Abstract
In Gram-positive bacteria, sortase enzymes assemble surface proteins and pili in the cell wall envelope. Sortases catalyze a transpeptidation reaction that joins a highly conserved LPXTG sorting signal within their polypeptide substrate to the cell wall or to other pilin subunits. The molecular basis of transpeptidation and sorting signal recognition are not well understood, because the intermediates of catalysis are short lived. We have overcome this problem by synthesizing an analog of the LPXTG signal whose stable covalent complex with the enzyme mimics a key thioacyl catalytic intermediate. Here we report the solution structure and dynamics of its covalent complex with the Staphylococcus aureus SrtA sortase. In marked contrast to a previously reported crystal structure, we show that SrtA adaptively recognizes the LPXTG sorting signal by closing and immobilizing an active site loop. We have also used chemical shift mapping experiments to localize the binding site for the triglycine portion of lipid II, the second substrate to which surface proteins are attached. We propose a unified model of the transpeptidation reaction that explains the functions of key active site residues. Since the sortase-catalyzed anchoring reaction is required for the virulence of a number of bacterial pathogens, the results presented here may facilitate the development of new anti-infective agents.
Bacterial surface proteins function as virulence factors that enable pathogens to adhere to sites of infection, evade the immune response, acquire essential nutrients, and enter host cells (1). Gram-positive bacteria use a common mechanism to covalently attach proteins to the cell wall. This process is catalyzed by sortase transpeptidase enzymes, which join proteins bearing a highly conserved Leu-Pro-X-Thr-Gly (LPXTG, where X is any amino acid) sorting signal to the cross-bridge peptide of the peptidylglycan (2–4). Sortases also polymerize proteins containing sorting signals into pili, filamentous surface exposed structures that promote bacterial adhesion (5, 6). The search for small molecule sortase inhibitors is an active area of research, since these enzymes contribute to the virulence of a number of important pathogens, including among others Staphylococcus aureus, Listeria monocytogenes, Streptococcus pyogenes, and Streptococcus pneumoniae (reviewed in Refs. 7 and 8). Sortase enzymes are also promising molecular biology reagents that can be used to site-specifically attach proteins to a variety of biomolecules (9–14, 72).
The sortase A (SrtA)7 enzyme from S. aureus is the prototypical member of the sortase enzyme family (15, 16). It anchors proteins to the murein sacculus that possess a COOH-terminal cell wall sorting signal that consists of a LPXTG motif, followed by a hydrophobic segment of amino acids and a tail composed of mostly positively charged residues (17). SrtA is located on the extracellular side of the membrane. After partial secretion of its protein substrate across the cell membrane, SrtA cleaves the LPXTG motif between the threonine and glycine residues, forming a thioacyl-linked protein-sortase intermediate (16). It then catalyzes the formation of an amide bond between the carboxyl group of the threonine and the cell wall precursor molecule lipid II (undecaprenyl-pyrophosphate-MurNAc(-l-Ala-d-iGln-l-Lys(NH2-Gly5)-d-Ala-d-Ala)-β1–4-GlcNAc)), creating a protein-lipid II-linked product that is incorporated into the peptidylglycan via the transglycosylation and transpeptidation reactions of bacterial cell wall synthesis (18–20). Over 900 sortase-attached proteins in 72 different strains of bacteria have thus far been identified (21, 22). The vast majority of these proteins contain a COOH-terminal sorting signal harboring an LPXTG motif and are anchored to the cell wall by enzymes closely related to SrtA.
In vitro studies of SrtA have begun to define the mechanism of transpeptidation. SrtA consists of two parts: an unstructured amino-terminal tail that contains a stretch of nonpolar residues that embed it in the membrane and an autonomously folded catalytic domain that competently performs the transpeptidation reaction in vitro (SrtAΔN59, residues 60–206) (16, 23–25). Catalysis occurs through a ping-pong mechanism that is initiated when the thiol group of amino acid Cys184 nucleophilically attacks the carbonyl carbon of the threonine residue within the LPXTG sorting signal (16, 23–25). This forms a transient tetrahedral intermediate that, upon breakage of the threonine-glycine peptide bond, rearranges into a more stable thioacyl enzyme-substrate linkage. SrtA then joins the terminal amine group within the pentaglycine branch of lipid II to the carbonyl carbon of the threonine, creating a second tetrahedral intermediate that is resolved into the lipid II-linked protein product (23).
Sortase enzymes contain three conserved residues within their active sites: His120, Cys184, and Arg197 (SrtA numbering). These residues play a critical role in catalysis, since their mutation in SrtA causes severe reductions in enzyme activity (16, 26–30). Although it is well established that Cys184 forms a covalent linkage to the sorting signal, the functions of His120 and Arg197 are controversial. A variety of disparate functions have been ascribed to Arg197. These include deprotonating Cys184 (28), deprotonating lipid II (31), or stabilizing the binding of either the LPXTG sorting signal (28, 32) or oxyanion intermediates (31, 32). Different functions have also been proposed for His120. Originally, it was suggested that it activated Cys184 by forming an imidazolium-thiolate ion pair (26). However, subsequent pKa measurements revealed that both His120 and Cys184 are predominantly uncharged at physiological pH values, leading to the suggestion that His120 functions as a general base during catalysis (33). Most recently, it has been proposed that the most active form of the enzyme contains His120 and Cys184 in their charged states but that only a small fraction of SrtA exists in this form (∼0.06%) prior to binding to the sorting signal (25).
NMR and crystal structures of SrtAΔN59 have revealed that it adopts an eight-stranded β-barrel fold (31, 34). Other sortase enzymes have also been shown to possess a similar overall structure, including SrtB from S. aureus (27, 35), SrtB from Bacillus anthracis (27, 36), SrtA from S. pyogenes (37), and the SrtC-1 and SrtC-3 enzymes from S. pneumoniae (38). However, the molecular basis of substrate recognition remains poorly understood, because all of the structures reported to date have not contained a sorting signal bound to the enzyme. The lone exception is the crystal structure of SrtAΔN59 bound to an LPETG peptide (31). However, in this structure the peptide substrate is bound nonspecifically (see below) (32, 39).
In this paper, we report the structure and dynamics of SrtA covalently bound to an analog of the LPXTG sorting signal. The structure of the complex resembles the thioacyl intermediate of catalysis, providing insights into the molecular basis of binding of the LPXTG sorting signal and the functions of key active site residues. Notably, the mechanism of substrate binding visualized in the NMR structure differs substantially from a previously reported crystal structure of SrtAΔN59 non-covalently bound to a LPETG peptide (31). We have also used NMR chemical shift mapping experiments to localize the binding site for a triglycine cell wall substrate analog. A mechanism of transpeptidation compatible with these new data is proposed.
EXPERIMENTAL PROCEDURES
Preparation of the Covalent Complex for NMR Studies
Wild-type SrtA from S. aureus containing amino acid residues 60–206 (SrtAΔN59) was produced as described previously (29). Uniformly 15N- and 13C- or 15N-labeled SrtAΔN59 protein was covalently attached to an analog of the LPXTG sorting signal, Cbz-LPAT* (where T* is (2R,3S)-3-amino-4-mercapto-2-butanol, and Cbz is a carbobenzyloxy protecting group). The methods used to synthesize the analog and to prepare its covalent complex with SrtAΔN59 have been described previously (40). Three ∼1 mm samples of the complex were studied. Each was dissolved in 50 mm Tris-HCl (pH 6.0), 100 mm NaCl, 20 mm CaCl2, and 0.01% NaN3. The complexes contained either 1) 15N-labeled SrtAΔN59 bound to the unlabeled peptide dissolved in H2O (7% 2H2O), 2) 13C,15N-labeled protein bound to the unlabeled peptide dissolved in H2O (7% 2H2O), or 3) 13C,15N-labeled protein bound to the unlabeled peptide dissolved in 2H2O.
NMR Spectroscopy and Structure Determination
NMR spectra were acquired at 302 K on Bruker Avance 500-, 600-, and 800-MHz spectrometers equipped with triple resonance cryogenic probes. 1H, 13C, and 15N protein chemical shift assignments were obtained using standard methods (41, 42). Chemical shift assignments for the sorting signal were obtained by analyzing two-dimensional (F1,F2) 13C-filtered NOESY (43) and (F1) 13C-filtered TOCSY (44) spectra. Distance restraints to define the structure of the protein were obtained from three-dimensional 15N- and 13C-edited NOESY spectra (mixing time 100 ms), and intramolecular restraints for the sorting signal analog were obtained by analyzing two-dimensional (F1,F2) 13C-filtered NOESY spectra. Intermolecular distance restraints between SrtAΔN59 and the bound peptide were identified in three-dimensional (F1) 13C,15N-filtered (F2) 13C-edited NOESY-HSQC and (F1) 13C,15N-filtered (F2) 15N-edited NOESY-HSQC spectra (45) and in a two-dimensional (F1) 13C-filtered NOESY spectrum (43). 3JHN-Hα values were measured from a water flip-back three-dimensional HNHA spectrum (46), and backbone ψ and ϕ dihedral angles were obtained using the program TALOS (47). The NMR data were processed using NMRPipe (48) and analyzed using the PIPP (49) and CARA (version 1.4.1) (50) software packages.
Structure calculations were performed using the ATNOS/CANDID and NIH-XPLOR programs (51–53). Three three-dimensional NOESY data sets were used as input for ATNOS/CANDID: three-dimensional 13C-edited NOESY-HSQC and 15N-edited NOESY-HSQC spectra of the complex dissolved in H2O and a 13C-edited NOESY-HSQC spectrum of the complex dissolved in 2H2O. These data were supplemented with restraints for the backbone and side chain dihedral angles. Seven cycles of ATNOS/CANDID calculations yielded a converged ensemble of the protein in the complex. The structure was then refined in an iterative manner by manually checking all of the NOEs assigned by CANDID and by including 3JHN-Hα couplings and carbon chemical shifts in the calculations. New manually identified intra- and intermolecular distance restraints were also added, and at the final stages of refinement, hydrogen bonds were identified and included as distance restraints. The latter were obtained by inspecting the structures of the complex and by identifying NOE patterns characteristic of distinct secondary structures. All of the NOEs within the active site and binding pocket were manually checked. Residual dipolar couplings were measured by taking the difference in J couplings between partially aligned and unaligned protein samples. The protein was aligned using 15% (w/v) charged bicelles (30:10:1 molar ratio of dimyristoylphosphatidylcholine/dihexanoylphosphatidylcholine/hexadecyl(cetyl)trimethylammonium bromide). The programs MOLMOL (54) and PyMOL (55) were used to generate figures.
NMR Relaxation Studies
NMR data were collected using the 15N-labeled sample of the complex at 600 MHz. The strategy used to collect and analyze the relaxation data has been described previously (56, 57). The well resolved 1H-15N HSQC spectrum enabled the reliable measurement of relaxation parameters for 86 of a total of 148 residues. The average R1, R2, and 15N{1H} NOE values for the backbone nitrogen atoms in the complex are 1.50 ± 0.02 s−1, 12.28 ± 0.16 s−1, and 0.62 ± 0.10, respectively. The tensor parameters were calculated using the program Quadric_Diffusion, which follows the approach outlined by Bruschweiler et al. (58, 59). This yielded a correlation time of 8.56 ns, and the axial symmetric model was preferred over the isotropic model. A total of 76 of 86 quantifiable residues could be fit satisfactorily. The data from Ala81, Val87, Tyr88, Arg99, Ser102, Asn132, Ala135, Lys137, Met155, and Asp165 could not be fit to any model, possibly because they undergo more complicated motions. Residues were classified as follows: model 1 (S2-only) was selected for 52 residues, 2 residues fit to model 2 (S2 and τe), 15 residues fit to model 3 (S2 and Rex), 1 residue fit to model 4 (S2, τe, and Rex), and 6 residues fit to model 5 (Sf2, Ss2, and τe). For residues located in regions of regular secondary structure, the average order parameter is 0.93 ± 0.01. The relaxation data were analyzed using the suite of analysis programs kindly provided by Prof. Arthur G. Palmer III (60–63).
Localization of the Lipid II Binding Pocket
These studies made use of a 1 mm sample of the complex containing 15N-labeled SrtAΔN59 covalently attached to the sorting signal analog (NMR buffer: 50 mm Tris-HCl, 100 mm NaCl, 20 mm CaCl2, 0.01% NaN3, and 7% 2H2O, pH 6.0). Triglycine (Gly3) was obtained from Sigma. A 500 mm concentrated stock solution of Gly3 dissolved in NMR buffer was used. A series of two-dimensional 1H,15N HSQC spectra were recorded at 302 K after the addition of small aliquots of Gly3. A total of 12 spectra were acquired with the following molar ratios of Gly3 to the SrtAΔN59-LPAT* complex: 0:1, 0.5:1, 1:1, 2:1, 3:1, 4:1, 8:1, 16:1, 40:1, 50:1, 80:1, and 100:1. No significant chemical shift changes were observed after a 40:1 molar ratio was achieved. A normalized chemical shift change (Δδ) was calculated as Δδ = ((ΔδH)2 + (ΔδN/6.49)2)½, where ΔδN and ΔδH are, respectively, the amide nitrogen and proton chemical shift difference for a given residue in the presence and absence of Gly3. The titration experiments using the apo-form of the enzyme were performed in an identical manner.
Site-directed Mutagenesis and Enzyme Assays
Five single amino acid mutants of SrtAΔN59 containing a COOH-terminal six-histidine tag were produced in Escherichia coli from a pET15b expression vector. The presence of the histidine tag does not affect the enzymatic activity of the protein (9, 56, 64). Mutations were made using the QuikChange® method (Stratagene) and confirmed by DNA sequencing. Mutant and wild-type enzymes were purified as described previously (56), and their proper folding was confirmed by one-dimensional 1H NMR. The enzyme kinetic parameters of five SrtAΔN59 mutants (L97A, A104G, E105A, D112A, and A118G) were measured as described previously (24, 56). Briefly, A self-quenched fluorescent peptide, o-aminobenzoyl-LPETG-2,4-dinitrophenyl, was used as a substrate in the cleavage reaction containing 1.5 μm SrtA enzyme dissolved in assay buffer (20 mm HEPES, pH 7.5, with various concentrations of CaCl2). The o-aminobenzoyl-LPETG-2,4-dinitrophenyl substrate was dissolved in dimethyl sulfoxide and added to the reaction to a final concentration between 6.25 and 25 μm, for a total reaction volume of 200 μl. The increase in fluorescence intensity was monitored at room temperature using excitation at 335 nm and recording the emission maximum at 420 nm on a Spectramax M5 spectrofluorometer (Molecular Devices). The steady-state velocities (Vs) from the biphasic progress curves were calculated. Data sets were collected in triplicate and were corrected for inner filter effects (65).
Substrate Specificity Assay
An 18-member peptide library containing alterations in the LPXTG sorting signal was purchased from Biopeptide Co., Inc. Each peptide in the library contains the amino acid sequence from the sorting signal of protein A (LPETG) but randomizes the leucine position (SKRQAXPETGEESTE; where X can represent any amino acid except for Ile or Cys). The ability of the SrtAΔN59 enzyme to selectively process peptides within the library was ascertained by mass spectrometry. A 40-μl reaction containing SrtAΔN59 and the library was incubated at 37 °C for 16 h (final reaction concentrations: 0.1 mg/ml peptide library, 15 μm SrtAΔN59 dissolved in 20 mm HEPES, 5 mm CaCl2, and 2 mm Gly3, pH 7.5). A 4-μl aliquot from the reaction was then quenched by the addition of 4 μl of 0.2% trifluoroacetic acid. After mixing with an equal amount of α-cyano-4-hydroxycinnamic acid, the products and reactants were analyzed by MALDI-TOF using a Voyager-DE STR Biospectrometry Work station (Applied Biosystems) under the positive ion mode. Both substrates and transpeptidation products can be simultaneously observed in the mass spectrum.
RESULTS AND DISCUSSION
Structure of the Covalent Complex between Sortase and an Analog of the Sorting Signal
The molecular basis of sorting signal recognition and transpeptidation is not well understood, because the reaction intermediates of catalysis are short lived and thus difficult to visualize by crystallography or NMR spectroscopy. To overcome this problem, we synthesized a peptide analog of the sorting signal that covalently modifies the enzyme. The peptide contains the amino acid sequence Cbz-LPAT*, where Cbz is a carbobenzyloxy protecting group and T* is a threonine derivative that replaces the carbonyl group with -CH2-SH (Fig. 1a) (40). Via its T* moiety, the peptide forms a disulfide bond with the active site Cys184 thiol generating a covalent SrtAΔN59-LPAT* complex that structurally mimics the thioacyl intermediate of catalysis (compared in Fig. 1b).
FIGURE 1.
The Cbz-LPAT* modifier and NMR data of its complex with SrtAΔN59. a, chemical structure of the Cbz-LPAT* peptide analog. b, comparison of the chemical structure of the thioacyl enzyme-substrate intermediate of the anchoring reaction (top) and the disulfide-linked covalent complex between the Cbz-LPAT* peptide and Cys184 of SrtAΔN59 (bottom). c, selected panels showing intermolecular NOEs between the SrtAΔN59 protein and the sorting signal peptide. The panels are taken from a three-dimensional (F1) 13C, 15N-filtered, (F2) 13C-edited NOESY-HSQC spectrum of the SrtAΔN59-LPAT* complex dissolved in 2H2O. The identity of the proton from SrtAΔN59 and its chemical shift are shown at the top and bottom of each panel, respectively. On the right side of each cross-peak the sorting signal peptide proton that is proximal to the protein is indicated.
The structure of the SrtAΔN59-LPAT* complex was determined using heteronuclear NMR methods (41). Previously, we assigned the backbone chemical shifts of the SrtAΔN59 protein in the complex (29). In this report, to solve the structure of the complex, we assigned nearly all of the 1H, 13C, and 15N chemical shifts of the protein and the 1H chemical shifts of the bound peptide. As shown in Fig. 1c, the covalent SrtAΔN59-LPAT* complex exhibits good quality NMR spectra, enabling 36 intermolecular NOE distance restraints between the protein and peptide to be identified. The structure of the complex was calculated using 3,186 experimental restraints: 2,454 intraprotein distances, 36 intermolecular distances, 94 hydrogen bonds, 88 3JHNα couplings, 254 carbon chemical shifts, and 260 dihedral angles. This produced an ensemble of 20 conformers that possess good covalent geometry with no NOE, dihedral angle, or scalar coupling violations greater than 0.5 Å, 5°, or 2 Hz, respectively (Fig. 2a). The amino acids of the peptide analog and residues Pro63–Ile207 of SrtAΔN59 are well defined by the NMR data and have backbone and heavy atom coordinate root mean square deviations to the mean structure of 0.28 ± 0.06 and 0.87 ± 0.08 Å, respectively. Complete structure and restraint statistics are presented in Table 1.
FIGURE 2.
NMR solution structure of the SrtAΔN59-LPAT* complex. a, stereo image showing the ensemble of 40 lowest energy structures of the SrtAΔN59-LPAT* complex. The protein backbone heavy atoms (blue) and the covalently linked peptide (red) are shown. A yellow sphere represents the calcium ion. b, ribbon drawing of the structure of the SrtAΔN59-LPAT* complex. The covalently bound peptide is shown in a red ball-and-stick representation with its amino acids labeled. c, expanded view of the SrtAΔN59-substrate interface showing how the leucine and proline residues of the sorting signal are recognized. A ribbon drawing of the SrtA protein (white) with select side chains (yellow) is shown. d, expanded view of the positioning of the threonine and alanine residues of the sorting signal. The side chains of His120 and Arg197 are also shown, demonstrating their roles in catalysis. The panel is colored as in c. The -CH2-SH group of the peptide is covalently bonded with the side chain thiol group of Cys184 through a disulfide bridge (pale yellow spheres).
TABLE 1.
Structural statistics for the NMR structure of the SrtAΔN59-LPAT* complex
The notation of the NMR structures is as follows: 〈SA〉 are the final 20 simulated annealing structures; (S̄A)r is the average energy-minimized structure. The number of terms for each restraint is given in parentheses.
| 〈SA〉 | (S̄A)r | |
|---|---|---|
| r.m.s. deviations from NOE interproton distance restraints (Å)a | ||
| All (2454) | 0.027 ± 0.011 | 0.039 |
| Intermolecular (36) | 0.030 ± 0.017 | 0.054 |
| r.m.s. deviations from hydrogen-bonding restraints (Å)b (94) | 0.023 ± 0.012 | 0.024 |
| r.m.s. deviations from dihedral angle restraints (degrees)c (260) | 0.304 ± 0.141 | 0.712 |
| r.m.s. deviations from 3JHNα coupling constants (Hz)d (88) | 0.579 ± 0.241 | 0.726 |
| r.m.s. deviations from secondary 13C shifts (p.p.m.) | ||
| 13Cα (127) | 0.91 ± 0.04 | 1.38 |
| 13Cβ (127) | 1.05 ± 0.15 | 1.54 |
| Residual dipolar coupling R-factorse (%) | ||
| DNH (66) | 4.6 ± 0.1 | 4.4 |
| DNC' (61) | 6.4 ± 0.2 | 5.0 |
| Deviations from idealized covalent geometry | ||
| Bonds (Å) | 0.002 ± 0.001 | 0.006 |
| Angles (degrees) | 0.355 ± 0.148 | 0.609 |
| Impropers (degrees) | 0.418 ± 0.174 | 0.566 |
| PROCHECK-NMRf | ||
| Most favored regions (%) | 71 ± 1 | 77.5 |
| Additionally allowed regions (%) | 27 ± 1 | 20.8 |
| Generously allowed regions (%) | 2.5 ± 0.8 | 1.7 |
| Disallowed regions (%) | 0.0 ± 0.0 | 0.0 |
| Coordinate precision (Å)g | ||
| Protein and peptide backbone | 0.28 ± 0.06 | |
| All protein and peptide heavy atoms | 0.87 ± 0.08 | |
a None of the structures exhibited distance violations greater than 0.5 Å, dihedral angle violations greater than 5°, or coupling constant violations greater than 2 Hz.
b Two distance restraints were employed for each hydrogen bond (rNH···O < 2.5 Å and rN···O < 3.5 Å).
c The experimental dihedral angle restraints comprised 132 φ, 98 ψ, and 21 χ1.
d The coupling constants were back-calculated from the structures using the following equation, J(φ) = A cos2(φ − 60) + B cos(φ − 60) + C, where the values for A, B, and C, are 6.98, −1.38, and 1.72, respectively.
e The dipolar coupling R-factor ranges between 0 and 100% and is defined as the ratio of the r.m.s. deviation between observed and calculated values to the expected r.m.s. deviation if the vectors were randomly distributed (69). The values of DaNH and η are −14.2 Hz and 0.55, respectively.
f Determined as described in Ref. 70.
g The coordinate precision is defined as the average atomic r.m.s. deviation of the 40 individual SA structures and their mean coordinates. The reported values are for residues Pro63 to Thr203 of SrtAΔN59 and all of the LPAT* peptide. The backbone value refers to the N, Cα, and C′ atoms.
Structural Basis of LPXTG Binding
SrtA recognizes the LPXTG sorting signal through a large groove that leads into the active site (Fig. 2b). Residues in strands β4 and β7 form the floor of the groove, whereas the walls are formed by surface loops that connect strand β6 to strand β7 (β6/β7 loop), strand β7 to strand β8 (β7/β8 loop), strand β3 to strand β4 (β3/β4 loop), and strand β2 to helix H1 (β2/H1 loop). The leucine residue of the signal rests against the β6/β7 loop, where residues Val166–Leu169 adopt a 310 helix that only forms when the substrate is bound (Fig. 2c). Helix formation enables the leucine methyl groups of the analog to be partially encircled by hydrophobic contacts. From above, the leucine side chain is in close proximity to the α-protons of Thr164 and Val166, whereas from below, it is contacted by the side chains of Val168 and Arg197. The proline ring of the sorting signal is buried in the binding groove by contacts from the side chains of Ile182 (β7) and Ala118 (β4) that project from the underlying sheet and by contacts from residues within both walls of the groove (Leu169 (310 helix), Ala92 (β2/H1 loop), and Ala104 (β3/β4 loop)). This latter interaction is supported by the observation of strong intermolecular NOEs between the methyl groups of the alanine residues and protons within the proline ring.
The LPAT* peptide adopts an “L-shaped” structure as a result of a ∼90° kink at the alanine-proline peptide bond, which is in a trans conformation. The kink redirects the trajectory of the signal, enabling it to approach the active site in parallel with the underlying β-strands of the binding groove. In the LPAT* peptide, an alanine residue mimics the X position of the LPXTG motif. In the structure, the alanine is packed against the side chain of Leu97 located in helix H1 as a result of several strong NOEs to the Leu97 Hδ protons (Fig. 2d). This explains the demonstrated promiscuity of SrtA for this site within the sorting signal, since modeling studies suggest that larger side chains could project away from the enzyme via a cleft located between helix H1 and His120 (39). Recognition is completed by packing of the threonine γ-methyl group beneath the indole ring of Trp194, as substantiated by several NOEs between the methyl and the Hϵ1 proton of the tryptophan. These contacts partially shield the active site from the solvent and help to project the -CH2-SH portion of the threonine analog toward Cys184 for disulfide bond formation.
Sorting Signal Binding Closes and Immobilizes the β6/β7 Loop
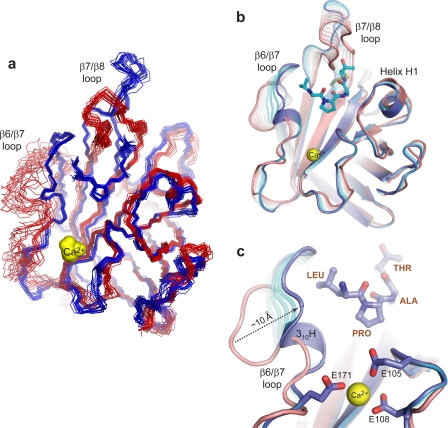
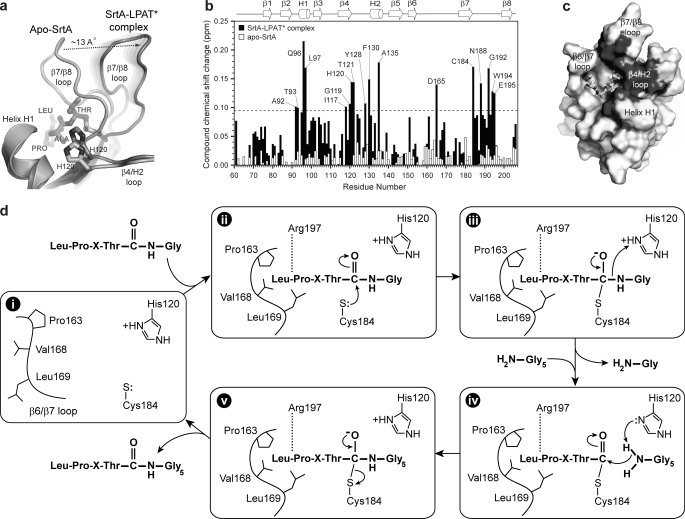
Based on biochemical studies, the active site β6/β7 loop plays an important role in catalysis, since amino acid mutations in this structural element significantly impair enzyme activity and alter the substrate specificity of the enzyme (32, 66). This is compatible with the structure of the complex, since residues from the NH2-terminal portion of the loop directly contact the leucine and proline residues of the sorting signal (Fig. 2, b and c). NMR and x-ray studies of apo-SrtAΔN59 indicate that the β6/β7 loop is unstructured and mobile in the absence of the sorting signal (31, 34). Interestingly, a comparison of the NMR structures of SrtAΔN59 solved in the presence and absence of the sorting signal reveals that substrate binding causes the loop to transition from a structurally disordered and open conformation to an ordered “closed” conformation containing a 310 helix (Fig. 3, a and b). The substrate-induced structural changes are extensive and involve a ∼10-Å displacement of the loop toward the active site (Fig. 3c). The loop is also shortened, as residues Thr156 to Val161 become incorporated into strand β6 by forming a network of new hydrogen bonds to residues within strand β8. Notably, this large structural change was not observed in a previously reported crystal structure of the SrtAΔN59-LPETG complex (see below) (31).
FIGURE 3.
Sorting signal binding induces changes in the structure of SrtA. a, overlay of the ensemble of NMR structures of apo-SrtAΔN59 (34) (Protein Data Bank code 1ija; red) and the SrtAΔN59-LPAT* complex (blue). The comparison shows that the structurally disordered β6/β7 loop becomes ordered upon binding the sorting signal. b, superposition of the average NMR structures of apo-SrtAΔN59 (34) (pink) and the SrtAΔN59-LPAT* complex (blue). Each structure is presented as a schematic diagram with relevant loops labeled. The largest substrate-induced conformational changes occur in residues located within the β6/β7 and β7/β8 loops. This transition is accentuated in the figure by showing hypothetical structural intermediates calculated using the Yale Morph Server (available on the World Wide Web) (68). c, as in b but expanded to show the shift of the β6/β7 loop over the sorting signal and the role that calcium plays in stabilizing the closed conformation of the β6/β7 loop.
To determine whether loop closure over the sorting signal analog quenches its mobility, we measured the R1, R2, and 15N{1H} NOE relaxation parameters of the protein backbone nitrogen atoms in the complex and interpreted these data using the model-free formalism (60–63). This analysis yields the order parameter (S2), which gives a concise account of the backbone amide's mobility on the picosecond time scale. It ranges from 0 to 1, with values of 1 indicating that the amide is completely immobilized. The model-free analysis also yields an Rex term that is diagnostic for the presence of slower micro- to millisecond time scale motions. Fig. 4 compares the S2 and Rex values of SrtAΔN59 in the SrtAΔN59-LPAT* complex with similar data reported for the apo-form of the enzyme (56). In the SrtAΔN59-LPAT* complex, the β6/β7 loop is rigid on fast time scales, as evidenced by S2 values that are on average 0.90 ± 0.01 (Fig. 4a, black). It is also immobile on slower time scales, since only a few residues distributed throughout the protein exhibit small magnitude Rex terms (Fig. 4b, black). This is in marked contrast to the apoenzyme, since many of the residues in the β6/β7 exhibit elevated Rex values and/or weak NMR resonances that indicate that they undergo slow micro- to millisecond time scale motions (Fig. 4b, white and gradient bars). Interestingly, in the apoenzyme, several residues in the loop have S2 values of >0.7, demonstrating that they do not participate in large amplitude picosecond time scale motions (Fig. 4a, gray). Combined, the structural and relaxation data suggest that the loop in the apoenzyme adopts a semirigid state that undergoes micro- to millisecond segmental motions that toggle it between open and closed conformations. Substrate binding quenches these motions, locking the loop in a closed state for productive interactions with the sorting signal.
FIGURE 4.
The β6/β7 loop is immobilized when the enzyme binds to the sorting signal. a, the order parameters (S2) for backbone amides plotted as a function of residue number. Data for apo-SrtAΔN59 (gray) (56) and the SrtAΔN59-LPAT* complex (black) are shown. Data for each residue are indicated by a filled circle and are connected by lines to emphasize trends. Secondary structural elements present in SrtAΔN59 are shown above the graph. b, plot of Rex values determined from the model-free analysis as a function of residue number. A gradient shaded bar indicates residues whose resonances are very weak in the NMR spectra of apo-SrtAΔN59, presumably because of line broadening caused by conformational exchange (Arg159 to Val161). The data are color-coded as in a.
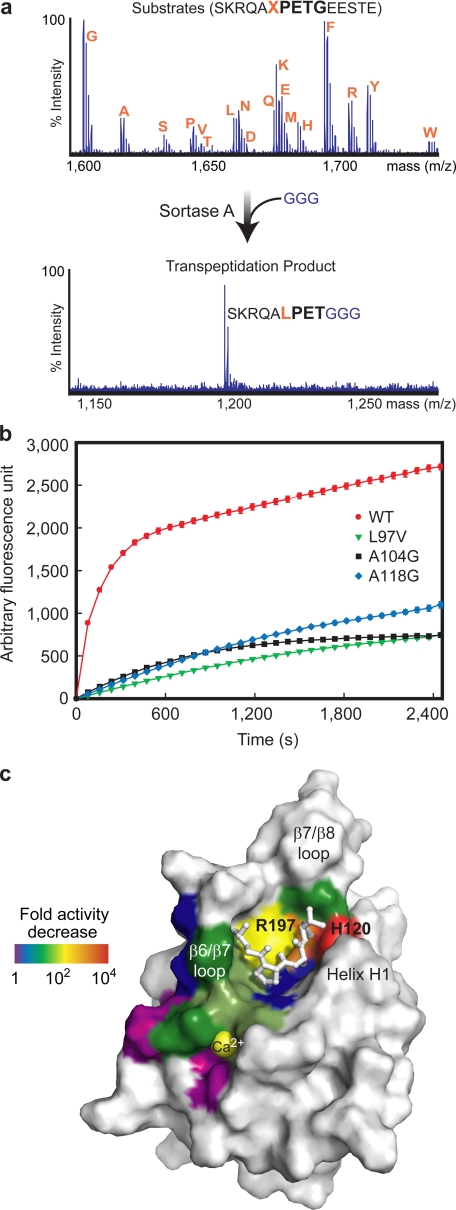
Closure and immobilization of the β6/β7 loop enables extensive enzyme contacts to the leucine residue within the sorting signal. We tested whether these interactions confer specificity for this site by challenging SrtAΔN59 with a peptide library containing the LPETG sequence randomized at the leucine position (SKRQAXPETGEESTE, where X represents any amino acid except for Ile or Cys). Monitoring product formation using mass spectrometry reveals that only peptides bearing a leucine amino acid at this site are effectively processed by the enzyme (Fig. 5a). This supports the notion that loop closure over the signal plays a key role in substrate recognition.
FIGURE 5.
The structure of the SrtAΔN59-LPAT* complex is compatible with biochemical data. a, data showing that SrtA only recognizes sorting signals that contain a leucine residue at the first position of the LPXTG motif. Shown are MALDI-TOF spectra of a peptide library containing a sorting signal randomized at the leucine position before (top) and after (bottom) incubation with SrtA and its second substrate Gly3. Each peptide contains the sequence SKRQAXPETGEESTE, where X is any amino acid except for Ile or Cys. The results of this assay show that SrtA specifically recognizes leucine within the sorting signal. b, enzyme reaction progress curves for wild-type SrtAΔN59 (WT) and L97V, A104G, and A118G mutants. The mutations disrupt protein-sorting signal contacts observed in the solution structure of the SrtAΔN59-LPAT* complex. The data show that each mutant has reduced activity relative to the wild-type enzyme and are compatible with the solution structure of the complex but incompatible with a previously reported crystal structure of the complex (31). The reaction progress curves monitor the transpeptidation of a d-QALPATGEE-e sorting signal substrate using a fluorescence resonance energy transfer assay. Analysis of the kinetics data yielded the following steady-state parameters: wild type, kcat = 5.7 × 10−3 ± 1.5 × 10−4 min−1, Km = 57 ± 9 μm; L97V, kcat = 5.6 × 10−4 ± 1.6 × 10−5 min−1, Km = 2.9 × 102 ± 1 × 102 μm; A104G, kcat = 6.7 × 10−4 ± 8.0 × 10−5 min−1, Km = 1.8 × 102 ± 26 μm; A118G, kcat = 9.0 × 10−4 ± 1.9 × 10−4 min−1, Km = 3.0 × 102 ± 73 μm. It is important to note that the reported kinetic parameters are for the isolated catalytic domain of sortase catalyzing the transpeptidation of a peptide fragment that mimics the intact protein substrate. Different kinetic parameters may be obtained if the intact protein substrate is used or if sortase is located in its native environment, the bacterial cell surface. c summarizes the compatibility of the structure of the SrtAΔN59-LPAT* complex with previously reported biochemical data. The solvent-accessible surface of SrtAΔN59 in the complex is color-coded to show the effects of amino acid mutations on enzyme activity. The -fold reduction of the catalytic activity of each mutant relative to the wild-type protein is shown. A visual spectrum color code is used and ranges from violet (no effect on catalysis) to red (most severe, >2000-fold reduction in activity).
Ca2+ Stabilizes the Closed Conformation of the β6/β7 Loop
In vitro, Ca2+ increases the enzymatic activity of SrtAΔN59 ∼8-fold by lowering the Km of the enzyme for the sorting signal (34, 56). This adaptation may enable S. aureus to increase the number of displayed proteins if elevated concentrations of Ca2+ are encountered at sites of infection. The atomic basis of divalent ion binding is incompletely understood, because all previously reported crystal structures of SrtAΔN59 were solved in the metal-free state (31). Moreover, the binding mechanism of Ca2+ cannot easily be defined by NMR methods, since Ca2+ lacks proton atoms needed to identify NOE distance restraints. In structure calculations of the complex, we employed three artificial distance restraints between a single Ca2+ ion and the side chains of residues Glu105, Glu108, and Glu171. These were included because mutant proteins that replace these residues with alanine are insensitive to Ca2+ and because the chemical shifts of these residues are significantly perturbed when Ca2+ is added (56). Importantly, structures calculated with the artificial restraints are compatible with all of the experimental data. In the structure of the complex, Ca2+ binds to a pocket formed by the β3/β4 (Glu105 and Glu108) and β6/β7 loops (Glu171) (Fig. 3c). As compared with the crystal structure of the apoenzyme solved in the absence of Ca2+, ion contacts from the side chain of Glu171 appear to stabilize the closed, binding-competent conformation of the β6/β7 loop. This conclusion is supported by relaxation data, which indicate that Ca2+ binding to the enzyme retards motions in the loop (56).
Compatibility of the SrtAΔN59-LPAT* Complex with Biochemical Data
The structure of the complex explains the enzymatic properties of 35 previously described amino acid mutants of the S. aureus SrtAΔN59 protein (26, 28–30). These data are not fully described here but are summarized in Fig. 5c. As expected, the most severe effects are observed when residues His120, Cys184, and Arg197 in the active site are mutated, consistent with their direct participation in the transpeptidation reaction. Outside of the enzyme active site, some of the most severe effects occur when the β6/β7 loop is mutated, with V168A and L169A mutations reducing enzymatic activity 5.5- and 93-fold, respectively (32). This is consistent with the structure, since in it, these side chains directly contact the sorting signal (Fig. 2c). Mutations within the Ca2+ binding site also disrupt activity, consistent with the ion acting to stabilize the closed state of the substrate contacting β6/β7 loop (Fig. 3c).
To further substantiate the mode of binding observed in the NMR structure, we created A104G, A118G and L97V mutants of SrtAΔN59 and tested their enzymatic activity using a fluorescence resonance energy transfer assay. Based on the structure, the A104G and A118G mutations should remove methyl groups from the floor and the sides of the binding groove that contact the proline ring (Fig. 2c). This is consistent with our results, since these mutants are 27–34-fold less active than the wild-type protein (Fig. 5b). The side chain of Leu97 is conserved in >97% of all sortase enzymes, and in the structure of the complex, its side chain packs against the backbone of the sorting signal (Fig. 2d). Our results indicate that these contacts play an important role in stabilizing the positioning of the sorting signal, since even a conservative L97V mutant of SrtAΔN59 is 10-fold less active than the wild-type protein (Fig. 5b). It is important to note that the structure of the complex reported in this paper is not a perfect mimic of the thio-acyl intermediate, since the peptide contains an extra methylene group that separates the Cys184 thiol from the threonine. However, the spacer only adds ∼2 Å, which is unlikely to significantly alter the way in which the peptide is recognized by the enzyme. This is especially true for recognition of the leucine residue of the peptide, which is positioned distal to the active site.
Some Sortase Enzymes May Contain a Preformed Binding Pocket for the LPXTG Sorting Signal
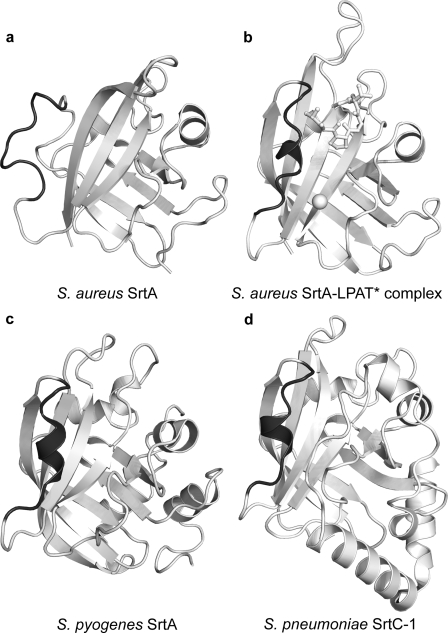
In addition to the SrtA enzyme from S. aureus, the structures of three sortase enzymes that recognize the LPXTG sorting signal have now been determined (Fig. 6) (37, 38). The structures were solved in the absence of the sorting signal and are of enzymes that share only limited sequence homology with S. aureus SrtA. They include S. pyogenes SrtA (26% sequence identity) and the SrtC-1 (25% sequence identity) and SrtC-3 (27% sequence identity) enzymes from S. pneumoniae. Interestingly, these enzymes adopt the same β-barrel fold observed in the S. aureus enzyme; the backbone coordinates of the S. pyogenes SrtAΔN81, and the S. pneumoniae SrtC-1ΔN16 and SrtC-3ΔN31 enzymes can be superimposed with the coordinates of the S. aureus SrtAΔN59 enzyme with an r.m.s. deviation of 1.8, 1.6, and 1.7 Å, respectively. Additionally, the arginine, cysteine, and histidine active site are positioned similarly.
FIGURE 6.
Some sortase enzymes contain a preformed binding pocket for the LPXTG sorting signal. The figure compares the three-dimensional structures of sortase enzymes that recognize LPXTG sorting signals. Four structures of SrtA-type sortases are shown. a, the crystal structure of S. aureus apo-SrtAΔN59 (Protein Data Bank code 1t2p) (31); b, the NMR structure of the S. aureus SrtAΔN59-LPAT* complex; c, the crystal structure of S. pyogenes apo-SrtAΔN81 (Protein Data Bank code 3fn5) (37); d, the crystal structure of S. pneumoniae apo-SrtC-1ΔN16 (Protein Data Bank code 2w1j) enzyme (38). The β6/β7 loop in each structure is highlighted in dark gray to emphasize differences and similarities. The comparison reveals that the β6/β7 loops in the S. pneumoniae and S. pyogenes enzymes adopt a closed helical conformation similar to the substrate-bound form of the S. aureus SrtA enzyme in the SrtAΔN59-LPAT* complex. This is in marked contrast to the S. aureus apoenzyme, which adopts an open conformation. Note that the structure of S. pneumoniae apo-SrtC-3ΔN31 has also been determined and is not shown here, because it is generally similar to the structure of S. pneumoniae apo-SrtC-1ΔN16 (38).
The S. pyogenes and S. pneumoniae enzymes may contain a preformed binding pocket for the sorting signal. In this paper, we have shown that binding of the sorting signal to the S. aureus SrtAΔN59 enzyme causes major changes in the structure and dynamics of the β6/β7 loop. It transitions from a disordered open state to an ordered closed conformation that contains a 310 helix (compared in Fig. 6, a and b). Intriguingly, although the structures of the S. pyogenes and S. pneumoniae enzymes were solved in the absence of the sorting signal, their β6/β7 loops also adopt a closed conformation that contains the 310 helix (Fig. 6, c and d). This conformation is similar to the loop structure in the SrtAΔN59-LPAT* complex, suggesting that in the S. pyogenes and S. pneumoniae enzymes, only modest changes in the structure of the β6/β7 loop are needed to bind the sorting signal.
In the crystal structures of the S. pneumoniae SrtC-1 and SrtC-3 enzymes, an additional polypeptide segment is inserted into the active site and has been proposed to act as a “lid” that opens and closes during pilus assembly (38). Remarkably, two of the residues in the lid are embedded in the sorting signal binding pocket (Pro59-Trp60 in SrtC-1 or Pro74-Phe75 in SrtC-3) in a similar position as the proline and alanine residues of the sorting signal in the NMR structure of the SrtAΔN59-LPAT* complex (data not shown). This suggests that the lid in these proteins occludes the substrate binding site by mimicking the PX portion of the LPXTG sorting signal.
The NMR and Crystal Structures of the Sortase-Substrate Complex Differ Significantly
The crystal structure of a non-covalent complex between C184ASrtAΔN59 (SrtAΔN59 containing a C184A mutation) and an LPETG peptide has been determined (Protein Data Bank code 1t2w) (31). Interestingly, it differs substantially from the structure of the covalent SrtAΔN59-LPAT* complex reported in this paper. In the crystal structure, the peptide adopts an extended conformation, whereas in the covalent SrtAΔN59-LPAT* complex, a ∼90° kink occurs between the alanine and proline residues (Fig. 7a). The peptide in the SrtAΔN59-LPAT* complex is also positioned closer to the side chain of Cys184 within the active site. These differences cause the leucine, proline and X positions within the sorting signals to be contacted by different amino acids in each complex. Indeed, none of the contacts between SrtA and the sorting signal substrate are conserved.
FIGURE 7.
The structures of the SrtAΔN59-LPAT* and C184ASrtAΔN59-LPETG complexes differ substantially. a, structures of the covalent SrtAΔN59-LPAT* complex (left) and the previously reported structure of the C184ASrtAΔN59-LPETG complex (right) (31). The solvent-accessible surface of the protein and the structure of the bound peptide are shown. The surface has been colored to indicate the electrostatic properties from acidic (red) to basic (blue). b, ribbon diagrams of the covalent SrtAΔN59-LPAT* complex (left, colored blue) and the C184ASrtAΔN59-LPETG complex (right, colored tan). The secondary structural elements, relevant substrate contacting loops and amino acids within each peptide are labeled. The Ca2+ ion in the covalent SrtAΔN59-LPAT* complex is represented by a yellow sphere and labeled. The images have been rotated by 60° relative to a.
The conformations of the β6/β7 and β7/β8 loops differ substantially. In the non-covalent C184ASrtAΔN59-LPETG complex, the loop adopts an “open” conformation in which it extends away from the body of the protein. In contrast, the loop in the covalent SrtAΔN59-LPAT* complex is in a “closed” conformation that places it closer to the active site (Fig. 7b). The largest differences in loop positioning occur at residue Val166, whose α-carbon is shifted by ∼10 Å toward the active site in the covalent complex. There are also substantial differences in the structure of the β6/β7 loop itself. In the NMR structure, it contains a 310 helix at its center and is shorter in length by approximately 6 residues. The length difference is caused by different conformations of residues Thr156–Val161. In the crystal structure, they adopt an unusual bulge structure, whereas in the covalent SrtAΔN59-LPAT* complex, they are incorporated into strand β6. The positioning of the β7/β8 loop in the two complexes also differs. In the non-covalent complex, the COOH-terminal end of the β7/β8 loop is positioned adjacent to residues at the NH2-terminal end of helix H1. However, in the covalent SrtAΔN59-LPAT* complex, attachment of the peptide to the Cys184 thiol causes these structural elements to separate from one another (Figs. 7b and 8a).
FIGURE 8.
Localization of the lipid II binding site and proposed mechanism of transpeptidation. a, superposition of the NMR structures of apo-SrtAΔN59 (34) (Protein Data Bank code 1ija) and the SrtAΔN59-LPAT* complex. The image shows an expanded view of the substrate-dependent structural change in the β7/β8 loop that unmasks a groove leading into the active site. The image was generated in a similar manner as the one shown in Fig. 3b. b, histogram plot of the compound chemical shift changes for the backbone amide hydrogen and nitrogen atoms of SrtAΔN59 after the addition of the Gly3 tripeptide. Chemical shift changes after the addition of the peptide to either the SrtAΔN59-LPAT* complex (black) or SrtAΔN59 in its apo-state (white) are shown. The secondary structure of the enzyme and amino acids experiencing the largest changes are labeled. The data indicate that only SrtA in the context of the SrtAΔN59-LPAT* complex binds to the Gly3 tripeptide. The dashed line represents one S.D. above the average chemical shift perturbation of all amino acids. c, solvent-accessible surface of SrtAΔN59 in the complex with residues that are significantly perturbed by the addition of the Gly3 peptide colored dark gray. The majority of the chemical shift changes occur near the active site around the surface uncovered when the β7/β8 is displaced from helix H1. d, transpeptidation mechanism based on the structure of the SrtAΔN59-LPAT* complex and biochemical data. i, apoenzyme containing a flexible β6/β7 loop; ii, sorting signal binding closes the β6/β7 loop. Arg197 hydrogen-bonds to the backbone, and the threonine carbonyl carbon is attacked by the thiolate of Cys184. iii, the displaced β7/β8 loop serves as an exit point for residues COOH-terminal to the LPXTG motif. This enables His120 to protonate the amide of the glycine residue as the scissile bond is broken. iv, the cross-bridge peptide of lipid II enters the active site via the surface unmasked by the movement of the β7/β8 loop. v, a second tetrahedral intermediate forms, followed by the rupture of the Cys184-sorting signal linkage and product release.
Several lines of evidence suggest that the LPXTG sorting signal in the crystal structure of the non-covalent C184ASrtAΔN59-LPETG complex is nonspecifically bound to the enzyme. First, in the structure of the C184ASrtAΔN59-LPETG complex, the leucine side chain in the sorting signal projects into the solvent and is not contacted by the enzyme (Fig. 7b). This contradicts biochemical data that indicate that the leucine residue of the signal is specifically recognized by the enzyme (Fig. 5a), and it does not explain why the leucine is completely conserved in the sorting signals of S. aureus proteins anchored by SrtA (22, 39). Second, the structure of the non-covalent C184ASrtAΔN59-LPETG complex is incompatible with the enzymatic properties of several amino acid mutants of SrtA. For example, recently reported E171A and V168A mutations reduce activity 5.4- and 5.5-fold, respectively (32). However, in the C184ASrtAΔN59-LPETG complex, the side chains of these residues project into the solvent and do not contact the peptide. Another discrepancy is the positioning of the side chain of Gln172. Based on the structure of the C184ASrtAΔN59-LPETG complex, it should be important for catalysis, because it forms a hydrogen bond to the backbone of the sorting signal, but a Q172A mutant surprisingly retains wild-type activity (32, 56). The reduced enzymatic activities of the A104G, A118G, and L97V mutants of SrtAΔN59 reported in this paper are also incompatible, since in the crystal structure, the side chains of these residues do not contact the sorting signal (Fig. 5b). Finally, the structure of the C184ASrtAΔN59-LPETG complex is incompatible with intermolecular NOE data (Fig. 1a). The most significant discrepancies occur at the protein interface that interacts with the proline and leucine residues of the sorting signal. For example, unambiguous NOEs are observed between the proline side chain of the sorting signal and the methyl groups of Ala118 and Val166, indicating that they are separated by <5 Å (Fig. 1c). However, in the crystal structure, these hydrogen atoms are no closer than 11 Å apart. In sum, the previously published crystal structure of the non-covalent C184ASrtAΔN59-LPETG complex is incompatible with our NMR data and the enzymatic properties of several amino acid mutants, and it does not explain why the leucine residue is highly conserved in LPXTG sorting signals.
It seems likely that the sorting signal in the C184ASrtAΔN59-LPETG complex is nonspecifically bound to the enzyme, because the β6/β7 loop failed to undergo a disordered to ordered transition. The x-ray structure of the complex was solved by molecular replacement using crystals of apo-C184ASrtAΔN59 soaked with the LPETG peptide (31). Three molecules of SrtAΔN59 were present in the asymmetric unit of the crystal, and in each, the β6/β7 loop adopted fundamentally distinct conformations. This structural heterogeneity may be caused by protein-protein interactions within the crystal as the β6/β7 loop interacts with other proteins in the unit cell. In addition, the loop may be mobile in the crystal, since in each of the three SrtAΔN59 models, residues Lys162 to Lys175 have B-factors in excess of 40 Å2, or their electron density is missing. Moreover, NMR relaxation studies indicate that the loop is mobile in the absence of the sorting signal (56). Notably, when the peptide is added to the crystal, it binds to only one of the SrtAΔN59 enzymes in the asymmetric unit, and the β6/β7 loop in this protein does not change its structure. Thus, it appears that the β6/β7 loop in the C184ASrtAΔN59-LPETG complex failed to undergo substrate-induced structural changes required to properly recognize the signal, presumably because SrtA only weakly binds the LPXTG signal and/or because of lattice packing interactions (25). This problem has been overcome in the NMR structure of the SrtAΔN59-LPAT* complex, because the peptide is covalently attached to the enzyme.
Function of Arg197
All sortases contain a highly conserved arginine residue within their active site (28). This residue corresponds to Arg197 in SrtA, which when mutated significantly reduces catalytic activity (28–30, 32). The function of Arg197 in catalysis has hitherto remained unclear, since it has been proposed to either deprotonate Cys184 (28) or lipid II (31) or, alternatively, to stabilize either the binding of the sorting signal (28, 32) or oxyanion intermediates (31, 32). In many of the structures in the ensemble, the side chain of Arg197 is wedged between Pro163 of the β6/β7 loop and the sorting signal (Fig. 2d). Here it presumably stabilizes the binding of the substrate in the active site by donating hydrogen bonds from its guanidine group to the backbone carbonyl oxygens of the leucine and proline residues of the sorting signal. It is important to note that the NMR data does not directly reveal the presence of these hydrogen bonds. Instead, they are inferred from the positioning of Arg197 relative to the substrate, which is defined by NOEs between the side chains of Arg197 and Val161 in the β6/β7 loop as well as NOEs between Arg197 and the leucine residue within the substrate. Contacts to the substrate require the specific chemical structure of the guanidine group, since mutations that replace Arg197 with alanine or lysine dramatically decrease activity, whereas replacement with an isosteric citrulline side chain has little impact on catalysis (32).
Location of the Binding Site for the Second Substrate of Catalysis, the Triglycine Portion of Lipid II
SrtA anchors proteins through a ping-pong mechanism in which the second substrate of catalysis, lipid II, nucleophilically attacks the thioacyl-linked complex between SrtA and the LPXTG sorting signal (24, 25). The structure of the SrtAΔN59-LPAT* complex mimics this thioacyl intermediate and thereby sheds light onto how lipid II might be recognized by the enzyme (Fig. 1b). A comparison of the substrate-free and -bound forms of the enzyme reveals that attachment of the sorting signal to Cys184 causes a large ∼13-Å displacement of the β7/β8 loop (Figs. 3b and 8a). The displacement is required in order to accommodate the insertion of the threonine side chain of the sorting signal underneath the indole ring of Trp194 (Fig. 2d). This acts as a wedge at the base of the loop that shifts it as a rigid unit away from helix H1, creating a new groove that leads into the active site. The floor of the exposed groove is formed by residues immediately following strand β4, and its walls are formed by residues in helix H1 and the β7/β8 loop. Interestingly, the center of the exposed groove contains the catalytically important His120 side chain, suggesting that it might serve as the entry point into the active site for lipid II.
To investigate whether the exposed groove serves as a binding site for lipid II, we performed chemical shift perturbation experiments. In vivo, SrtA joins the terminal amine group of the pentaglycine branch of lipid II to the threonine carbonyl carbon of the LPXTG sorting signal (18–20). A Gly3 peptide mimics this portion of lipid II and can be effectively used by the enzyme as a substrate in vitro (23). To locate the surface on the enzyme that interacts with Gly3, we titrated a sample of the SrtAΔN59-LPAT* complex with the peptide and used NMR to monitor the chemical shifts of the backbone amide atoms of the protein. A histogram plot of the chemical shift differences in the presence and absence of Gly3 reveals that the peptide selectively perturbs the NMR spectrum of the protein (Fig. 8b, black bars). Interestingly, when the most significantly perturbed residues are mapped onto the structure of the protein in the SrtAΔN59-LPAT*, they define a continuous surface that encompasses the groove that is unmasked when the sorting signal binds (Fig. 8c). When the apo-form of the enzyme is titrated with the Gly3 peptide, the backbone amide chemical shifts of residues within this surface are not perturbed (Fig. 8b, white bars). Combined, these data suggest that sorting signal-induced displacement of the β7/β8 loop unmasks the binding site for the Gly3 portion of lipid II and may thereby direct catalysis toward product formation.
Mechanism of Catalysis
The structure of the complex enables a more detailed mechanism of the SrtA-catalyzed transpeptidation reaction to be proposed (Fig. 8d). The apoenzyme is dynamic, with residues in the β6/β7 loop undergoing motions that periodically displace them from the active site (56) (step i in Fig. 8d). Substrate binding nucleates the folding and closure of the loop over the sorting signal, enabling recognition (step ii). Based on NMR and enzyme kinetic studies of the apo-form of sortase, the pKa values of His120 and Cys184 are ∼6.3–7 and ∼9.4, respectively (25, 33). Therefore, at physiological pH values, the predominant form of the enzyme contains His120 and Cys184 in their uncharged states. Interestingly, solvent isotope effect measurements have led to the suggestion that this form of the enzyme is inactive. Instead, the active form of sortase has been proposed to be sparsely populated (0.06% of the total protein) and to contain His120 and Cys184 both in their ionized states (25). Therefore, the incoming threonine carbonyl carbon of the sorting signal is probably attacked by the thiolate of Cys184, resulting in the concerted displacement of the β7/β8 loop (step iii). This opens a large groove leading out of the active site that accommodates residues positioned on the COOH-terminal side of the threonine residue of the sorting signal in the surface protein precursor. The groove contains the imidazolium side chain of His120 at its center, which is poised to protonate the amide leaving group as the scissile bond is broken (67). NMR chemical shift experiments indicate that the groove also functions as the entry point for the Gly5 portion of lipid II, suggesting that the newly deprotonated His120 side chain activates the incoming terminal amine of lipid II for nucleophilic attack on the enzyme-linked thioacyl intermediate (step iv). The dual function of His120 as a general acid and base is consistent with its measured pKa value of ∼6.3–7 and is functionally similar to histidine residues in a number of other enzymes, including serine proteases (33, 71). The reaction is then completed by formation of the second tetrahedral intermediate and subsequent breakage of the enzyme-substrate bond to liberate the protein-lipid II-linked product (step v). In all steps, Arg197 plays a key role in catalysis by stabilizing the positioning of the substrate through direct hydrogen bonding to its polypeptide backbone. In addition, the side chain of Arg197 is properly positioned in several conformers in the NMR ensemble to stabilize both tetrahedral intermediates of catalysis by interacting with the oxyanion.
SrtA is required for the virulence of multidrug-resistant methicillin resistant S. aureus, which in the United States kills an estimated 18,000 people annually (67). In addition, a number of other important human pathogens contain SrtA homologs that when genetically eliminated cause defects in virulence. The data presented here should therefore aid in the design of small molecule sortase inhibitors that are useful in treating a range of bacterial infections.
Supplementary Material
Acknowledgments
We thank Dr. Robert Peterson for assistance with the NMR experiments, Dr. Joseph A. Loo for mass spectrometry experiments, Drs. Mandar T. Naik and Rosemarie L. Pilpa for technical assistance with the NMR relaxation studies, and Drs. James U. Bowie and Scott A. Robson for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AI52217 (to R. T. C. and M. E. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- SrtA
- sortase A
- Cbz
- benzylozycarbonyl
- T*
- (2R,3S)-3-amino-4-mercapto-2-butanol analog of threonine that replaces the carbonyl group with -CH2-SH
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- r.m.s.
- root mean square.
REFERENCES
- 1.Navarre W. W., Schneewind O. (1999) Microbiol. Mol. Biol. Rev. 63, 174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marraffini L. A., Dedent A. C., Schneewind O. (2006) Microbiol. Mol. Biol. Rev. 70, 192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson G. K., Mitchell T. J. (2004) Trends Microbiol. 12, 89–95 [DOI] [PubMed] [Google Scholar]
- 4.Ton-That H., Marraffini L. A., Schneewind O. (2004) Biochim. Biophys. Acta 1694, 269–278 [DOI] [PubMed] [Google Scholar]
- 5.Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott J. R., Zähner D. (2006) Mol. Microbiol. 62, 320–330 [DOI] [PubMed] [Google Scholar]
- 7.Maresso A. W., Schneewind O. (2008) Pharmacol. Rev. 60, 128–141 [DOI] [PubMed] [Google Scholar]
- 8.Suree N., Jung M. E., Clubb R. T. (2007) Mini Rev. Med. Chem. 7, 991–1000 [DOI] [PubMed] [Google Scholar]
- 9.Mao H., Hart S. A., Schink A., Pollok B. A. (2004) J. Am. Chem. Soc. 126, 2670–2671 [DOI] [PubMed] [Google Scholar]
- 10.Chan L., Cross H. F., She J. K., Cavalli G., Martins H. F., Neylon C. (2007) PLoS ONE 2, e1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popp M. W., Antos J. M., Grotenbreg G. M., Spooner E., Ploegh H. L. (2007) Nat. Chem. Biol. 3, 707–708 [DOI] [PubMed] [Google Scholar]
- 12.Clow F., Fraser J. D., Proft T. (2008) Biotechnol. Lett. 30, 1603–1607 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T., Yamamoto T., Tsukiji S., Nagamune T. (2008) ChemBioChem 9, 802–807 [DOI] [PubMed] [Google Scholar]
- 14.Samantaray S., Marathe U., Dasgupta S., Nandicoori V. K., Roy R. P. (2008) J. Am. Chem. Soc. 130, 2132–2133 [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian S. K., Liu G., Ton-That H., Schneewind O. (1999) Science 285, 760–763 [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H., Liu G., Mazmanian S. K., Faull K. F., Schneewind O. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12424–12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneewind O., Model P., Fischetti V. A. (1992) Cell 70, 267–281 [DOI] [PubMed] [Google Scholar]
- 18.Perry A. M., Ton-That H., Mazmanian S. K., Schneewind O. (2002) J. Biol. Chem. 277, 16241–16248 [DOI] [PubMed] [Google Scholar]
- 19.Ruzin A., Severin A., Ritacco F., Tabei K., Singh G., Bradford P. A., Siegel M. M., Projan S. J., Shlaes D. M. (2002) J. Bacteriol. 184, 2141–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneewind O., Fowler A., Faull K. F. (1995) Science 268, 103–106 [DOI] [PubMed] [Google Scholar]
- 21.Comfort D., Clubb R. T. (2004) Infect. Immun. 72, 2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pallen M. J., Lam A. C., Antonio M., Dunbar K. (2001) Trends Microbiol. 9, 97–102 [DOI] [PubMed] [Google Scholar]
- 23.Ton-That H., Mazmanian S. K., Faull K. F., Schneewind O. (2000) J. Biol. Chem. 275, 9876–9881 [DOI] [PubMed] [Google Scholar]
- 24.Huang X., Aulabaugh A., Ding W., Kapoor B., Alksne L., Tabei K., Ellestad G. (2003) Biochemistry 42, 11307–11315 [DOI] [PubMed] [Google Scholar]
- 25.Frankel B. A., Kruger R. G., Robinson D. E., Kelleher N. L., McCafferty D. G. (2005) Biochemistry 44, 11188–11200 [DOI] [PubMed] [Google Scholar]
- 26.Ton-That H., Mazmanian S. K., Alksne L., Schneewind O. (2002) J. Biol. Chem. 277, 7447–7452 [DOI] [PubMed] [Google Scholar]
- 27.Zhang R., Wu R., Joachimiak G., Mazmanian S. K., Missiakas D. M., Gornicki P., Schneewind O., Joachimiak A. (2004) Structure 12, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marraffini L. A., Ton-That H., Zong Y., Narayana S. V., Schneewind O. (2004) J. Biol. Chem. 279, 37763–37770 [DOI] [PubMed] [Google Scholar]
- 29.Liew C. K., Smith B. T., Pilpa R., Suree N., Ilangovan U., Connolly K. M., Jung M. E., Clubb R. T. (2004) FEBS Lett. 571, 221–226 [DOI] [PubMed] [Google Scholar]
- 30.Frankel B. A., Tong Y., Bentley M. L., Fitzgerald M. C., McCafferty D. G. (2007) Biochemistry 46, 7269–7278 [DOI] [PubMed] [Google Scholar]
- 31.Zong Y., Bice T. W., Ton-That H., Schneewind O., Narayana S. V. (2004) J. Biol. Chem. 279, 31383–31389 [DOI] [PubMed] [Google Scholar]
- 32.Bentley M. L., Lamb E. C., McCafferty D. G. (2008) J. Biol. Chem. 283, 14762–14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly K. M., Smith B. T., Pilpa R., Ilangovan U., Jung M. E., Clubb R. T. (2003) J. Biol. Chem. 278, 34061–34065 [DOI] [PubMed] [Google Scholar]
- 34.Ilangovan U., Ton-That H., Iwahara J., Schneewind O., Clubb R. T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6056–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong Y., Mazmanian S. K., Schneewind O., Narayana S. V. (2004) Structure 12, 105–112 [DOI] [PubMed] [Google Scholar]
- 36.Maresso A. W., Wu R., Kern J. W., Zhang R., Janik D., Missiakas D. M., Duban M. E., Joachimiak A., Schneewind O. (2007) J. Biol. Chem. 282, 23129–23139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Race P. R., Bentley M. L., Melvin J. A., Crow A., Hughes R. K., Smith W. D., Sessions R. B., Kehoe M. A., McCafferty D. G., Banfield M. J. (2009) J. Biol. Chem. 284, 6924–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manzano C., Contreras-Martel C., El Mortaji L., Izoré T., Fenel D., Vernet T., Schoehn G., Di Guilmi A. M., Dessen A. (2008) Structure 16, 1838–1848 [DOI] [PubMed] [Google Scholar]
- 39.Kruger R. G., Otvos B., Frankel B. A., Bentley M., Dostal P., McCafferty D. G. (2004) Biochemistry 43, 1541–1551 [DOI] [PubMed] [Google Scholar]
- 40.Jung M. E., Clemens J. J., Suree N., Liew C. K., Pilpa R., Campbell D. O., Clubb R. T. (2005) Bioorg. Med. Chem. Lett. 15, 5076–5079 [DOI] [PubMed] [Google Scholar]
- 41.Cavanagh J., Fairbrother W. J., Palmer A. G., Skelton N. J. (2006) Protein NMR spectroscopy, 2nd Ed., Elsevier Science and Technology, San Diego, CA [Google Scholar]
- 42.Iwahara J., Iwahara M., Daughdrill G. W., Ford J., Clubb R. T. (2002) EMBO J. 21, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwahara J., Wojciak J. M., Clubb R. T. (2001) J. Biomol. NMR 19, 231–241 [DOI] [PubMed] [Google Scholar]
- 44.Ogura K., Terasawa H., Inagaki F. (1996) J. Biomol. NMR 8, 492–498 [DOI] [PubMed] [Google Scholar]
- 45.Zwahlen C., Legault P., Vincent S. J., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 46.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 47.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 49.Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 50.Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, Cantina Verlag, Goldau, Switzerland [Google Scholar]
- 51.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 52.Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 53.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 54.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 55.DeLano W. L. (2006) The PyMOL Molecular Graphics System, DeLano Scientific, LLC, Palo Alto, CA [Google Scholar]
- 56.Naik M. T., Suree N., Ilangovan U., Liew C. K., Thieu W., Campbell D. O., Clemens J. J., Jung M. E., Clubb R. T. (2006) J. Biol. Chem. 281, 1817–1826 [DOI] [PubMed] [Google Scholar]
- 57.Iwahara J., Peterson R. D., Clubb R. T. (2005) Protein Sci. 14, 1140–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brüschweiler R. (2003) Curr. Opin. Struct. Biol. 13, 175–183 [DOI] [PubMed] [Google Scholar]
- 59.Brüschweiler R., Liao X., Wright P. E. (1995) Science 268, 886–889 [DOI] [PubMed] [Google Scholar]
- 60.Lipari G., Szabo A. (1982) J. Am. Chem. Soc. 104, 4559–4570 [Google Scholar]
- 61.Lipari G., Szabo A. (1982) J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 62.Mandel A. M., Akke M., Palmer A. G., 3rd (1995) J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 63.Mandel A. M., Akke M., Palmer A. G., 3rd (1996) Biochemistry 35, 16009–16023 [DOI] [PubMed] [Google Scholar]
- 64.Mao H. (2004) Protein Expr. Purif. 37, 253–263 [DOI] [PubMed] [Google Scholar]
- 65.Kruger R. G., Dostal P., McCafferty D. G. (2004) Anal. Biochem. 326, 42–48 [DOI] [PubMed] [Google Scholar]
- 66.Bentley M. L., Gaweska H., Kielec J. M., McCafferty D. G. (2007) J. Biol. Chem. 282, 6571–6581 [DOI] [PubMed] [Google Scholar]
- 67.Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., Fridkin S. K. (2007) JAMA 298, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 68.Krebs W. G., Gerstein M. (2000) Nucleic Acids Res. 28, 1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clore G. M., Garrett D. S. (1999) J. Am. Chem. Soc. 121, 9008–9012 [Google Scholar]
- 70.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 71.Hedstrom L. (2002) Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 72.Parthasarathy R., Subramanian S., Boder E. T. (2007) Bioconjugate Chem. 18, 469–476 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.