari et al. (36) with the same sequences. The annealing was carried out in a water bath within ∼5 h by slowly cooling from 85 to 20 °C. The quality of annealing was monitored by native gel electrophoresis. Proper annealing was verified by the mobility of a corresponding substrate (e.g. static Holliday junction moves slowest because of its largest size). DNA-binding electrophoretic mobility shift assay (EMSA) analyses were performed in 10-μl reactions containing 25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm EDTA, 1 mm DTT, 6% glycerol, 1 nm 5′-32P-labeled oligonucleotide substrates, and the indicated amounts of protein. The reactions were incubated at 18 °C for 45 min, followed by the addition of 4 μl of 50% (w/v) sucrose. The reaction mixtures were resolved by electrophoresis through a 4% nondenaturing polyacrylamide gel in 1× TAE (40 mm Tris-acetate, pH 7.6, 10 mm EDTA) buffer with 6% glycerol and visualized by autoradiography. With the Owl P9DS (Thermo Scientific) electrophoresis system, the setting was 100 V (∼1.5 watts/gel) for 40 min. The supershift assay was conducted using 80 nm FANCI. 1 μl of anti-FANCI antibody (courtesy of Weidong Wang, National Institutes of Health) was incubated with FANCI on ice in the above reaction buffer for 2.5 h prior to the addition of 32P-labeled ssDNA substrate. After 45 min of incubation at 18 °C, the reaction mixture was resolved by electrophoresis for 1 h at 100 V and visualized by autoradiography.
Co-Immunoprecipitation Assay
6 pmol of the purified non-tagged FANCD2 was mixed with 6 pmol of the purified His6-tagged FANCI, FANCI-R1285Q, FANCI-D1301A, FANCI-(1–1000), or FANCI-(1001–1328) proteins as indicated in 100 μl of reaction buffer (150 mm KCl, 20 mm Tris-HCl, pH 7.6, 250 μg/ml BSA, 0.45% Nonidet P-40). After incubation on ice for 10 min, an anti-His6 antibody (Calbiochem) was added, and the mixture was incubated on a nutator at 4 °C overnight. Agarose A beads were then included to precipitate the tagged FANCI and interacting FANCD2 at 4 °C for 1 h. After the beads were washed in the lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA) twice with 300 mm NaCl and twice with 150 mm NaCl, the pellet was resuspended in 8 μl of the 150 mm NaCl lysis buffer and subject to Western blot analysis with a FANCD2-specific antibody.
Immunofluorescence and Confocal Microscopy
Subconfluent HeLa-S3 cells grown on coverslips, in RPMI 1640 medium supplemented with 10% fetal bovine serum, were treated or mock-treated with 1 μm mitomycin C (Sigma) or H2O, respectively, for 24 h. Cells were then rinsed with PBS buffer at pH 7.4 and fixed in ice-cold methanol for 10 min at −20 °C (37). After a 45-min incubation in 10% fetal bovine serum to block nonspecific protein binding, the fixed cells were incubated with the following primary antibody: affinity-purified rabbit anti-KIAA1794/FLJ10719 (FANCI) antibody (1:300; Bethyl) diluted in PBS containing 3% low fat milk at 37 °C for 2 h. After washing with PBS containing 3% low fat milk, the fixed cells were incubated with fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody (1:150; Sigma) diluted in PBS containing 3% low fat milk at 37 °C for 1 h. After washing, the fixed cells were incubated again with the following primary antibody: monoclonal mouse anti-PCNA antibody (1:500; Sigma) or polyclonal goat anti-FANCD2 antibody (1:50; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) diluted in PBS containing 3% low fat milk at 37 °C for 1 h. Then, after another wash, the fixed cells were incubated with TRITC-conjugated anti-mouse IgG antibody (1:50; Sigma) or anti-goat IgG antibody (1:400; Santa Cruz Biotechnology) diluted in PBS containing 3% low fat milk at 37 °C for 1 h. After yet another wash, the fixed cells were stained with 4′,6-diamidino-2-phenylindole (Sigma) at 0.1 μg/ml to confirm nuclear localization and washed again, and the coverslips were mounted on slides using Prolong Gold antifade reagent (Invitrogen). Triton pre-extraction was performed by incubating cells for 5 min at room temperature with 0.05% (v/v) Triton X-100 in PBS (38). Cells were fixed and processed as above. Co-staining experiments included proper controls to exclude crossing of signal between channels by omitting primary antibodies. When photographed under the same conditions as the samples, the controls did not show any fluorescence above background. Fluorescence microscopy was performed using a Zeiss LSM510/UV confocal microscope with a ×63 oil immersion objective. Immunolabeled slides (n = 4–5 representative fields/slide) were sectioned optically at 0.5-μm intervals (one focal plane) through the cell monolayer to obtain the appropriate focal depth. Images were captured and collected using the Axiovision version 4.7 program.
RESULTS
FANCI Is a DNA-binding Protein
In response to DNA-damaging agents, FANCI, together with FANCD2, forms damage-induced nuclear foci (11, 13). In addition, FANCD2 was shown to bind to double-stranded DNA ends and Holliday junctions (34). These reports raised our interest in finding out if FANCI could bind to DNA, although no sufficient information could be obtained from its primary structure (11) (Fig. 1A).
Full-length human FANCI with an N-terminal hexahistidine tag was purified to near homogeneity from the Bac-to-Bac baculovirus system (Fig. 1B). The purified FANCI did not seem to be modified by either phosphorylation or ubiquitination, as indicated by a single FANCI band in SDS-PAGE and Western blot analysis using a FANCI-specific antibody and an anti-His6 antibody (Fig. 1B) (data not shown). Increasing amounts of purified FANCI were then incubated with nine different oligonucleotide DNA substrates, including ssDNA, dsDNA, 5′-tailed, 3′-tailed, splayed arm, 5′-flap, 3′-flap, static fork, and static Holliday junction (Fig. 1C, top). An EMSA indicated that FANCI binds to all DNA structures in a concentration-dependent manner (Fig. 1C). It seemed that FANCI by itself does not have significantly higher affinity toward any particular structure. The FANCI concentration was between 40 and 60 nm when roughly half of DNA substrates were shifted. A supershift assay with a FANCI-specific antibody, ssDNA, and purified FANCI confirmed that the shift was specifically caused by FANCI (Fig. 1D). These results indicate that human FANCI is a promiscuous DNA-binding protein.
DNA Binding Domain of FANCI
To define the DNA binding domain of FANCI, we created a series of N- and C-terminal truncation mutants and purified them to near homogeneity (Fig. 1, A and B). Removal of the N-terminal 800 amino acids (FANCI-(801–1328)) did not reduce the DNA binding activity of FANCI (Fig. 2, compare B with A), although it changed the interacting behavior with DNA. The mobility of the DNA-protein complex declined as the protein amount increased, indicating that the complex formation could be relatively unstable when less protein is included.
FIGURE 2.
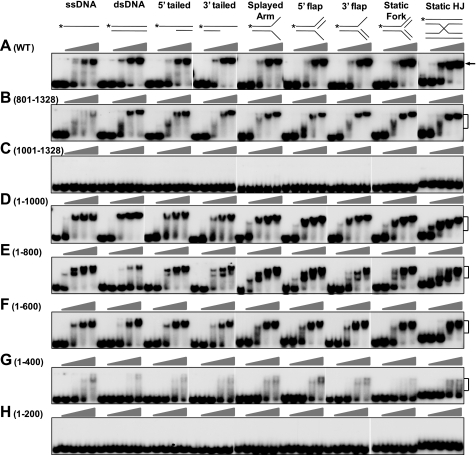
DNA binding domain of FANCI. EMSAs were performed with titration of the indicated FANCI truncation mutants. Concentrations of the mutants were 0, 20, 40, 60, and 80 nm (ascending triangles). The substrate concentration was 1 nm. Diagrams of the DNA substrates are shown at the top of each set of reactions. *, 32P-labeled 5′-end. Brackets, protein-DNA complex. A, WT-FANCI was included for convenient comparison. B, FANCI-(801–1328). C, FANCI-(1001–1328). D, FANCI-(1–1000). E, FANCI-(1–800). F, FANCI-(1–600). G, FANCI-(1–400). H, FANCI-(1–200). WT, wild type.
Further removal of 200 additional N-terminal amino acids from FANCI (FANCI-(1001–1328)) completely abolished its DNA binding activity (Fig. 2C). On the contrary, the N-terminal FANCI-(1–1000) moiety of FANCI retained robust binding activity to DNA (Fig. 2D). These results suggest that the C-terminal 328-aa fragment is dispensable for DNA binding, and the C-terminal boundary of the DNA binding domain is around amino acid 1000.
To define the N-terminal boundary of the DNA binding domain, we created and purified a series of C-terminal truncation mutants (Fig. 1, A and B). Compared with FANCI-(1–1000), the N-terminal FANCI-(1–800) fragment had the same robust binding activity toward all DNA structures except for the dsDNA, where a very significant reduction in DNA binding was steadily observed with the aa 1–800 fragment (Fig. 2, compare E with D). This result indicates that the area from residue 801 to residue 1000 on FANCI could contain a regulatory element for selective dsDNA binding. Multiple shift bands on most substrates (except for dsDNA) and an unusually fast moving band for Holliday junction seemed to be unique to this mutant when a 40–60 nm concentration of the protein was used (Fig. 2, compare E with other panels; discussion below).
Further shortening the N-terminal fragment to 600 aa (FANCI-(1–600)) did not seem to reduce its affinity to DNA significantly (Fig. 2F). The overall activity of FANCI-(1–600) is very close to wild-type FANCI (Fig. 2, compare F with A). Removal of an additional 200 amino acids (FANCI-(1–400)) significantly reduced DNA binding activity and drastically increased instability of the interaction between FANCI and DNA substrates, as indicated by the smear shift bands (Fig. 2G). The purified extreme N terminus of FANCI (FANCI-(1–200)), which contains a leucine zipper, did not show any affinity to all DNA substrates in the titration range (Fig. 2H). In summary, these results indicate the DNA binding domain of FANCI is roughly from residue 200 to 1000. The size of the FANCI DNA binding domain resembles the one for FANCD2, where the full-length protein is required for DNA binding (34).
Two C-terminal FANCI Point Mutations Cause Altered DNA Binding
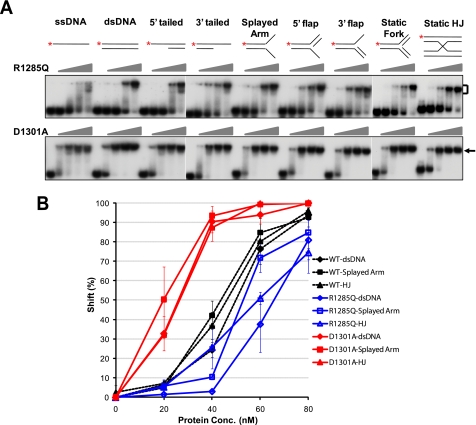
An arginine to glutamine point mutation at the C terminus of FANCI (FANCIR1285Q) is the disease-causing mutation (11, 12). FANCI with the R1285Q mutation failed to restore MMC resistance and was unable to localize to damage-induced foci despite strong protein expression (11). Because the DNA binding ability could be one of the contributing factors to foci formation, we investigated whether the R1285Q mutation has any effect on DNA binding of FANCI. The purified FANCIR1285Q protein (Fig. 1B) indeed showed slightly reduced DNA binding activity toward all DNA substrates when compared with the wild-type FANCI (Fig. 3A, R1285Q panel). Quantitative analyses of the representative shifts caused by dsDNA, splayed arm, and Holliday junction further confirmed the moderate but consistent reduction in DNA binding activity (Fig. 3B).
FIGURE 3.
DNA binding activity of FANCI R1285Q and D1301A. A, EMSAs of FANCI R1285Q and D1301A. Concentrations of the mutants were 0, 20, 40, 60, and 80 nm (ascending triangles). The substrate concentration was 1 nm. Diagrams of the DNA substrates are shown at the top of each set of reactions. *, 32P-labeled 5′-end. Protein-DNA complex is indicated by a bracket or an arrow. B, quantitation of the DNA binding activity of WT-FANCI, R1285Q, and D1301A to dsDNA, splayed arm, and Holliday junction (HJ). All experiments were repeated at least three times. Error bars, S.D. values. Broken lines with filled markers, WT-FANCI; blue solid lines with open markers, R1285Q; red solid lines with filled markers, D1301A. WT, wild type.
FANCI has a conserved EDGE motif at its C terminus, as does FANCD2. The FANCD2 EDGE motif is not required for monoubiquitination but is required for MMC sensitivity (28). To test if this motif is involved in the regulation of DNA binding, we created a mutant by changing aspartic acid 1301 into alanine (D1301A). The purified FANCID1301A protein (Fig. 1B) showed dramatically enhanced DNA binding for all substrates (Fig. 3, A and B, D1301A panel). In summary, these results indicate that although the C terminus of FANCI is dispensable for the direct DNA binding activity, it probably regulates the dynamics of DNA binding.
FANCI Interacts with FANCD2 through its C Terminus to Recognize Branched Structures
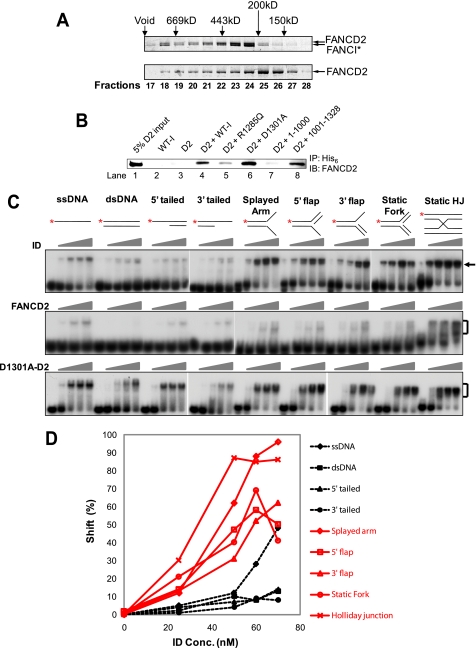
Because FANCI forms an interdependent complex with FANCD2 and this complex form is required for the monoubiquitination and chromatin association (11), we next ask what the effect of FANCD2 is on the DNA binding activity of FANCI. We co-transfected the High Five insect cells with both hexahistidine-tagged FANCI and non-tagged FANCD2 viruses for complex formation. After nickel column, a small portion (∼5%) of FANCD2 was co-purified with the tagged FANCI, indicating the weak interaction between FANCI and FANCD2. Further purification with the Superdex 200 gel filtration column did not separate the two proteins. FANCI (150 kDa with tag) and FANCD2 (164 kDa) probably formed a 1:1 complex, since the elution peak position on the gel filtration column matched the calculated molecular mass of the heterodimeric ID complex, which is 314 kDa (Fig. 4A). We also observed that the ID complex was eluted immediately after the void volume of the column, indicating either a higher order of complex formation or an aggregate formation (Fig. 4A). The complex formation was also verified by the distinct behavior of FANCD2 alone on the gel filtration. The elution peak of FANCD2 on the gel filtration column was at fractions 25 and 26 (∼150–200 kDa), indicating that the monomeric FANCD2 was the major form (Fig. 4A).
FIGURE 4.
FANCI interacts with FANCD2 to recognize branched structures. A, SDS-PAGE of the ID complex (314 kDa) and FANCD2 (164 kDa) alone on the Superdex 200 column. The column calibration was performed with blue dextran for void volume and a series of molecular weight markers (Sigma). The peaks of each marker in kilodaltons are approximately marked at the top. Fraction numbers (0.5 ml/tube) are marked at the bottom. FANCI and FANCD2 are indicated by arrows. *, His6 tag. B, co-immunoprecipitation of FANCI mutants with FANCD2. All FANCI mutants were tagged with His6. The antibody used for pull-down (IP) was an anti-His6 antibody (Calbiochem). The antibody used for detection (IB) was a FANCD2-specific antibody. WT-I, wild-type FANCI; D2, FANCD2. R1285Q, D1301A, 1–1000, and 1001–1328 are different mutants of FANCI. C, DNA binding activity of the ID complexes and FANCD2. EMSAs were performed with titration of the indicated protein mutants. Concentrations of the wild-type ID complex and D1301A-FANCD2 complex were 0, 25, 50, 60, and 70 nm (ascending triangles). Concentrations of FANCD2 were 0, 50, 100, and 150 nm (ascending triangles). The substrate concentration was 1 nm. Diagrams of the DNA substrates are shown at the top of each set of reactions. *, 32P-labeled 5′-end. Protein-DNA complex is indicated by a bracket or an arrow. D, quantitation of the DNA binding by the wild-type ID complex in C. Red solid lines with open markers, branched structures. Broken lines with filled markers, non-branched structures.
To further verify the FANCI-FANCD2 interaction and to map out the FANCD2-interacting domain of FANCI, we performed a co-immunoprecipitation assay using purified proteins (Fig. 4B). An anti-His6 antibody was able to pull down a very small portion of the input FANCD2 when it was incubated with the His6-tagged wild-type FANCI and non-tagged FANCD2 (Fig. 4B, lane 4). This is specifically due to the interaction between His6-FANCI and FANCD2, since the same antibody was incapable of pulling down FANCD2 in the absence of FANCI (Fig. 4B, lane 3). Co-immunoprecipitation analysis with the truncated FANCI indicated that FANCI interacts with FANCD2 through its C terminus (Fig. 4B, compare lanes 7 and 8). Although the EDGE mutant FANCID1301A was able to interact with FANCD2 efficiently, the patient-oriented mutant FANCIR1285Q showed dramatically reduced interaction with FANCD2 (Fig. 4B, lanes 5 and 6). This observation was further enhanced by the facts that FANCID1301A can be co-purified with FANCD2, whereas FANCIR1285Q cannot (Fig. 1B) (data not shown).
The EMSA with our purified FANCD2 showed a previously reported Holliday junction binding activity (34), but we were unable to observe the reported dsDNA binding activity (Fig. 4C, FANCD2 panel). One obvious difference is that our purified FANCD2 protein does not contain any tag or additional amino acids, whereas the FANCD2 purified by Park et al. contains 9 additional amino acids at its N terminus. When we tested for the DNA binding activity of the purified ID complex (Figs. 1B and 4A), surprisingly different dynamics were observed when compared with either FANCI alone or FANCD2 alone (Fig. 4C, ID panel). The ID complex had dramatically lower binding activity toward dsDNA, 5′-tailed, 3′-tailed, and, to a lesser extent, ssDNA than FANCI alone (compare Fig. 1C with Fig. 4C, ID panel). But the ID complex retained robust affinity for the splayed arm and Holliday junction and good binding activity to the 5′-flap, 3′-flap, and static fork structures (Fig. 4, C and D). One common feature of the latter substrates is that they are branched structures. These results indicate that the purified ID complex preferentially recognizes branched DNA structures.
The EDGE mutant FANCID1301A interacts and forms a complex with FANCD2 (Figs. 1B and 4B), but the FANCID1301A-FANCD2 complex showed dramatically lower substrate discrimination than the wild-type ID complex (Fig. 4C, compare ID panel with D1301A-D2 panel). Although the FANCID1301A-FANCD2 complex showed some preference toward branched structures (compare Fig. 4C (D1301A-D2 panel) with Fig. 3A (D1301A panel)), it bound to non-branched structures (ssDNA, dsDNA, 5′-tailed, and 3′-tailed) much better than wild-type ID complex (Fig. 4C).
Since the R1285Q mutant of FANCI has weak interaction and failed to be co-purified with FANCD2 (Fig. 4B) (data not shown), we examined how FANCD2 affects the DNA binding activity of R1285Q by adding the individually purified R1285Q and FANCD2 together in the EMSA. Results indicated that FANCD2 did not have any effect on the DNA binding activity of R1285Q (data not shown). In fact, even gel shift assays performed by adding individually purified WT-FANCI and FANCD2 together did not show preferential binding to the branched structures (data not shown). This is not surprising when the strength of the interaction between FANCI and FANCD2 is considered (Fig. 4B). Although R1285Q showed slightly reduced DNA binding activity, we think that the lack of complex formation with FANCD2 is more likely to be the reason why R1285Q fails to form nuclear foci with the MMC treatment (11).
FANCI Forms Nuclear Foci and Colocalizes with PCNA and FANCD2
Since FANCI and the ID complex bind to undamaged DNA and branched structures in vitro (Figs. 1–4), we next examined if endogenous FANCI associates with chromatin and replication sites in intact cells in the absence of DNA-damaging agents. For this purpose, we assessed FANCI localization in HeLa S3 cells by immunofluorescence microscopy using an anti-FANCI antibody used by Smogorzewska et al. (11). The HeLa cells were fixed by cold methanol instead of formaldehyde, because methanol is also considered as a permeabilizing and partial extraction agent, and it is particularly useful to study proteins associated with replication sites (39, 40).
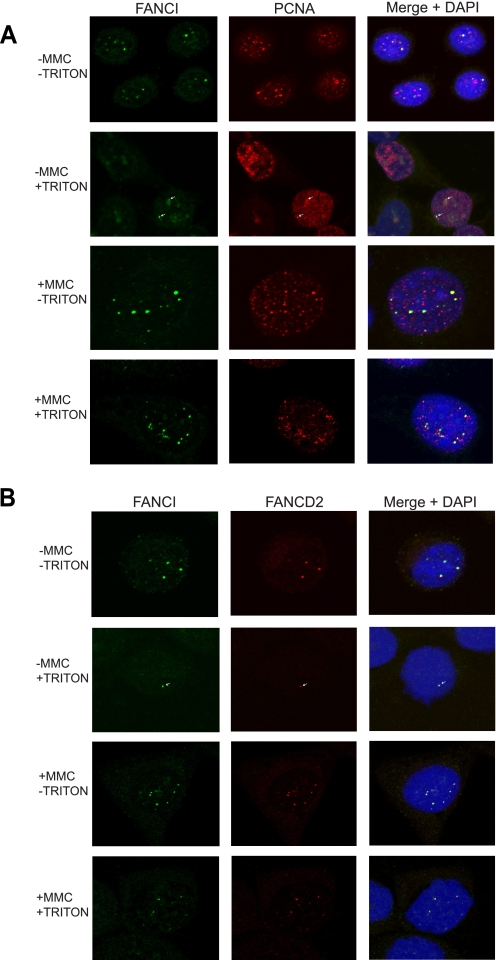
Analysis of the acquired confocal microscopy images revealed FANCI foci in nearly all of the untreated HeLa-S3 cells. Interestingly, most of these foci colocalized with endogenous PCNA foci in undamaged cells (Fig. 5A, −MMC −Triton). PCNA is well known as a DNA sliding clamp for DNA polymerases and as a replication and repair marker that is abundantly expressed in proliferating cells (37, 41, 42). It has been previously reported that PCNA foci after methanol extraction actually represent the PCNA associated with replication sites (40). Thus, our data indicate that FANCI is likely to be associated with DNA replication. When untreated cells were also stained with a FANCD2 antibody, FANCI was found to perfectly colocalize with FANCD2 in the absence of exogenous DNA-damaging agents (Fig. 5B, −MMC −Triton).
FIGURE 5.
FANCI forms nuclear foci and colocalizes with PCNA and FANCD2 in the absence or presence of DNA damage. Methanol-fixed cells with or without Triton X-100 pre-extraction were stained pairwise with either FANCI- and PCNA-specific antibodies (A), or FANCI- and FANCD2-specific antibodies (B). The stained cells were subject to confocal microscopy. Because the Triton X extraction dramatically reduced the fluorescence signal of FANCI in the absence of MMC treatment, we presented a two-dimensional view of a three-dimensional reconstruction of multifocal plane images to enhance the signal. The images in the −MMC +Triton panels of both A and B reflect overall signal level of the whole cell. Other panels represent a two-dimensional view of one focal plane image. Green, FANCI. Red, PCNA or FANCD2. Yellow and arrows, colocalization. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue).
After further extraction with Triton X-100, the number of FANCI foci was dramatically reduced. But we still found FANCI colocalized with PCNA and FANCD2 in a subset of the treated HeLa-S3 cells (Fig. 5, A and B, −MMC +Triton). This observation leads us to conclude that FANCI-FANCD2 is indeed associated with DNA and replication in intact cells, although the association is weak in the absence of exogenous DNA damage.
In the presence of MMC treatment, significantly more FANCI foci were observed (Fig. 5, A and B, compare +MMC −Triton with −MMC −Triton). FANCI still colocalized perfectly with FANCD2 and partially with PCNA (Fig. 5, A and B, +MMC −Triton). With further Triton extraction, the colocalization between FANCI, PCNA, and FANCD2 remained. Intriguingly, both FANCI and FANCD2 foci became more resistant to Triton extraction in response to MMC treatment, indicating stronger association of FANCI-FANCD2 with chromatin and replication sites (Fig. 5, A and B, compare +MMC +Triton with −MMC +Triton).
DISCUSSION
Inspired by the DNA binding activity of FANCD2 and the sequence similarity between FANCI and FANCD2 (11, 13, 34), we discovered the DNA binding activity of FANCI. Unlike FANCD2, which binds to Holliday junction (34),4 FANCI by itself is a very promiscuous DNA-binding protein. It has affinity to all structures we tested. The DNA binding activity of FANCI is also much stronger than that of FANCD2 in our study (Fig. 4). Physical mapping of the protein indicates that, very similar to FANCD2, FANCI has a relatively large DNA binding domain ranging from residue 200 to residue 1000. One has to keep in mind that a caveat about comparing the DNA binding activities of various FANCI truncation mutants is that they may not all be equally pure and properly folded. To reduce this possibility, all proteins were purified in 4 °C in a relatively short period of time (2 days). Additionally, we only used the proportional Coomassie Brilliant Blue staining to guarantee purity of the proteins. The similar and relatively high affinity of most FANCI truncation mutants with various DNA substrates also helps to show that these truncated proteins may be in their correct conformations (Fig. 2, A, B, D, E, and F; 40–60 nm for 50% shift).
Both FANCI and FANCD2 are leucine-rich proteins with 14.8 and 13.4% leucine residues, respectively. Leucine-rich proteins usually contain ARM repeats, HEAT repeats, leucine-rich repeats, or leucine zipper structures that are generally related to protein-protein interaction, DNA binding, and RNA binding (30, 43–46). These repeated superhelical assemblies of α helices and/or β sheets provide larger accessible surface for interactions than a more globular protein with comparable molecular weight (44). It is likely that the leucine-rich property of FANCI and FANCD2 enables these two proteins to interact with DNA and other interstrand cross-link repair proteins. According to a virtual structure simulation conducted using the YASARA structure program (available on the World Wide Web), the full-length FANCI could indeed form a relatively stable superhelical structure consisting of HEAT repeats and ARM repeats that might provide a platform for DNA interaction (data not shown).
The leucine zipper in the N terminus of FANCI does not bind to DNA directly (Fig. 2H), but it could contribute to the protein stability. A L137A/L144A double mutant on the conserved leucines makes FANCI very unstable and difficult to express (data not shown). Very interestingly, we found that the C terminus of FANCI is dispensable for DNA binding, but it plays a very important regulatory role for the DNA binding. Two point mutations (R1285Q and D1301A) at the C terminus altered the DNA binding activity of FANCI. Coincidentally, FANCD2 interacts with FANCI through this C terminus and channels the DNA binding specificity of the FANCI (or the ID complex) toward branched structures (Fig. 4). The ARM repeats located at the C terminus of FANCI could mediate the interaction between FANCI and FANCD2, but the extreme C terminus of FANCI is very important too. The R1285Q mutation, outside of the ARM repeats, disrupts the interaction between FANCI and FANCD2 (this study) and therefore explains its inability to induce FANCD2 monoubiquitination and nuclear foci formation (11). It is conceivable that FANCD2 would change the conformation of FANCI or both FANCI and FANCD2 reciprocally through interaction and therefore alter their DNA binding specificity. The altered DNA binding specificity (less dsDNA affinity) of the N-terminal aa 1–800 fragment (FANCI-(1–800)) provided the evidence that N terminus of FANCI is sensitive to conformational changes that cause alteration of binding specificity as a consequence. The multiple shift bands and the unusually high mobility for Holliday junction caused by this mutant may also be due to the altered dynamics of DNA binding (Fig. 2E), although multiple molecule binding is certainly another possibility. The EDGE motif in FANCD2 is dispensable for monoubiquitination of FANCD2 but not for MMC sensitivity (28). Our data on the FANCID1301A mutant (Figs. 3 and 4C) suggest that the EDGE motif could affect dynamics of chromatin association or DNA substrate specificity of the ID complex during ICL repair. The enhanced DNA binding activity of FANCID1301A and decreased substrate selectivity of FANCID1301A-FANCD2 complex could make FANCI and FANCD2 associate with DNA rigidly and non-specifically and therefore hinder the subsequent repair attempt. This scenario helps explain the defective ICL repair of the FANCD2 EDGE mutant even in the presence of normal FANCD2 monoubiquitination (28).
A very interesting observation of this study is the preferential binding activity of the ID complex to the branched structure. This property of FANCI-FANCD2 is surprisingly close to the branch-specific DNA binding specificity of FANCM (36, 47) and explains why the FA pathway of ICL repair is specific to the S phase. The preferential recognition of branched structures by the ID complex, but not by FANCI or FANCD2 alone, also supports the observation that FANCD2 is hardly detectable in the chromatin fraction in FANCI-deficient cells and the notion that FANCI could be a localizer of FANCD2 to chromatin (2, 48). Alpi and Patel (49) recently proposed a very refreshing FANCD2/FANCI receptor hypothesis for how the ID complex is recruited to the damage site in order to promote DNA repair. Since the ID complex preferentially binds to branched (fork) structures, a stalled replication fork (branched) seems to be the best candidate as a “receptor.” It will be interesting to determine if the phosphorylation and monoubiquitination of FANCI and FANCD2 would serve as the accessory factors to effect the complex formation and the recruitment to replication forks when DNA damage is present. The fork-targeting activity of the ID complex does not rule out the possibility that other factors, like FANCE, help to recruit the ID complex to the damage forks (50).
Remarkably, we also observed that FANCI forms nuclear foci and colocalizes with PCNA and FANCD2 in the absence of exogenous DNA damage and synchronization with or without Triton X pre-extraction. We selected methanol instead of formaldehyde to fix cells because of the partial extraction property of methanol and the advantage of using methanol fixation for study of replication sites, whereas the protein-protein cross-linking property of formaldehyde made it less supportive to our protein-DNA interaction study. The partial colocalization of FANCI and PCNA, a DNA replication and repair marker, further supports the preferential binding activity of branched structures (replication forks) of the ID complex. These data serve as the in vivo evidence that the ID complex binds to DNA and the branched structures. Enhanced FANCI and FANCD2 foci formation and resistance of these foci to Triton extraction after MMC treatment supports the concept that FANCI and FANCD2 work as a complex to execute their repair functions.
Smogorzewska et al. (11) observed FANCI foci formation in a subset of untreated cells and in nearly all cells after DNA damage. Using the same antibody, we observed that FANCI formed nuclear foci in more untreated cells (Fig. 5) (data not shown). The discrepancy can be explained by usage of different type of cells and different fixation. Even in the absence of exogenous DNA damaging agents, cultured cells are under constant attack spontaneously by many genotoxic agents (51). The spontaneous DNA damage or secondary structures on DNA could block replication, activate FANCI-FANCD2 proteins, and therefore account for the FANCI and FANCD2 foci formation in unperturbed cells.
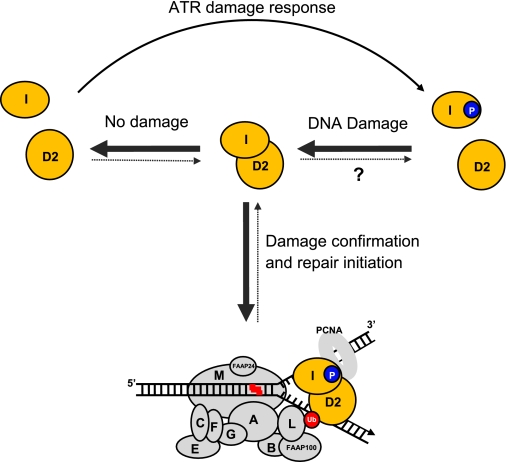
The DNA binding activity of FANCI, branch recognition activity of the ID complex, and colocalization of the ID complex with PCNA in the absence of exogenous DNA damage also support the observation that FA proteins are involved in stabilizing replication forks during unperturbed S phase and DNA replication (8, 11, 52–54). Based on the preferential binding activity of the ID complex toward branched structures and previous knowledge (9), a working model is proposed (Fig. 6). We hypothesize that the ID complex is the major form of FANCI and FANCD2 for repair of damaged replication forks. The ID complex formation is regulated by DNA damage level. When DNA damage level is high enough to affect the progression of replication forks, FANCI will be phosphorylated by ATR kinase, and the phosphorylated FANCI could shift the equilibrium of FANCI and FANCD2 interaction toward more ID complex formation. The ID complex will then be recruited to stalled replication forks through its preferential fork recognition activity. FANCI is functionally acting as a localizer of FANCD2 to replication forks through its robust DNA binding activity and its interaction with FANCD2 during the damage response (2) (this study). This hypothesis is supported by the fact that the phosphorylation of FANCI is required for FANCD2 foci formation and monoubiquitination (16). At the same time of FANCI phosphorylation and recruitment, the FA core complex is activated by ATR-mediated checkpoint response. Through its FANCM-FAAP24 translocase component, the activated FA core translocates along DNA to the damaged fork, where it meets and monoubiquitinates the ID complex present at the site (9) (Fig. 6). The monoubiquitinated FANCD2 will then recruit downstream repair factors, including ICL unhooking endonucleases (e.g. XPF-ERCC1 and/or MUS81/EME1), translesion synthesis polymerases (e.g. REV1/Polζ, Polκ, and/or Polθ), double strand break repair factors (e.g. BRCA1, FANCD1/BRCA2, FANCJ/BRIP1, FANCN/PALB2, and RAD51), and replication restart factors (e.g. BRAFT complex) (9).
FIGURE 6.
A working model for the role of FANCI DNA binding in ICL repair. An equilibrium of FANCI and FANCD2 interaction exists to sense DNA damage level that stalls replication forks in a cell. When DNA damage level is high, more ID complex will be formed and bound to stalled replication forks. The phosphorylation of FANCI could promote ID complex formation. The ID complex bound to the stalled fork will then be monoubiquitinated by the translocating FA core and therefore recruits downstream factors to initiate repair. When the damage is repaired or when the cells are free of damage, FANCI and FANCD2 in the ID complex tend to dissociate from each other because of dephosphorylation and deubiquitination. The hypothesized tendency of balance shift is indicated by either boldface solid arrows (association) or broken thin arrows (dissociation). A question mark denotes that this step is fully hypothetical without any direct evidence. FANCI and FANCD2 are highlighted with orange. The FA core complex with 10 subunits is depicted using gray ovals with their FA group letters. FANCL is the ubiquitin ligase. Red zigzag line, interstrand cross-link. Blue circle with the letter P, phosphorylation. Red circle with Ub, monoubiquitination.
When the damage becomes low after efficient repair, deubiquitination and dephosphorylation events dominate. Deubiquitination of FANCD2 will dismiss the repair regiments, and dephosphorylation of FANCI will shift the balance of FANCI and FANCD2 interaction toward dissociation from each other and therefore less recruitment to forks (Fig. 6). Our observation that the unmodified FANCI and FANCD2 are very inefficient in forming the ID complex supports this notion. This scenario is also applicable to the unperturbed cell cycle, when the damage is low and limited to endogenous resources and thus the phosphorylation level is low.
Clearly, a lot more work is required to prove the validity of our working model. For example, it will be very interesting to find out if the modification on FANCI and FANCD2, particularly the phosphorylation of FANCI and/or the monoubiquitination of FANCD2 (16, 49), would shift the equilibrium of FANCI and FANCD2 interaction toward more ID complex formation. Measurement of the affinity of FANCI and the ID complex to DNA damage, including single strand base modifications and ICLs, should be crucial to understanding if the ID complex is specialized in dealing with ICLs only or all replication-stalling lesions. It will also be critical to evaluate how the phosphorylation and monoubiquitination affects the DNA binding activity of FANCI and the ID complex. Apparently, another interesting challenge is to determine if the DNA binding activity of the ID complex has anything to do with the ICL unhooking, translesion synthesis, and double strand break repair directly. Since the ID complex binds to the Holliday junction very well, it deserves further investigation to find out what the ID complex would do to help resolve the Holliday junction.
Acknowledgments
We appreciate critical reading and comments by Drs. Peggy Hsieh, Guo-Min Li, and Weidong Wang.
This work was supported in part by a University of Miami/Sylvester Pap Corps Cancer Research Development Grant and the Florida Bankhead-Coley Cancer Research Program.
F. Yuan, J. El Hokayem, W. Zhou, and Y. Zhang, unpublished observations.
- FA
- Fanconi anemia
- ID
- FANCI-FANCD2
- ICL
- interstrand cross-link
- PCNA
- proliferating cell nuclear antigen
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- EMSA
- electrophoretic mobility shift assay
- MMC
- mitomycin C
- aa
- amino acid(s)
- WT-FANCI
- wild-type FANCI
- dsDNA
- double-stranded DNA
- PBS
- phosphate-buffered saline
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Alter B. P., Greene M. H., Velazquez I., Rosenberg P. S. (2003) Blood 101, 2072. [DOI] [PubMed] [Google Scholar]
- 2.de Winter J. P., Joenje H. (2009) Mutat. Res. 668, 11–19 [DOI] [PubMed] [Google Scholar]
- 3.Joenje H., Patel K. J. (2001) Nat. Rev. Genet. 2, 446–457 [DOI] [PubMed] [Google Scholar]
- 4.Kennedy R. D., D'Andrea A. D. (2005) Genes Dev. 19, 2925–2940 [DOI] [PubMed] [Google Scholar]
- 5.Kutler D. I., Singh B., Satagopan J., Batish S. D., Berwick M., Giampietro P. F., Hanenberg H., Auerbach A. D. (2003) Blood 101, 1249–1256 [DOI] [PubMed] [Google Scholar]
- 6.Niedernhofer L. J., Lalai A. S., Hoeijmakers J. H. (2005) Cell 123, 1191–1198 [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi T., D'Andrea A. D. (2006) Blood 107, 4223–4233 [DOI] [PubMed] [Google Scholar]
- 8.Thompson L. H., Hinz J. M., Yamada N. A., Jones N. J. (2005) Environ. Mol. Mutagen. 45, 128–142 [DOI] [PubMed] [Google Scholar]
- 9.Wang W. (2007) Nat. Rev. Genet. 8, 735–748 [DOI] [PubMed] [Google Scholar]
- 10.Cohn M. A., D'Andrea A. D. (2008) Mol. Cell 32, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsman J. C., Levitus M., Rockx D., Rooimans M. A., Oostra A. B., Haitjema A., Bakker S. T., Steltenpool J., Schuler D., Mohan S., Schindler D., Arwert F., Pals G., Mathew C. G., Waisfisz Q., de Winter J. P., Joenje H. (2007) Cell. Oncol. 29, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sims A. E., Spiteri E., Sims R. J., 3rd, Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T. (2007) Nat. Struct. Mol. Biol. 14, 564–567 [DOI] [PubMed] [Google Scholar]
- 14.Andreassen P. R., D'Andrea A. D., Taniguchi T. (2004) Genes Dev. 18, 1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi T., Garcia-Higuera I., Xu B., Andreassen P. R., Gregory R. C., Kim S. T., Lane W. S., Kastan M. B., D'Andrea A. D. (2002) Cell 109, 459–472 [DOI] [PubMed] [Google Scholar]
- 16.Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T., Tashiro S., Elledge S. J., Takata M. (2008) Nat. Struct. Mol. Biol. 15, 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D'Andrea A. D. (2001) Mol. Cell 7, 249–262 [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) Blood 100, 2414–2420 [DOI] [PubMed] [Google Scholar]
- 19.Hussain S., Wilson J. B., Medhurst A. L., Hejna J., Witt E., Ananth S., Davies A., Masson J. Y., Moses R., West S. C., de Winter J. P., Ashworth A., Jones N. J., Mathew C. G. (2004) Hum. Mol. Genet. 13, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Andreassen P. R., D'Andrea A. D. (2004) Mol. Cell. Biol. 24, 5850–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogliolo M., Lyakhovich A., Callén E., Castellà M., Cappelli E., Ramírez M. J., Creus A., Marcos R., Kalb R., Neveling K., Schindler D., Surrallés J. (2007) EMBO J. 26, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. (2001) Cell 105, 149–160 [DOI] [PubMed] [Google Scholar]
- 23.Reid S., Schindler D., Hanenberg H., Barker K., Hanks S., Kalb R., Neveling K., Kelly P., Seal S., Freund M., Wurm M., Batish S. D., Lach F. P., Yetgin S., Neitzel H., Ariffin H., Tischkowitz M., Mathew C. G., Auerbach A. D., Rahman N. (2007) Nat. Genet. 39, 162–164 [DOI] [PubMed] [Google Scholar]
- 24.Xia B., Dorsman J. C., Ameziane N., de Vries Y., Rooimans M. A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., Wang W., Livingston D. M., Joenje H., de Winter J. P. (2007) Nat. Genet. 39, 159–161 [DOI] [PubMed] [Google Scholar]
- 25.Folias A., Matkovic M., Bruun D., Reid S., Hejna J., Grompe M., D'Andrea A., Moses R. (2002) Hum. Mol. Genet. 11, 2591–2597 [DOI] [PubMed] [Google Scholar]
- 26.Meetei A. R., Yan Z., Wang W. (2004) Cell Cycle 3, 179–181 [PubMed] [Google Scholar]
- 27.Alpi A. F., Pace P. E., Babu M. M., Patel K. J. (2008) Mol. Cell 32, 767–777 [DOI] [PubMed] [Google Scholar]
- 28.Montes de Oca R., Andreassen P. R., Margossian S. P., Gregory R. C., Taniguchi T., Wang X., Houghtaling S., Grompe M., D'Andrea A. D. (2005) Blood 105, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 29.Xu W., Kimelman D. (2007) J. Cell Sci. 120, 3337–3344 [DOI] [PubMed] [Google Scholar]
- 30.Landschulz W. H., Johnson P. F., McKnight S. L. (1988) Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 31.Demuth I., Wlodarski M., Tipping A. J., Morgan N. V., de Winter J. P., Thiel M., Gräsl S., Schindler D., D'Andrea A. D., Altay C., Kayserili H., Zatterale A., Kunze J., Ebell W., Mathew C. G., Joenje H., Sperling K., Digweed M. (2000) Eur. J. Hum. Genet. 8, 861–868 [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A. D. (2000) Blood 96, 3224–3230 [PubMed] [Google Scholar]
- 33.Stone S., Sobeck A., van Kogelenberg M., de Graaf B., Joenje H., Christian J., Hoatlin M. E. (2007) Genes Cells 12, 841–851 [DOI] [PubMed] [Google Scholar]
- 34.Park W. H., Margossian S., Horwitz A. A., Simons A. M., D'Andrea A. D., Parvin J. D. (2005) J. Biol. Chem. 280, 23593–23598 [DOI] [PubMed] [Google Scholar]
- 35.Mao G., Pan X., Zhu B. B., Zhang Y., Yuan F., Huang J., Lovell M. A., Lee M. P., Markesbery W. R., Li G. M., Gu L. (2007) Nucleic Acids Res. 35, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A. (2008) Mol. Cell 29, 141–148 [DOI] [PubMed] [Google Scholar]
- 37.Kleczkowska H. E., Marra G., Lettieri T., Jiricny J. (2001) Genes Dev. 15, 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X., Geraldes P., Platt J. L., Cascalho M. (2005) J. Immunol. 174, 934–941 [DOI] [PubMed] [Google Scholar]
- 39.Koester S. K., Bolton W. E. (2001) Methods Cell Biol. 63, 253–268 [DOI] [PubMed] [Google Scholar]
- 40.Toschi L., Bravo R. (1988) J. Cell Biol. 107, 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maga G., Hubscher U. (2003) J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 42.Bravo R., Macdonald-Bravo H. (1987) J. Cell Biol. 105, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 44.Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 45.Tsuchiya N., Fukuda H., Nakashima K., Nagao M., Sugimura T., Nakagama H. (2004) Biochem. Biophys. Res. Commun. 317, 736–743 [DOI] [PubMed] [Google Scholar]
- 46.Andrade M. A., Perez-Iratxeta C., Ponting C. P. (2001) J. Struct. Biol. 134, 117–131 [DOI] [PubMed] [Google Scholar]
- 47.Xue Y., Li Y., Guo R., Ling C., Wang W. (2008) Hum. Mol. Genet. 17, 1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levitus M., Joenje H., de Winter J. P. (2006) Cell. Oncol. 28, 3–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alpi A. F., Patel K. J. (2009) DNA Repair 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 50.Pace P., Johnson M., Tan W. M., Mosedale G., Sng C., Hoatlin M., de Winter J., Joenje H., Gergely F., Patel K. J. (2002) EMBO J. 21, 3414–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2005) DNA Repair and Mutagenesis, 2nd Ed., pp. 9–69, American Society for Microbiology Press, Washington, DC [Google Scholar]
- 52.Mi J., Kupfer G. M. (2005) Blood 105, 759–766 [DOI] [PubMed] [Google Scholar]
- 53.Sobeck A., Stone S., Costanzo V., de Graaf B., Reuter T., de Winter J., Wallisch M., Akkari Y., Olson S., Wang W., Joenje H., Christian J. L., Lupardus P. J., Cimprich K. A., Gautier J., Hoatlin M. E. (2006) Mol. Cell. Biol. 26, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L. C., Stone S., Hoatlin M. E., Gautier J. (2008) DNA Repair 7, 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]