Abstract
Tailed bacteriophages and herpes viruses use powerful molecular motors to translocate DNA into a preassembled prohead and compact the DNA to near crystalline density. The phage T4 motor, a pentamer of 70-kDa large terminase, gp17, is the fastest and most powerful motor reported to date. gp17 has an ATPase activity that powers DNA translocation and a nuclease activity that cuts concatemeric DNA and generates the termini of viral genome. An 18-kDa small terminase, gp16, is also essential, but its role in DNA packaging is poorly understood. gp16 forms oligomers, most likely octamers, exhibits no enzymatic activities, but stimulates the gp17-ATPase activity, and inhibits the nuclease activity. Extensive mutational and biochemical analyses show that gp16 contains three domains, a central oligomerization domain, and N- and C-terminal domains that are essential for ATPase stimulation. Stimulation occurs not by nucleotide exchange or enhanced ATP binding but by triggering hydrolysis of gp17-bound ATP, a mechanism reminiscent of GTPase-activating proteins. gp16 does not have an arginine finger but its interaction with gp17 seems to position a gp17 arginine finger into the catalytic pocket. gp16 inhibits DNA translocation when gp17 is associated with the prohead. gp16 restricts gp17-nuclease such that the putative packaging initiation cut is made but random cutting is inhibited. These results suggest that the phage T4 packaging machine consists of a motor (gp17) and a regulator (gp16). The gp16 regulator is essential to coordinate the gp17 motor ATPase, translocase, and nuclease activities, otherwise it could be suicidal to the virus.
During the late stages in the life cycle of tailed bacteriophages and herpes viruses, a metabolically highly active viral genome is disengaged from processes such as transcription and recombination and packaged into a viral capsid to near crystalline density (1, 2). In phage T4, a ∼171-kb 56-μm long DNA molecule is compacted within a 120 nm × 86 nm capsid shell (3). Many viruses use powerful molecular motors assembled at a special portal vertex of the icosahedral capsid to accomplish this task (4). The DNA is translocated through a channel formed by the dodecameric portal protein, utilizing ATP as an energy source (5). The phage T4 motor, packaging up to ∼2000 bp/s and having a power density of ∼5,000 kilowatts/m3, is the fastest and most powerful molecular motor reported to date (6).
The structure of the phage T4 DNA packaging motor has been determined recently (7, 8). It is a pentamer of the large terminase,3 gene product (gp)4 17, which, when assembled on the prohead, forms a continuous channel with the portal. gp17 consists of two domains connected by a flexible hinge, an N-terminal ATPase domain that provides energy for pumping DNA and a C-terminal translocase domain that moves DNA (9, 10). The ATPase domain has two subdomains, a larger RecA-like nucleotide binding subdomain I and a smaller subdomain II, forming a cleft into which ATP binds. The C-domain has an RNase H fold that most closely resembles resolvases and a putative DNA translocation groove facing the motor channel.
T4 and most tailed phages and herpes viruses replicate their genomes as a head-to-tail concatamer. A sequence nonspecific nuclease activity associated with gp17 cuts the concatamer once at the beginning of DNA packaging (initiation cut) and again when one headful DNA, equivalent to a ∼1.02-length viral genome, is packaged into the capsid (termination cut) (11–13). The nuclease center consisting of a DNA binding groove and a Mg-coordinated catalytic triad of acidic amino acids is localized in the C-domain, facing the outside of the motor (8). Mutational and biochemical analyses established the catalytic residues in ATPase, translocase, and nuclease functional motifs that are critical for various steps of DNA packaging (14–18).
An electrostatic force-dependent packaging mechanism was proposed based on different conformational states of the motor (8). ATP hydrolysis causes rotation of subdomain II, aligning an array of complementary charged pairs and hydrophobic surfaces between the N- and C-domains. The electrostatic attractive force pulls the DNA-bound C-domain by ∼7 Å, translocating 2 bp of DNA into the capsid. The adjacent C-domain then comes into alignment with the DNA phosphates, binding DNA, and repeating the translocation cycle. After one headful DNA is packaged, a signal is presumably transmitted to gp17, causing it to dissociate from the capsid and make the termination cut (13). Thus the ATPase, translocase, and nuclease activities of the packaging motor are precisely coordinated to package one complement of viral genome into each capsid.
Tailed phages express another packaging/terminase protein, the small terminase, which is also essential for phage viability (1, 2). Mutations in the T4 small terminase, gp16, result in the production of empty proheads (19). The small terminase, unlike the large terminase, does not possess any enzymatic activities, although a weak low affinity ATPase activity was reported for the phage λ small terminase, gpNu1 (20–22). In cos phages (e.g. phage λ (23)) and pac phages (e.g. phages SPP1 (24) and P22 (25)), the small terminase recognizes a sequence in the viral genome and helps assemble a holoenzyme complex by interacting with the large terminase (1, 26). The nuclease activity of the large terminase then cleaves the DNA concatamer at a nearby site, and the complex then docks on to the portal to initiate translocation (1). On the other hand, the T4 gp16, unlike the cos and pac phage small terminases, shows weak nonspecific binding to DNA, that too only in a partially folded state (27), and the mature phage genomic ends are nearly random (28, 29). Evidence indicates that the small terminase may play broader roles in the post-DNA recognition steps of the packaging process (30). Thus there is much to be understood about the mechanism of action of the small terminase in DNA packaging.
Here we report mutational and biochemical analyses of gp16, which show that gp16 contains three functional domains, a central domain that is required for oligomerization, and N- and C-terminal domains that are required for gp17-ATPase stimulation.5 Our results further show that gp16 stimulates the gp17-ATPase activity by triggering the hydrolysis of gp17-bound ATP in a manner similar to that of the GTPase-activating proteins (GAPs). gp16 modulates gp17-dependent DNA translocation through interaction with the prohead-assembled gp17 complex. gp16 restricts gp17-nuclease such that the putative packaging initiation cut is made but extensive random DNA cutting is inhibited. These data suggest that the gp16 small terminase is a global regulator of the T4 DNA packaging motor, modulating the ATPase, translocase, and nuclease activities of the large terminase gp17. Such a regulator might be essential for orchestrating various activities of the packaging process. Hypotheses on how a small regulator molecule can modulate a complex motor are discussed.
EXPERIMENTAL PROCEDURES
Construction of g16 Mutant Clones
The g16 DNA fragments were amplified by PCR using purified phage T4 DNA as template and appropriate primers. The N- and C-terminal truncation clones were constructed by amplifying the desired part of g16. The single point mutants, R41A, R53E, R81A, Q61A, and D44A, were constructed by PCR-directed splicing-by-overlap-extension strategy that was described in detail elsewhere (32, 14). The amplified DNAs were purified by agarose gel electrophoresis, digested with appropriate restriction enzyme(s), and ligated to the linearized pET-15b or pET-28b plasmid DNA. Insertion in the right orientation results in the fusion of gp16 in-frame with a ∼25-amino acid vector sequence containing hexahistidine tag at the N terminus. The ligated DNAs were transformed into Escherichia coli XL10 Gold cells (Stratagene, La Jolla, CA), mini-prep plasmid DNAs were prepared by alkaline lysis procedure, and the presence and orientation of inserts were ascertained by restriction digestion and/or amplification with insert-specific primers. The entire insert of each clone was sequenced to ensure the incorporation of mutant sequence and absence of any nonspecific errors (Davis Sequencing, Davis, CA). The clones were then transferred into the expression strains, E. coli BL21(DE3) pLys-S or E. coli BL21 (DE3) RIPL (33).
Purification of gp16 Mutant Proteins
The E. coli cells containing a given gp16 mutant clone was induced with isopropyl-1-thio-β-d-galactopyranoside at 32 °C for 2 h to overexpress the His-tagged proteins. Cells were harvested by centrifugation at 8,200 × g for 10 min at 4 °C and lysed using a French Press. Cell-free extracts were prepared by centrifugation of the lysate at 17,000 × g for 15 min at 4 °C. All the mutants produced soluble proteins. The supernatant containing soluble gp16 was purified by successive chromatography on Histrap HP (affinity), Mono Q-5/50 GL (ion-exchange), and Hiload Superdex 200 prep-grade (size exclusion) columns using AKTA-PRIME and AKTA-FPLC systems (GE Healthcare, Piscataway, NJ) (see Ref. 9 for additional details). The purified proteins were concentrated by Amicon Ultra centrifugal filters (Millipore, Billerica, MA) and stored at −70 °C. The proteins were ∼95% pure as judged by SDS-PAGE and Coomassie Blue R staining.
ATPase
ATPase assays were performed as per the basic procedure described previously (31). The purified gp176 (0.2–0.5 μm) either alone or with gp16 (2–5 μm) were incubated in a reaction mixture (20 μl) containing 1 mm unlabeled ATP and 75 nm [γ32P]ATP (specific activity, 3000 Ci/mmol; GE Healthcare) at 37 °C in ATPase buffer (50 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 5 mm MgCl2) for 20 min. The reaction was terminated by adding EDTA to 50 mm final concentration, and the products were separated by thin layer chromatography on PEI-cellulose plates (Sigma-Aldrich). The amount of 32Pi was quantified by phosphorimaging (Storm 820, Molecular Dynamics, GE Healthcare), and the total Pi produced was calculated from the 32Pi value. The data shown are an average of duplicates from two or three independent experiments.
Oligomerization
Native gradient (4% to 20%, w/v) polyacrylamide gel electrophoresis (PAGE) was used under non-denaturing conditions to separate various species of the purified proteins (30). The gels were stained with Coomassie Blue R.
UV Cross-linking
The purified gp17 (1.4 μm) and gp16 (1 μm-18 μm) were incubated either alone, or as a mixture, with 3 μm of either [γ-32P]8N3ATP or [α-32P]8N3ATP, or with 5 μm [γ-32P]2N3ATP (Affinity Labeling Technologies, Lexington, KY) in a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.5, 0.1 m NaCl, and 5 mm MgCl2. Reaction mixtures were incubated for 5 min on ice and 1-μl aliquots were withdrawn for thin layer chromatography. The rest of the sample was subjected to cross-linking by exposing to UV light (365 nm) for 5 min at 4 °C at 5-cm height (15). SDS-sample buffer was added, and the samples were electrophoresed by SDS-PAGE (12% gel). The gels were stained with Coomassie Blue R, and the amount of bound nucleotide was quantified by phosphorimaging.
DNA Packaging
The purified gp17 (1.5 μm) was incubated either alone or in the presence of gp16 (0.067–0.333 μm) with purified proheads (6 × 109 particles) and 300 ng of 48-kb phage λ DNA (Fermentas, Glen Burnie, MD) in a reaction mixture (20 μl) containing 30 mm Tris-HCl, pH 7.5, 80 mm NaCl, 3 mm MgCl2, and 1 mm ATP for 45 min at room temperature. The reaction was terminated by adding DNase I (Sigma-Aldrich) to a final concentration of 0.5 μg/μl and incubated for 30 min at 37 °C. The reaction was deproteinized by the addition of 50 mm EDTA and 0.5 μg/μl of proteinase K (Fermentas) plus 0.2% SDS and incubation for 30 min at 65 °C. The samples were electrophoresed on an agarose gel, stained with ethidium bromide, and the amount of DNase-protected DNA was quantified by Gel DOC XR imaging system (Bio-Rad) (10).
Nuclease
The purified gp17 (0.2–1 μm) was incubated either alone or in the presence of gp16 (0.2–10 μm) with 100 ng of pAD10 plasmid DNA (29 kb) in a reaction mixture (25 μl) containing 5 mm Tris-HCl, pH 8.0, 6 mm NaCl, and 5 mm MgCl2 for 15–30 min at 37 °C. The reaction was terminated by adding EDTA to a final concentration of 50 mm, and the samples were electrophoresed on 1.0% (w/v) agarose gel followed by ethidium bromide staining (13).
RESULTS
Domain Organization of gp16
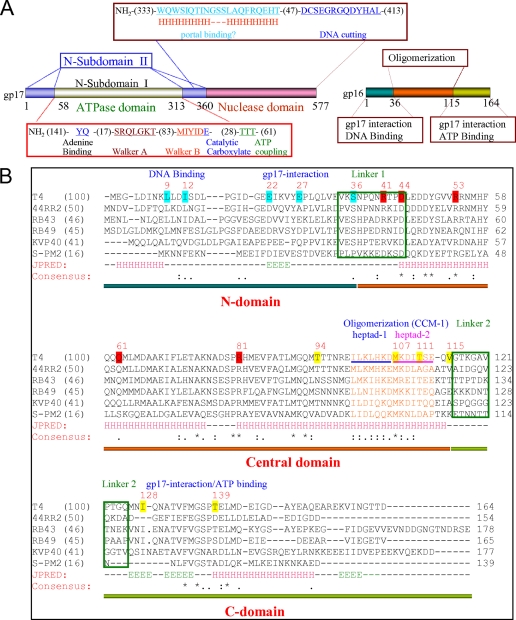
gp16 has a predicted helix-turn-helix motif at the N terminus that is presumably required for viral genome recognition, and 2–3 coiled-coil motifs (CCM) that are required for oligomerization into what appears to be octamers (Fig. 1) (27, 30, 36). The central CCM-1 containing two classic heptad sequences is especially critical as mutations in this motif disrupt coiled-coil interactions and impair gp16 oligomerization. The monomeric wild-type (WT) gp16, or the CCM-1 mutant (L103R-I110H), unlike the gp16 oligomer, is unable to stimulate gp17-ATPase (30). Programs that predict linker regions between domains, namely DLP-SVM, DisEMBEL, and DISOPRED2, predict two linkers between amino acids 34–44 and 116–125 with the highest probability for disorder at Pro-38 and Ala-120, respectively (Fig. 1). Functional analyses (see below) show that both the N- and C-terminal regions of gp16 are required for binding to gp17 or ATP. Thus gp16 appears to have three domains; a central oligomerization domain flanked by N-terminal and C-terminal domains that interact with DNA, ATP, and gp17. This domain organization is consistent with the proteolysis results that produced a protease-resistant 9.5-kDa core domain corresponding to amino acids 36–115.7
FIGURE 1.
Domain organization of phage T4 small terminase, gp16. A, domain end points of the large terminase, gp17, were determined by the x-ray structures of ATPase and nuclease domains and the full-length gp17 (7, 8). The functional motifs were determined by mutational and biochemical studies (12, 14–17, data not shown). The domain organization of the small terminase, gp16, is predicted by this study. Numbers represent the number of amino acids in the respective coding sequence. B, T4 family gp16 sequences with various degree of similarity to the T4 sequence were selected for multiple sequence alignment by ClustalW and secondary structure predictions by JPRED (accession numbers: T4, NP049775; 44RR2.8t, NP932507RB69, RB43, AAX78740;NP861868; RB49, NP891723; KVP40, NP899600; S-PM2, CAF34160 sequences were obtained from Phage Tulane website. Pairwise alignment scores of each gp16 with the T4 gp16 are shown in parentheses after the name of each phage. Amino acids highlighted in cyan represent the endpoints for N-terminal truncations. Amino acids highlighted in yellow represent the endpoints of C-terminal truncations. CCM-1 heptad residues are shown in orange. Heptad-1 and heptad-2 residues in T4 gp16 sequence are indicated by blue and pink underlines, respectively. Conserved arginines, aspartic acids, glutamine, and tyrosine that are mutated are highlighted in red. Linker regions predicted by DLP-SVM (34) and DISOPRED2 (35) are boxed in green. The sequences corresponding to N- (amino acids 1–35), central (amino acids 36–115), and C-domains (amino acids 116–164) are underlined by horizontal blue, orange, and green bars, respectively. Consensus sequence is shown below the secondary structure predictions (H residues correspond to α-helix, E residues to β-strand, and − residues to loop).
Both N- and C-domains Are Required for gp17-ATPase Stimulation
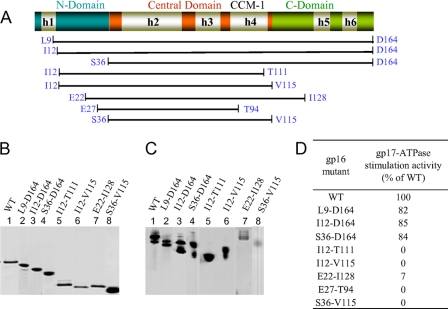
Progressive deletions at the C terminus of gp16, T139 (amino acids 1-T139), I128 (amino acids 1-I128), and V115 (amino acids 1-V115) (Fig. 2, A and B, see Fig. 1), did not affect oligomerization (Fig. 2C), although the gp17-ATPase stimulation was progressively reduced (Fig. 2D). On the other hand, the T111 (amino acids 1-T111) mutant in which the deletion extended into two of the seven heptad-2 residues of CCM-1 showed no discernable difference in oligomerization but showed greater than 50% loss of gp17-ATPase stimulation (Fig. 2D, lanes 21–24 in E). The M107 construct in which six of seven heptad-2 residues were deleted completely lost oligomerization (Fig. 2C, lane 7) as well as gp17-ATPase stimulation (Fig. 2D, lanes 25–28 in E).
FIGURE 2.
The gp16 C-domain truncation mutants show partial loss of gp17-ATPase stimulation. A, schematic of the gp16 polypeptide showing domain organization (N-domain in blue, central domain in orange, and C-domain in green), predicted helices (h1–h6), and endpoints of C-terminal truncation clones (e.g. T139 refers to truncation at the threonine 139 residue). Note that the secondary structure prediction of gp16 sequence, unlike that of the aligned T4 family gp16 sequences (Fig. 1), shows six helices (h1–h6). B, SDS-polyacrylamide gel (12%) showing the purity of mutant proteins. C, 4–20% native non-denaturing gradient gel showing the oligomeric state of mutants. Both the gels were stained with Coomassie Blue R. The WT gp16 migrates as two oligomeric species (labeled as O1 and O2). All the C-terminal truncation mutants except M107 in which the truncation extended into the CCM-1 motif produced oligomers. M107 (lane 7), like the CCM-1 double mutant L103R-I110H (lane 2) (30), produced monomeric gp16 (labeled as M). D, gp17-ATPase stimulation activity of gp16 mutants expressed as percentage of the WT gp16 activity. The amount of Pi produced was quantified by phosphorimaging and ImageQuant 5.2 software. Values represent average of duplicates from two independent experiments. E, autoradiogram showing the gp17-ATPase stimulation activity of various mutants. Each ATPase assay was done in duplicate as per the procedure described under “Experimental Procedures.”
Deletions at the N terminus, L9 (amino acids L9–164), I12 (amino acids I12–164), and S36 (amino acids S36–164) also retained oligomerization and gp17-ATPase stimulation (Fig. 3). But deletions at both N and C termini, I12-T111, I12-T115, E22-I128, and S36-V115 retained oligomerization but lost the ATPase stimulation. Only the E22-I128 mutant in which a small part of the C-domain is present showed very weak stimulation. Another N- and C-terminal mutant, E27-T94, in which the C-terminal deletion extended into CCM-1 completely lost both oligomerization and gp17-ATPase stimulation activities (Fig. 3D and data not shown).
FIGURE 3.
Both the N- and C-domains of gp16 are important for gp17-ATPase stimulation. A, schematic of the gp16 polypeptide showing the endpoints of the truncation clones (see Fig. 2 legend for description of the schematic). The truncations are of two types: (i) truncations in N-terminal domain (L9-D164, I12-D164, and S36-D164); and (ii) truncations in both N- and C-terminal domains (I12-T111, I12-V115, E22-I128, E27-T94, and S36-V115). The S36-V115 mutant also contained a T96L mutation that increases the length of CCM-1 from two to three heptads but is phenotypically similar to the WT (30). B, SDS-polyacrylamide gel (12%) showing the purity of mutant proteins. C, 4–20% native non-denaturing gel showing the oligomeric state of gp16 mutants. Both the gels were stained with Coomassie Blue R. All the mutants, like the WT gp16, produced oligomers. D, gp17-ATPase stimulation activity of gp16 mutants expressed as percentage of the WT gp16. The amount of Pi produced was quantified by phosphorimaging and ImageQuant 5.2 software. Panels B and C are composites from different experiments, and the values in the table represent average of duplicates from two independent experiments.
gp16 Has an ATP Binding Site with Broad Specificity
gp16 cross-linked to 8N3- ATP, GTP, ADP, or AMP with nearly the same affinity as gp17 (Kd = 1–5 mm), suggesting that gp16 has a nucleotide binding site with broad specificity (Ref. 37, Fig. 4, and data not shown). However, unlike gp17, gp16 showed no hydrolysis of ATP (or any other nucleotide). No canonical ATP binding motifs such as Walker A or Walker B are present in the gp16 sequence (36). The gp16 mutants tested, deletions in C-terminal domain or in both C- and N-terminal domains, lost ATP (Fig. 4A), or ADP (Fig. 4B) cross-linking activity. Several gp16 mutants that lost nucleotide binding (e.g. T139, I128, V115, T111) retained gp17-ATPase stimulation (see Fig. 2D). Furthermore, even the apparent differences in the basal level nonspecific cross-linking of various mutants did not correlate with their ability to stimulate ATPase. For instance, the I12-T111 mutant that lost gp17-ATPase stimulation showed similar basal level ATP binding as the T111 mutant which retained ATPase stimulation. In essence, the results show that ATP binding to gp16 is not essential for ATPase stimulation.
FIGURE 4.
gp16 has a nucleotide binding site that is inactivated in mutants. The ability of WT and gp16 mutants to bind ATP or ADP was determined by UV cross-linking in the presence of [γ-32P]8N3ATP (A) or [γ-32P]8N3ADP (B). Each sample was cross-linked in duplicate and electrophoresed in adjacent lanes. See “Experimental Procedures” for details.
gp16 Is Unlikely a Nucleotide Exchange Factor
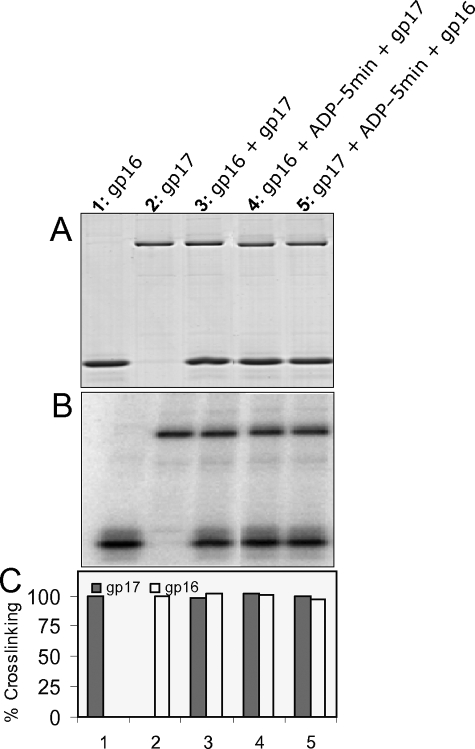
Stimulation of gp17-ATPase by gp16 is analogous to GTPase/ATPase stimulation by the nucleotide exchange factors (GEFs) (38) or the GTPase-activating proteins (GAPs) (39, 40). A hallmark of the nucleotide exchange factors is that they catalyze exchange of NDP product with NTP substrate in the catalytic site of GTPases and ATPases. One method to assess this activity is by showing reduced binding or increased release of nucleoside diphosphate in the presence of an exchange factor. Also, these factors sometimes require acidic residues (38).
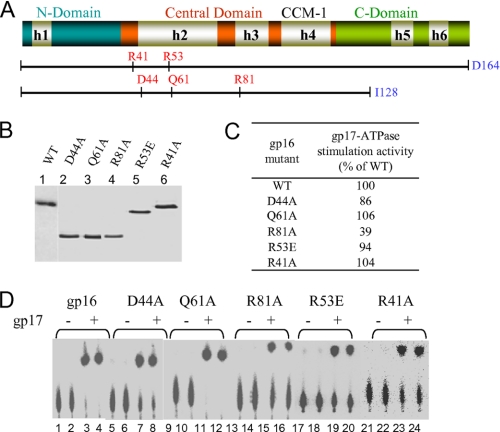
Mutation at the conserved acidic residue, Asp-44, retained gp17-ATPase stimulation as well as oligomerization (Fig. 5, C and D). Both gp16 and gp17 bind ADP (Fig. 6, lanes 1 and 2). gp17 binding to ADP was unaffected by gp16 (lane 3). Preloading of gp17 with ADP followed by addition of gp16 did not cause significant release of bound ADP (lane 5). Nor the preloading of gp16 with ADP affected the binding of ADP to gp17 (lane 4). Thus, under the conditions tested, ADP binding to, or release from, gp17-ATPase is not affected by the presence of gp16.
FIGURE 5.
None of the conserved arginines or the glutamine is essential for gp17-ATPase stimulation. A, schematic of the gp16 clones showing the positions of the mutated residues (see Fig. 2 legend for description of the schematic). B, Coomassie Blue R-stained SDS-polyacrylamide gel (12%) showing the purity of the mutant proteins. C, gp17-ATPase stimulation activity of mutants expressed as the percentage of WT gp16. D, autoradiogram showing the gp17-ATPase stimulation activity of various mutants. See legend to Fig. 2 and “Experimental Procedures” for more details.
FIGURE 6.
gp16 does not show nucleotide exchange activity. WT gp17 (1 μm) and WT gp16 (8.5 μm) either alone (lanes 1 and 2) or together (lane 3) were incubated with 5 μm [α-32P]8N3ADP for 5 min prior to UV cross-linking. For lane 4, gp16 was preincubated with [α-32P] 8N3ADP for 5 min before the addition of gp17. For lane 5, gp17 was preincubated with [α32P] 8N3ADP for 5 min before the addition of gp16. Panel A, Coomassie Blue R stained SDS-polyacrylamide gel, Panel B, phosphorimage of panel A. Panel C, histogram showing the quantification of cross-linking data from panel B.
gp16 Stimulates the Hydrolysis of gp17-bound ATP
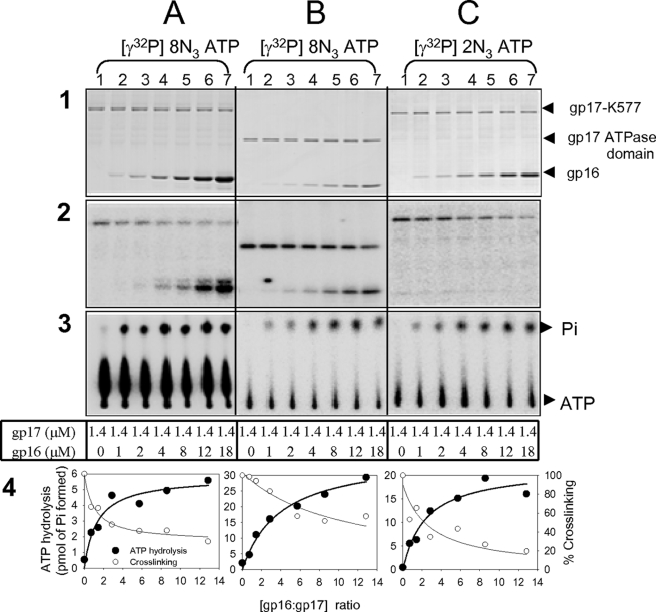
Both gp17 and gp16 cross-linked to 8N3-ATP (Fig. 7, columns 2A and 2B, lanes 2–7). However, no gp16 cross-linking was observed when 2N3-ATP was used (column 2C, lanes 2–7), which is likely due to the lack of a cross-linkable lysine in the gp16 structure near the 2-azido group of bound nucleotide. Addition of increasing amounts of gp16 to gp17 resulted in progressive decrease of gp17-bound ATP (columns 2 and 4) and a corresponding increase in the amount of ATP hydrolyzed (columns 3 and 4). Maximum hydrolysis was observed at a gp16 (oligomer):gp17 (monomer) ratio of ∼1:1 or higher. The same behavior was observed with 8N3-[γ-32-P]ATP, 8N3-[α-32P]ATP, and 2N3-[γ-32P]ATP, with the full-length gp17 or the gp17-ATPase domain, with or without preloading gp17 with ATP (Fig. 7; additional data shown in Ref. 37).
FIGURE 7.
gp16 stimulates the hydrolysis of gp17-bound ATP. Vertical columns: gp17-K577 (columns A and C) or gp17-ATPase domain (column B) (1.4 μm) was incubated either alone or in the presence of increasing concentration of gp16 (1 μm-18 μm) and 3 μm of [γ-32P]8N3ATP (columns A and B) or 5 μm of [γ-32P]2N3ATP (column C). Following the ATPase assay, 1-μl aliquots of the samples were removed, and thin layer chromatography was performed. The rest was exposed to UV light for 5 min at 4 °C and subjected to SDS-PAGE and phosphorimaging (see “Experimental Procedures” for details of assays). Horizontal columns: 1: Coomassie Blue R-stained SDS-polyacrylamide gel; 2: phosphorimage of column 1; 3: Autoradiogram of samples following thin layer chromatography; 4: quantification of cross-linking and ATP hydrolysis data from columns 2 and 3, respectively. The positions of gp17, gp17-ATPase domain, and gp16 bands are marked with arrows.
gp16 Does Not Provide an Arginine Finger for gp17-ATPase Stimulation
GAPs either provide an arginine finger, or their interaction with GTPase positions an arginine finger (or glutamine) into the catalytic site, triggering bond cleavage (41, 42). Several arginines (Arg-41, Arg-53, Arg-81) and a glutamine (Gln-61) that are conserved in the T4 family small terminases (see Fig. 1) were tested to see whether any of these are essential for gp17-ATPase stimulation (Fig. 5, A and B). All the mutants retained gp17-ATPase stimulation activity (Fig. 5, C and D), and none showed any loss of oligomerization (data not shown).
gp16 Modulates DNA Packaging
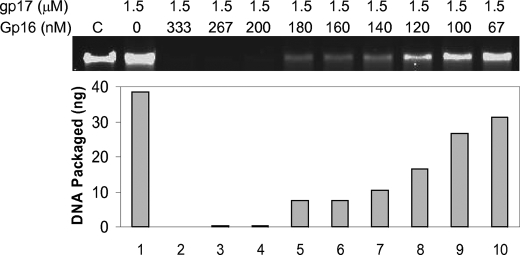
In a crude in vitro DNA packaging system, gp16 enhances packaging efficiency (27, 31, 43), whereas it inhibits packaging in a defined system (10, 44). The crude system contains all the components induced by phage infection that includes transcription, replication, and recombination proteins, whereas the defined system contains only two components, proheads and gp17. Surprisingly, very low gp16 concentrations, a ratio of 1 gp16 oligomer: 60 gp17 monomer, are sufficient to completely inhibit packaging (Fig. 8). This is in contrast to 1 gp16 oligomer:1 gp17 monomer ratio required for maximum stimulation of gp17-ATPase.
FIGURE 8.
gp16 interaction to prohead-assembled gp17 inhibits DNA translocation. Defined in vitro DNA translocation was performed in the presence of gp17 alone (1.5 μm; 1.8 × 1013 molecules) or in the presence of gp17 plus increasing concentrations of gp16 (67–333 nm; 0.5–3 × 1011 gp16 oligomers). The reaction mixture contained 6 × 109 proheads and 6 × 109 48-kb phage λ DNA molecules (see “Experimental Procedures” for assay details). The amount of packaged DNA was quantified using the control lane C where 30 ng (10% of the total DNA) of phage λ DNA was loaded, and the quantified data are shown in the histogram below.
The reaction mixture in which near complete inhibition was observed (Fig. 8, lane 4) contained 6 × 109 proheads, 6 × 109 DNA molecules, 3 × 1011 gp16 oligomers, and 1.8 × 1013 gp17 molecules. As gp17 assembles on the prohead portal as a pentamer (8), there would be ∼3 × 1010 prohead-bound gp17 molecules. As one gp16 oligomer is expected to interact with one gp17 monomer (9), it is unlikely that the gp16 inhibition was due to the titration of gp17 from binding to the portal because there would be a vast excess of free (unbound) gp17 molecules. Inhibition through titration of DNA ends where packaging presumably initiates is also unlikely because increasing the DNA ends by 200-fold (1012 molecules of 500 bp DNA as packaging substrate), which is nearly 7 times that of gp16 oligomers, did not relieve inhibition. On the other hand, addition of gp16 to the purified prohead-gp17-DNA complex (containing no free gp17 monomer) resulted in inhibition of DNA translocation as well as ATPase stimulation at a ratio of 1 gp16 oligomer:1 gp17 monomer.8
gp16 Regulates gp17 Nuclease
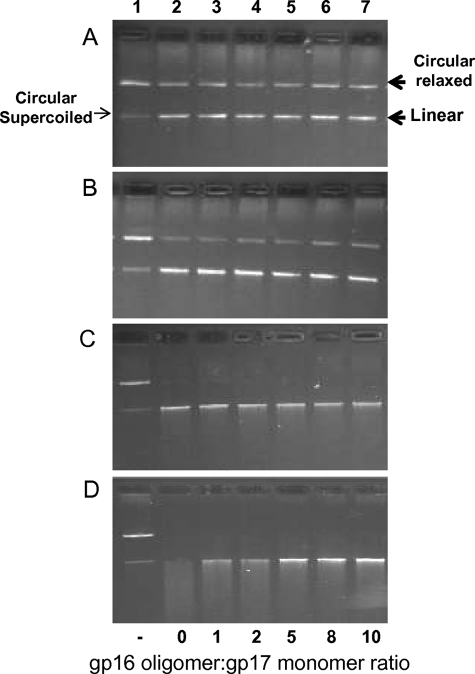
gp17 exhibits a nonspecific endonuclease activity that is presumably required for the cutting of concatemeric DNA before and after DNA packaging (12, 13, 16). When incubated with the pAd10 circular plasmid DNA (29 kb) (45), gp17 first converts it into linear DNA (first cut) (Fig. 9, panels A and B; e.g. lanes 2), and the linear DNA is then degraded to a series of short fragments (panels C and D, lanes 2). In the presence of gp16, gp17 nuclease can still make the first cut (panels A and B, compare lanes 6 and 7 with lane 2), but further cutting of linear DNA is inhibited (panel D, compare lanes 6 and 7 with lane 2). As with the gp17-ATPase stimulation, both the gp16 N- and C-domains are required for gp17-nuclease inhibition; the N- and C-domain mutants, I12-T111, I12-V115, and E22-I128, lost the ability to inhibit gp17-nuclease (data not shown).
FIGURE 9.
gp16 regulates gp17-nuclease. Four sets of reactions were carried out with four different concentrations of gp17 (A, 0.2 μm; B, 0.4 μm; C, 0.8 μm; D, 1.0 μm). In each set, the amount of gp17 was kept constant and incubated either alone (lanes 2) or in the presence of increasing concentrations of gp16 (lanes 3–7). The ratio of gp16 (oligomer):17 (monomer) is shown at the bottom of lanes. Lane 1 corresponds to control sample containing no gp16 or gp17. See “Experimental Procedures” for assay details.
DISCUSSION
Phage DNA packaging is a complex process involving the orchestration of a series of enzyme activities as well as protein-protein and protein-DNA interactions (1). In addition to the components that directly participate in packaging, the prohead, the portal, and the large terminase, a small terminase is also essential. Mutations in the phage T4 small terminase gp16, including a C-terminal truncation mutant (Q114am), are lethal. However, gp16 does not exhibit any enzymatic activities and is dispensable for DNA translocation in vitro (10, 44). The precise role(s) of the small terminase in the packaging process has remained poorly understood. Here we show that gp16 is a regulator of the packaging motor, modulating the ATPase, translocase, and nuclease activities associated with the motor protein, the large terminase gp17.
Sequence and mutagenesis analyses showed that the 18-kDa gp16 consists of three functional domains, a central domain that is essential for oligomerization (amino acids 36–115), an N-terminal domain (amino acids 1–35), and a C-terminal domain (amino acids 116–164) that interact with gp17. The C-terminal domain is also required for ATP binding, whereas the N-terminal domain is predicted to interact with DNA (27, 36). Either the N- or the C-domain can partially stimulate gp17-ATPase activity but both are essential for maximum stimulation. The gp16 mutants that do not contain both the domains (e.g. E22-I128) retained oligomerization but completely lost ATPase stimulation. These results suggest that the central domain forms the structural core whereas the N- and C-domains form the functional core. This domain organization is consistent with the greater divergence of the N- and C-domains, whereas the central domain is well conserved (see Fig. 1). Perhaps the N- and C-domains of gp16 provide specificity to capture the respective gp17 partner (and viral DNA) in mixed infections that frequently occur in nature.
Although gp16 lacks a canonical nucleotide binding motif, it binds ATP, GTP, and ADP at nearly the same affinity as gp17. There are several gp16 mutants that lost ATP binding activity, yet were able to stimulate gp17-ATPase. Thus ATP binding is not essential for ATPase stimulation. Analysis of a large number of mutants further showed that there is no correlation between gp16 ability to bind ATP and stimulation of gp17-ATPase, inhibition of gp17-nuclease, or DNA translocation. While it is unclear what the role of ATP binding is on gp16 function, it is possible that gp16 might act as a reservoir of ATP fuel, by capturing ATP and passing it on to gp17.
The gp17-ATPase is essentially inactive as a monomer but is stimulated during active packaging as a portal-assembled motor (10). The same ATPase is stimulated by gp16, albeit in the unassembled monomeric state, in the absence of packaging (31). Thus gp16 stimulation mimics the conformational changes occurring during translocation. Other small terminases show similar behavior (24, 46), although gp16 generates the most robust stimulation, as high as 400-fold (47). Our data show that the stimulation was due to hydrolysis of gp17-bound ATP. This implies that in the T4 packaging motor, unlike in some molecular motors such as F1 ATPase, kinesin, and UvrD helicase (48–50), the step that is coupled to DNA movement, the power stroke, is not ATP binding per se, but post-ATP binding. Because, the power stroke should be generated when the ATPase is part of an assembled motor and ready to fire, but not when it is a free molecule. This is consistent with the recent single molecule data using the phi29 packaging motor where “dwells” and “bursts” alternate during DNA translocation (51). Dwells were attributed to the ATP binding phase and bursts to the ATP hydrolysis phase. DNA movement occurred during the burst phase suggesting that the motor must be preloaded with ATP in order for the power stroke to occur.
The two well characterized mechanisms for stimulation of ATPases and GTPases are: (i) nucleotide exchange and (ii) arginine finger positioning. Our data show that the nucleotide exchange is unlikely. Otherwise gp16 as was shown in other systems (41, 52) would have catalyzed the exchange of ADP and ATP in the gp17 ATPase catalytic site. But the data show that ADP binding to gp17 was unaffected by gp16. Neither did the gp17 preloaded with ADP significantly released the nucleotide upon the addition of gp16.
In the arginine finger mechanism, an arginine (or glutamine) sidechain is precisely positioned into the GTPase (ATPase) catalytic center in response to a distant interaction (39). The arginine is either provided by a partner protein such as the GAP (41), or it is present in the GTPase itself but interaction with GAP causes conformational change, positioning the arginine finger into the nucleotide-binding pocket (42). The guanidino group forms hydrogen bonds with the βγ phosphates of bound ATP, leading to charge polarization and bond cleavage (53). Our mutational search turned up no arginine finger candidates in gp16; all the conserved arginine and glutamine mutants retained full ATPase stimulation. On the other hand, a potential arginine finger (Arg-162) was found in gp17, which is appropriately located at the junction of ATPase subdomains I and II (8). Not only was this arginine conserved among the T4-family gp17s but also that mutations at this residue resulted in a null phenotype and loss of both ATPase and DNA translocation activities (8, 14, 36). Evidence further suggests that the gp16 interaction sites are present in the ATPase domain of gp17 since the purified ATPase domain showed even better stimulation than the full-length gp17. Structural evidence showed that the position of this arginine finger is different in different conformational states and a 6° rotation of gp17 ATPase subdomain II was observed, consistent with that seen in GTPases/ATPases (8). Thus our data lead to the hypothesis that interaction of gp16 with the ATPase domain causes a conformational change in the ATP-bound gp17, orienting the Arg-162 residue into the catalytic site triggering ATP hydrolysis. This might be the same mechanism that, during active translocation, triggers ATP cleavage when a portal-assembled gp17 subunit is loaded with DNA and ATP, and is ready to fire.
gp16 shows differential effects on DNA translocation depending on the functional state of gp17. In the crude system which mimics in vivo packaging (31), gp16 interacts with gp17 and probably other phage factor(s) forming a “native” packaging machine, enhancing efficiency. On the other hand, in the defined system that contains only two components (10, 44), proheads, and gp17, gp16-gp17 interactions lead to inhibition of DNA translocation. The latter could be due to interference of gp17 binding either to the portal (motor assembly) or to the ends of DNA where packaging is presumably initiated. Our data argues against either because inhibition was observed even when gp17 and DNA were in excess, thus the alternative hypothesis that gp16 preferentially interacts with the prohead-assembled gp17 motor. In the absence of other phage factors, these interactions apparently trap the motor in an inactive conformation. Preliminary evidence, (i) inhibition of DNA translocation and packaging ATPase activities of the purified prohead-gp17-DNA complex by gp16, and (ii) reconstruction of prohead-assembled gp17 in the presence of gp16 showing substantial change in the gp17 cryoEM density,9 support the above hypothesis and further suggest that a major conformational change occurs in the gp17 motor following gp16 interaction.
Control of the gp17-nuclease such that it cuts concatemeric DNA once for packaging initiation and again for packaging termination after translocation of viral genome is critical for production of an infectious virus. While the termination cut is linked to headful packaging, the initiation cut is regulated by the gp16-gp17 terminase complex (1). Our data show that gp17, in the presence of gp16 (and no ATP), acts as a restrictive nuclease; it makes the first cut, converting circular DNA (super-coiled or relaxed) to linear form but further cutting of linear DNA is inhibited. It appears that once the cut is made, the terminase complex remains bound to the newly generated end. In the absence of gp16, this interaction is not stable. As a result, gp17 either dissociates from the DNA and makes random cuts upon reassociation, or translocates on the same DNA molecule and makes random cuts at a distance.
The importance of the small terminase in viral genome recognition has been well established in cos and pac phages, and similarly thought of in phage T4 (27–29). Our results are not in conflict with this conclusion, but further suggest that the small terminase plays broader roles throughout the packaging process. The activities of the packaging motor protein must be regulated and coordinated to produce an infectious phage. Otherwise, it could be suicidal to the virus. For instance, an unregulated gp17-ATPase capable of hydrolyzing at a rate of ∼200 ATPs/sec (6) could result in wastage of biochemical energy. Uncontrolled cutting of concatemeric DNA by gp17-nuclease could result in loss of genetic information and production of noninfectious virus particles. A regulator such as the small terminase gp16 is necessary to coordinate various functions of the motor. gp16 interaction causes conformational changes in gp17, resulting in different outcomes depending on the functional or quaternary state of gp17. When gp17 is present as a monomer, interaction with gp16 stimulates ATPase but when it is present as a portal-assembled motor, the ATPase and DNA translocation are inhibited. The latter in the presence of other phage factors such as transcription and recombination proteins leads to more efficient DNA packaging (31, 43).
How a small molecule such as gp16 regulates the various properties of a complex motor is an interesting biological question. Structural and biochemical analyses show that gp17 is a flexible molecule with a number of binding sites, undergoing many transitions during the packaging process (1, 8, 18). We propose that the nucleotide state of gp17 (e.g. apo, ATP-, ADP-, and/or Pi-bound) is central to these transitions. Dynamic changes at the ATPase center are communicated to the distant sites and vice versa, turning on/off of activities and coordinating the packaging process. There is ample evidence that subtle differences in the orientation of gp17-bound ATP can cause profound effects on the motor function. For instance, the gp17 D255E-E256D mutant in which the sequence of Walker B aspartate and catalytic glutamate is flipped binds ATP, but the ATPase and DNA translocation activities are completely lost and the nuclease activity is greatly reduced (17). On the other hand, the E256Q mutant that also binds ATP shows loss of ATPase and DNA translocation but this mutant shows greater nuclease activity than the WT (15). In the ATPase coupling mutant, T287D, ATP is hydrolyzed once, but the products apparently remain bound to the active site; thus it cannot turn over (18). This mutant loses all three activities; ATPase, translocation, and nuclease. Thus it seems that the nucleotide state and the functional state of the packaging motor are intimately linked. The mechanism by which gp16 regulates motor function may be by altering the nucleotide state of gp17. The precise alteration depends on the oligomeric and/or ligand (DNA, portal) bound state of gp17, resulting in different outcomes with respect to ATPase, translocase, and nuclease properties of gp17. Our data demonstrate that at least in one instance (gp17 bound to ATP) gp16 interaction alters the nucleotide state, by hydrolyzing the bound ATP.
In conclusion, we suggest that the phage T4 DNA packaging machine consists of a motor and a regulator. Although the latter can be dispensable for DNA packaging in vitro, it is not so for production of infectious virions in vivo. The motor and regulator must co-evolve to generate desired motor behaviors that fit the “life-style” of a given virus. Indeed, most phage genomes encode both small and large terminases, and their order on the genome is well conserved (54, 55). These might be common themes in phage/virus DNA packaging motors as well as other complex molecular motors.
Acknowledgments
We thank Dr. Bonnie Draper (The Catholic University of America), Michael Rossmann, and Siyang Sun (Purdue University) for stimulating discussions throughout this investigation and for critical review of the manuscript, Karoly Viragh for constructing the gp16 mutant, E27-T94, and Vishal Kottadiel, Zhihong Zhang, and Shylaja Hegde (The Catholic University of America) for sharing data on the biochemical characterization of prohead-gp17 complex.
This work was supported by Grants MCB-423528 and MCB-0923873 (to V. B. R.) from the National Science Foundation.
The term “terminase” originally referred to the enzyme activity that generated the termini of virion DNA. Subsequently, it was found that it contains two subunits, which were named as large terminase and small terminase.
gp17 is a weak ATPase (kcat = ∼1–2 ATPs hydrolyzed/min/gp17 molecule). The gp17 rate of ATP hydrolysis is increased by ∼50-fold in the presence of gp16 (31).
gp17-K577 (amino acids 1–577) in which the C-terminal 33 amino acids were deleted was used in all the experiments reported here. This protein retained all the biological functions of the full-length gp17, including DNA packaging in a defined system.
S. Sun, personal communication.
V. Kottadiel, Z. Zhang, and S. Hegde, unpublished data.
S. Sun and M. Rossmann, personal communication.
- gp
- gene product
- CCM
- coiled-coil motif
- GAP
- GTPase-activating protein
- PAGE
- polyacrylamide gel electrophoresis
- WT
- wild-type.
REFERENCES
- 1.Rao V. B., Feiss M. (2008) Annu. Rev. Genet. 42, 647–681 [DOI] [PubMed] [Google Scholar]
- 2.Black L. W. (1989) Annu. Rev. Microbiol. 43, 267–292 [DOI] [PubMed] [Google Scholar]
- 3.Fokine A., Chipman P. R., Leiman P. G., Mesyanzhinov V. V., Rao V. B., Rossmann M. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 20, 6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith D. E., Tans S. J., Smith S. B., Grimes S., Anderson D. L., Bustamante C. (2001) Nature 413, 748–752 [DOI] [PubMed] [Google Scholar]
- 5.Simpson A. A., Tao Y., Leiman P. G., Badasso M. O., He Y., Jardine P. J., Olson N. H., Morais M. C., Grimes S., Anderson D. L., Baker T. S., Rossmann M. G. (2000) Nature 408, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller D. N., Raymer D. M., Kottadiel V. I., Rao V. B., Smith D. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16868–16873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., Rao V. B. (2007) Mol. Cell. 25, 943–949 [DOI] [PubMed] [Google Scholar]
- 8.Sun S., Kondabagil K., Draper B., Alam T. I., Bowman V. D., Zhang Z., Hegde S., Fokine A., Rossmann M. G., Rao V. B. (2008) Cell 135, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 9.Kanamaru S., Kondabagil K., Rossmann M. G., Rao V. B. (2004) J. Biol. Chem. 279, 40795–40801 [DOI] [PubMed] [Google Scholar]
- 10.Kondabagil K. R., Zhang Z., Rao V. B. (2006) J. Mol. Biol. 363, 786–799 [DOI] [PubMed] [Google Scholar]
- 11.Streisinger G., Emrich J., Stahl M. M. (1967) Proc. Natl. Acad. Sci. U.S.A. 57, 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuebler D., Rao V. B. (1998) J. Mol. Biol. 281, 803–814 [DOI] [PubMed] [Google Scholar]
- 13.Alam T. I., Draper B., Kondabagil K., Rentas F. J., Ghosh-Kumar M., Sun S., Rossmann M. G., Rao V. B. (2008) Mol. Microbiol. 69, 1180–1190 [DOI] [PubMed] [Google Scholar]
- 14.Rao V. B., Mitchell M. S. (2001) J. Mol. Biol. 314, 401–411 [DOI] [PubMed] [Google Scholar]
- 15.Goetzinger K. R., Rao V. B. (2003) J. Mol. Biol. 331, 139–154 [DOI] [PubMed] [Google Scholar]
- 16.Rentas F. J., Rao V. B. (2003) J. Mol. Biol. 334, 37–52 [DOI] [PubMed] [Google Scholar]
- 17.Mitchell M. S., Rao V. B. (2006) J. Biol. Chem. 281, 518–527 [DOI] [PubMed] [Google Scholar]
- 18.Draper B., Rao V. B. (2007) J. Mol. Biol. 369, 79–94 [DOI] [PubMed] [Google Scholar]
- 19.Luftig R. B., Ganz C. (1972) J. Virol. 10, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumner-Smith M., Becker A., Gold M. (1981) Virology 111, 642–646 [DOI] [PubMed] [Google Scholar]
- 21.Feiss M., Catalano C. E. (2005) in Viral Genome Packaging Machines: Genetics, Structure and Mechanis (Catalano C. E. ed) pp. 5–39, Landes Biosci, Georgetown, TX [Google Scholar]
- 22.Yang Q., Berton N., Manning M. C., Catalano C. E. (2005) Biochemistry 38, 14238–14247 [DOI] [PubMed] [Google Scholar]
- 23.Becker A., Gold M. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 4199–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho A. G., Gual A., Lurz R., Tavares P., Alonso J. C. (2003) J. Biol. Chem. 278, 23251–23259 [DOI] [PubMed] [Google Scholar]
- 25.Nemecek D., Lander G. C., Johnson J. E., Casjens S. R., Thomas G. J., Jr. (2008) J. Mol. Biol. 383, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catalano C. E., Cue D., Feiss M. (1995) Mol. Microbiol. 16, 1075–1086 [DOI] [PubMed] [Google Scholar]
- 27.Lin H., Simon M. N., Black L. W. (1997) J. Biol. Chem. 272, 3495–3501 [DOI] [PubMed] [Google Scholar]
- 28.Lin H., Black L. W. (1998) Virology 242, 118–127 [DOI] [PubMed] [Google Scholar]
- 29.Rao V. B., Black L. W. (2005) in Viral Genome Packaging Machines: Genetics, Structure and Mechanism (Catalano C. E. ed) pp. 40–58, Landes Biosci, Georgetown, TX [Google Scholar]
- 30.Kondabagil K. R., Rao V. B. (2006) J. Mol. Biol. 358, 67–82 [DOI] [PubMed] [Google Scholar]
- 31.Leffers G., Rao V. B. (2000) J. Biol. Chem. 275, 37127–37136 [DOI] [PubMed] [Google Scholar]
- 32.Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 33.Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990) Methods Enzymol. 185, 60–89 [DOI] [PubMed] [Google Scholar]
- 34.Miyazaki S., Kuroda Y., Yokoyama S. (2002) J. Struct. Funct. Genomics 2, 37–51 [DOI] [PubMed] [Google Scholar]
- 35.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 36.Mitchell M. S., Matsuzaki S., Imai S., Rao V. B. (2002) Nucleic Acids Res. 30, 4009–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Zahrani A. (2006) Biochemical Mechanism of the DNA Packaging ATPase from Bacteriophage T4. Ph.D. Thesis, Catholic University of America [Google Scholar]
- 38.Wieden H. J., Gromadski K., Rodnin D., Rodnina M. V. (2002) J. Biol. Chem. 277, 6032–6036 [DOI] [PubMed] [Google Scholar]
- 39.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 40.Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J. (1997) Nature 389, 758–762 [DOI] [PubMed] [Google Scholar]
- 41.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 42.Zhang B., Zhang Y., Collins C. C., Johnson D. I., Zheng Y. (1999) J. Biol. Chem. 274, 2609–2612 [DOI] [PubMed] [Google Scholar]
- 43.Rao V. B., Black L. W. (1988) J. Mol. Biol. 200, 475–488 [DOI] [PubMed] [Google Scholar]
- 44.Black L. W., Peng G. (2006) J. Biol. Chem. 281, 25635–25643 [DOI] [PubMed] [Google Scholar]
- 45.Skorupski K., Pierce J. C., Sauer B., Sternberg N. (1992) J. Mol. Biol. 223, 977–989 [DOI] [PubMed] [Google Scholar]
- 46.Hwang Y., Feiss M. (1996) J. Mol. Biol. 261, 524–535 [DOI] [PubMed] [Google Scholar]
- 47.Baumann R. G., Black L. W. (2003) J. Biol. Chem. 278, 4618–4627 [DOI] [PubMed] [Google Scholar]
- 48.Menz R. I., Walker J. E., Leslie A. G. W. (2001) Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 49.Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D. (1999) Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 50.Lee J. Y., Yang W. (2006) Cell 127, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moffitt J. R., Chemla Y. R., Aathavan K., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2009) Nature 457, 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. (1997) Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 53.Allin C., Ahmadian M. R., Wittinghofer A., Gerwert K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burroughs A. M., Iyer L. M., Aravind L. (2007) Genome Dyn. 3, 48–65 [DOI] [PubMed] [Google Scholar]
- 55.Casjens S. R. (2005) Curr. Opin. Microbiol. 8, 451–458 [DOI] [PubMed] [Google Scholar]