Abstract
The neuroinvasive pathogen Neisseria meningitidis has 13 capsular serogroups, but the majority of disease is caused by only 5 of these. Groups B, C, Y, and W-135 all display a polymeric sialic acid-containing capsule that provides a means for the bacteria to evade the immune response during infection by mimicking host sialic acid-containing cell surface structures. These capsules in serogroups C, Y, and W-135 can be further acetylated by a sialic acid-specific O-acetyltransferase, a modification that correlates with decreased immunoreactivity and increased virulence. In N. meningitidis serogroup Y, the O-acetylation reaction is catalyzed by the enzyme OatWY, which we show has clear specificity toward the serogroup Y capsule ([Glc-(α1→4)-Sia]n). To understand the underlying molecular basis of this process, we have performed crystallographic analysis of OatWY with bound substrate as well as determined kinetic parameters of the wild type enzyme and active site mutants. The structure of OatWY reveals an intimate homotrimer of left-handed β-helix motifs that frame a deep active site cleft selective for the polysialic acid-bearing substrate. Within the active site, our structural, kinetic, and mutagenesis data support the role of two conserved residues in the catalytic mechanism (His-121 and Trp-145) and further highlight a significant movement of Tyr-171 that blocks the active site of the enzyme in its native form. Collectively, our results reveal the first structural features of a bacterial sialic acid O-acetyltransferase and provide significant new insight into its catalytic mechanism and specificity for the capsular polysaccharide of serogroup Y meningococci.
The bacterial pathogen Neisseria meningitidis is a major cause of life-threatening neuroinvasive meningitis in humans (1). In the United States, 75% of bacterial meningitis infections are caused by serogroup C, Y, or W-135 (2). In particular, the proportion of meningococcal infection occurrences in the United States caused by the group Y meningococci has increased significantly from 2% during 1989–1991 to 37% during 1997–2002 (2). Vaccines based on the capsular polysaccharide have been developed for groups A/C/Y/W-135 (2), and the introduction of a group C conjugate vaccine has reduced the incidence and carriage of the C serogroup significantly (3). Although these vaccines are working, they do not yet provide complete protection from meningococcal disease (4).
The capsular polysaccharides of N. meningitidis are classified into 13 distinct serogroups based on their chemical structures (5). The capsules of serogroup B and C are homopolymers composed of α-2,8- or α-2,9-linked sialic acid, respectively, whereas serogroup Y and W-135 are heteropolymers of an α-2,6-linked sialic acid on glucose (Y) or galactose (W-135) (6, 7). N. meningitidis group B polysialic acid shares a biochemical epitope with the polysialylated form of the neural cell adhesion molecule of humans (8, 9). Because of this molecular mimicry of the polysialic acid-neural cell adhesion molecule, the bacterial capsular polysaccharide is thus considered a major virulence factor of N. meningitidis (5, 10).
Serogroup C, Y, and W-135 of N. meningitidis modify their sialic acid capsules by O-acetylation of the sialic acid (11). Sialic acid is acetylated at the C-7 or C-8 position hydroxyl group in serogroup C, whereas the C-7 or C-9 position is acetylated in serogroup W-135 and Y (11). The O-acetylation of sialic acids is known to alter the physicochemical properties of the polysaccharide capsule (12). In addition, there is growing evidence that O-acetylation of the polysaccharide enhances bacterial pathogenesis by masking the protective epitope in the polysaccharide (13–16). For these reasons, considerable effort has been expended to identify and characterize sialic acid O-acetyltransferases in pathogenic bacteria.
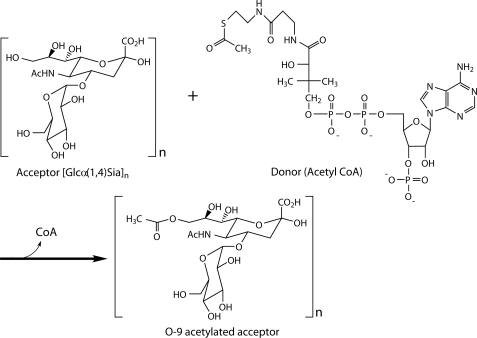
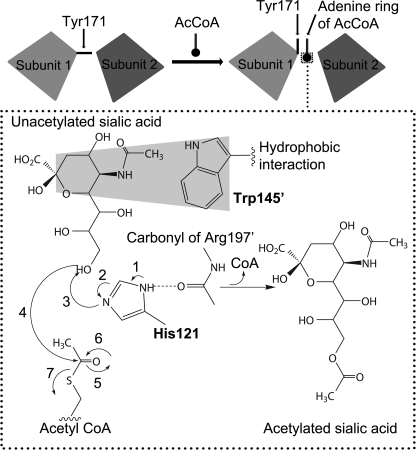
Recently, the sialic acid-specific O-acetyltransferases from group B Streptococcus, Campylobacter jejuni, Escherichia coli K1, and N. meningitidis serogroup C have been identified (17–20) with the latter two variants being the only ones to be characterized biochemically (21–23). These studies showed that bacterial sialic acid-specific O-acetyltransferases utilize an acetyl-CoA cofactor as a donor for the acetylation of their capsular sialic acid acceptor substrates (Fig. 1) and identified essential amino acid residues for potential catalytic roles in activity (22, 23). Although the gene encoding the capsule-specific O-acetyltransferase in N. meningitidis serogroup Y (known as OatWY) has been identified, biochemical characterization of the enzyme has not yet been reported. Furthermore, the lack of structural information on a sialic acid O-acetyltransferase from any bacterial species has hampered our ability to further understand the mode of substrate binding, specificity, and catalytic mechanism of this important sialic acid-modifying family.
FIGURE 1.
Reaction scheme of the OatWY-catalyzed O-acetyltransferase. Although acetylation of both the O-7 and O-9 hydroxyl group of the N. meningitidis serogroup Y polysialic acid has been implied through NMR analysis of the corresponding bacterial capsule (11), for simplicity only the O-9 transfer is shown here.
Here we report the first kinetic and structural analysis of polysialic acid O-acetyltransferase OatWY from N. meningitidis serogroup Y in complex with either CoA, acetyl-CoA, or S-(2-oxopropyl)-CoA, which is a nonhydrolyzable acetyl-CoA substrate analog. Collectively, this study significantly contributes to our understanding of bacterial polysialic acid O-acetyltransferases, providing valuable insight into how capsular polysaccharide is acetylated in pathogenic bacteria.
EXPERIMENTAL PROCEDURES
Cloning of the Polysialic Acid O-Acetyltransferase OatWY from N. meningitidis Y
The oatWY gene (GenBankTM accession number Y13969) coding for the polysialic acid O-acetyltransferase was directly amplified by PCR from genomic DNA of the N. meningitidis serogroup Y using the Pwo DNA polymerase (Stratagene) with the forward primer OATWY-F (5′-CCGACGCATATGGGAACTCACATGTATTCTGAACAGGGAATTAATAATAC-3′), which introduced an NdeI site (underlined) in the 5′ end, and the reverse primer OATWY-R (5′-CGTCGGAAGCTTTTATTTATAAAATTCATTTAAAGTAGGGTCATCATACATAGTTGAATG-3′) (Integrated DNA Technologies), which introduced a HindIII site (underlined). Amplified PCR products were digested by NdeI/HindIII (New England Biolabs) and subsequently ligated using T4 DNA ligase (Invitrogen) into pET-28 vector (Novagen) containing the cleavable hexahistidine tag at the N terminus. The properly cloned gene, which encodes the OatWY enzyme, was transformed into electrocompetent E. coli cells (BL21 λDE3, Novagen) for expression.
Cloning of OatWY Mutants by Site-directed Mutagenesis
Site-directed mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's guideline with the following primers. OATWY-H121A-F (5′-GAAATACTGATATGGCTCCAATTTATTCTTTAGAAAATGGCGAACG-3′) and OATWY-H121A-R (5′-CGTTCGCCATTTTCTAAAGAATAAATTGGAGCCATATCAGTATTTC-3′) were used to generate the H121A mutant. OATWY-W145A-F (5′-CGGTAATCACGTGGCGCTTGGCCGCAATGTTAC-3′) and OATWY-W145A-R (5′-GTAACATTGCGGCCAAGCGCCACGTGATTACCG-3′) were used to generate the W145A mutant enzyme. Furthermore, OATWY-Y171A-F (5′-GTTGTAGGTTCTCATACTGTGCTAGCTAAAAGTTTTAAAGAACC-3′) and OATWY-Y171A-R (5′-GGTTCTTTAAAACTTTTAGCTAGCACAGTATGAGAACCTACAAC-3′) were used to generate the Y171A mutant. All mutations were confirmed by DNA sequencing, and genes coding the mutant enzymes were transformed into electrocompetent E. coli cells as described above.
Expression and Purification of OatWY
The transformed cells were incubated overnight at 37 °C with shaking at 225 rpm in the presence of 50 μg/ml kanamycin. The cell cultures were grown until A600 reached ∼0.6 and were induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside at 20 °C with further overnight incubation. Cells were harvested by centrifugation at 5,000 rpm for 15 min and then either used immediately or frozen at −80 °C for storage. The cells were resuspended in lysis buffer (20 mm HEPES, pH 7.5, 500 mm NaCl, 5 mm imidazole) and lysed in the presence of an EDTA-free protease inhibitor mixture tablet (Roche Applied Science) at 20,000 p.s.i. using a high pressure homogenizer (Avestin). The lysate was centrifuged at 40,000 rpm for 40 min, and the resulting supernatant was loaded onto a pre-equilibrated HiTrap Chelating HP column (GE Healthcare) charged with NiCl2. The column was washed with buffer containing 100 mm imidazole and eluted with 100 mm EDTA in the buffer described above. All eluting fractions were monitored by SDS-PAGE. Fractions containing the OatWY protein were collected and dialyzed against 20 mm MES,4 pH 6.0, 100 mm NaCl with thrombin at 4 °C overnight. The dialyzed proteins were further purified on a Mono S cation exchange column (GE Healthcare) using a linear gradient of 100 mm to 1 m NaCl in 20 mm MES, pH 6.0, and concentrated using an Amicon concentrator (Millipore). The molar mass of the protein was monitored by matrix-assisted laser desorption/ionization-time of flight mass spectrometry.
In Vitro Activity Assay of OatWY
Activities of the wild type OatWY enzyme and mutants were determined in a spectrophotometric assay using the procedures reported previously (24). The common reaction mixture (200 μl final volume) was composed of 20 mm Tris-HCl, pH 7.5, 25 mm EDTA, 4 mm 5,5′-dithiobis(2-nitrobenzoic acid), 5.5 mg/ml Y meningococcal specific polysaccharide ([→6)-Glc-(α1→4)-Sia-(α2→]n) in its deacetylated form, 1 mm acetyl-CoA, and the enzymatic reaction was initiated by adding 34 μm purified OatWY enzyme at 25 °C. The change of absorbance per min was monitored continuously at 412 nm using a Cary 300 spectrophotometer (Varian) regulated by a temperature controller. For further investigations regarding acceptor specificity, CMP-Neu5Ac, Neu5Ac, glucose, colominic acid (Sigma), and [Glc-(α1→4)-Sia] disaccharide were standardized to an equal concentration of sialic acid (5 mm), ensuring equal saturation of the acceptor site by these various sialylated substrates. In addition, one assay mixture contained 1 mm S-(2-oxopropyl)-CoA instead of acetyl-CoA in the presence of the Y polysaccharide to investigate its reactivity toward the OatWY enzyme.
Kinetic Characterization
Kinetic parameters for acceptor (deacetylated Y polysaccharide) and donor (acetyl-CoA) were determined with OatWY native enzyme as follows. Assays were performed at 25 °C in a total volume of 200 μl containing acceptor and donor with enzyme (34 μm) in 20 mm Tris-HCl, pH 7.5, 25 mm EDTA, plus 4 mm 5,5′-dithiobis(2-nitrobenzoic acid) holding one substrate at a saturating concentration, although the other substrate concentration was varied. For donor kinetics, a range of different concentrations of acetyl-CoA (0.025–0.8 mm) was used, at a saturating concentration of the Y polysaccharide ([→6)-Glc-(α1→4)-Sia-(α2→]n, 5.5 mg/ml). For acceptor kinetics, varied concentrations of the Y polysaccharide (1.0–15.0 mg/ml) were used, at a saturating concentration of acetyl-CoA (1 mm). The kinetic parameters were calculated from the best fit of the data to the Michaelis-Menten equation using nonlinear regression analysis with GraFit 5.0 software (Erithacus Software).
Multiangle Light Scattering
Purified OatWY enzyme (5 mg/ml) was loaded onto a Superdex 200 10/300 GL gel filtration column (GE Healthcare), equilibrated with buffer (20 mm Tris, pH 7.5, 50 mm NaCl), and connected in line with miniDAWN multiangle light-scattering equipment coupled to an interferometric refractometer (Wyatt Technologies). Data analysis was done in real time using ASTRA (Wyatt Technologies), and molecular masses were calculated using the Debye fit method.
Synthesis of S-(2-Oxopropyl)-CoA
The synthesis of S-(2-oxopropyl)-CoA was performed according to the method described previously (25). Briefly, 100 mg (0.13 mmol) of CoA trilithium salt (Sigma) was dissolved in 5 ml of water, followed by addition of dithiothreitol (11 mg, 0.07 mmol) and Li2CO3 (60 mg, 0.81 mmol) under argon atmosphere. To the resulting mixture was then added chloroacetone (5.3 mg, 0.06 mmol) in 12 ml of 95% ethanol. The reaction mixture was stirred at room temperature for 24 h, and then the solution was filtered and concentrated in vacuo affording a yellow oil. Reverse phase HPLC purification using Luna C18(2) column (Phenomenex), with HPLC grade acetonitrile, 0.1% trifluoroacetic acid, and HPLC grade water, 0.1% trifluoroacetic acid, using a 40–60% acetonitrile/water gradient over 60 min, yielded 30 mg (29%) of S-(2-oxopropyl)-CoA.
Deacetylation of Y Meningococcal Polysaccharide
The deacetylation of N. meningitidis serogroup Y polysaccharide was carried out according to previously published procedures (6). Polysaccharide (70 mg) was dissolved in 0.1 m NaOH (30 ml), and the reaction mixture was stirred for 4 h at 30 °C. The solution was neutralized to pH 7.0 with acetic acid and then filtered through a 5-kDa molecular mass cutoff filter (Millipore). Flow-through was discarded, and the supernatant was concentrated to 2 ml of syrup. 1 ml of this syrup was lyophilized, and 1 ml was further used in the synthesis of 4-O-α-d-glucopyranosyl-β-N-acetylneuraminic acid.
Preparation of the [Glc-(α1→4)-Sia] Disaccharide
1 ml of de-O-acetylated Y polysaccharide was added to Amberlite IR-120 cation exchange resin (H+ form) until pH 3.0 was achieved, and the reaction mixture was then incubated for 30 min at 100 °C. The resin was filtered off, and the reaction mixture was loaded onto Dowex® 1 × 2 ion exchange resin (Sigma) and eluted using a linear gradient of 0–1 m formic acid. Low resolution mass spectrometry showed the desired peaks in both positive and negative ion modes (M + 1,472 and M − 1,470). The resulting compound was characterized by proton NMR spectroscopy.
Crystallization and Data Collection
Crystals of OatWY were grown by the hanging drop vapor diffusion technique at 21 °C. The first condition that produced well ordered crystals contained 100 mm sodium acetate trihydrate, pH 4.6, 1.0 m ammonium phosphate monobasic, 100 mm lithium sulfate monohydrate and contained one molecule per asymmetric unit with unit cell dimensions of a = 199.635, b = 199.635, c = 199.635 Å in space group F4132. In the second condition, the reservoir solution contained 100 mm sodium acetate, pH 4.6, 2.25 m ammonium acetate. This OatWY crystal belonged to space group P222, with unit cell dimensions of a = 79.06, b = 94.83, c = 101.06 Å, and contained three molecules per asymmetric unit. The crystals were cryoprotected in mother liquor containing 25% ethylene glycol and directly plunged into liquid nitrogen prior to data collection. Three co-substrate structures were solved from apo-OatWY crystals (P222) soaked in either the presence of 1 mm coenzyme A, acetyl-CoA, or S-(2-oxopropyl)-CoA. The same reservoir solution as described above was used, and the soaking times of either 4 h or overnight before flash-freezing in liquid nitrogen. X-ray diffraction data were collected at 100 K under a nitrogen stream using an in-house CuKα rotating anode x-ray generator coupled to a Mar345 detector or using synchrotron sources (beamlines 8.2.2 and 4.2.2 at the Advanced Light Source (Berkeley, CA) using an ADSC Q315 and NOIR-1 CCD detector, respectively, and the beamline 08ID-1 at the Canadian Light Source (Saskatoon, Saskatchewan, Canada) using a Marmosaic CCD225 detector). All diffraction data were processed and scaled by HKL or MOSFLM (26, 27).
Structure Determination, Refinement, and Modeling
The structure of OatWY was solved by the method of single isomorphous replacement with anomalous scattering. Crystals were soaked into the previously described reservoir solution containing the ethylene glycol cryoprotectant as well as the derivatizing agent 1 m NaI for 30 s and flash-frozen in liquid nitrogen. Positions of 11 iodide sites were found and refined by SHELX-C (28). Phase calculation and solvent flattening were carried out by using SHELX-DE (29, 30), which resulted in an interpretable electron density map. Subsequent automatic model building by RESOLVE (31) generated ∼60% of the structure followed by further manual building using COOT (32). Refinements were performed using REFMAC5 (33) with exclusion of 5% of the reflections for the Rfree calculation. Finally, Translation/Libration/Screw motion determination was incorporated into the procedure for further rounds of crystallographic refinement (34). All the ligand models, including CoA, acetyl-CoA, and S-(2-oxopropyl)-CoA, were generated using the PRODRG server (35). We found two partially occupied molecules (0.7) of CoA and S-(2-oxopropyl)-CoA, and three fully occupied molecules of acetyl-CoA at the interface of adjacent monomers of the P222 crystal form. The lack of occupancy of one interface of the homotrimeric subunits in the P222 crystal form may be due to the crystal packing and/or soaking method used for CoA incorporation (4 h for CoA or S-(2-oxopropyl)-CoA and overnight for acetyl-CoA, respectively). Final models were validated with MOLPROBITY (36). Modeling of Y meningococcal polysialic acid acceptor was performed using AutoDock Vina (37) with AutoDockTools (38) and the necessary ligand file prepared using the PRODRG server (35). A linear form of the polysialic acid, [→6)-Glc-(α1→4)-Sia-(α2→]4, was generated and tested for docking to the general area of potential acceptor binding between two adjoining monomers as suggested by our structural data. Subsequently, this roughly localized model [→6)-Glc-(α1→4)-Sia-(α2→]4 was utilized as the starting point in the docking procedure, which applied torsional rearrangements for the acceptor model to generate the lowest energy docking trials. Figures were generated with PyMOL (39), and electrostatic surface calculations were carried out with the APBS plugin (40).
RESULTS
Overall Architecture of OatWY
The crystal structure of the OatWY apoenzyme was solved by single isomorphous replacement with anomalous scattering utilizing phases from the sodium iodide soak. Co-substrate structures of OatWY were determined using the native model in a molecular replacement procedure. The high resolution edge of the diffraction data varied from 1.90 to 2.35 Å. Rwork values ranged from 17.8 to 20.1% and Rfree values from 20.7 to 24.7% with full details of the data collection and refinement summarized in Tables 1 and 2. Of the two distinct crystal forms resulting from our crystallization, the structure derived from the crystal defined by space group P222 contained three molecules per asymmetric unit. The structure derived from the crystal with space group F4132 (one molecule per asymmetric unit) adopts a nearly identical trimeric arrangement around the 3-fold symmetry axis of the crystal. The crystallographic models derived from the two unique crystal forms of OatWY are highly similar and can be overlapped with a root mean square deviation of 0.72–0.88 Å on all 210 C-α atoms. We believe the observed quaternary structure of OatWY as a homotrimer is physiologically relevant as supported by our multiangle light-scattering analysis in solution (supplemental Fig. 1) and earlier work describing the catalytically essential homotrimerization of other acetyltransferases of the LβH family (41–43).
TABLE 1.
Data collection and refinement statistics for OatWY
| OatWY NaI | OatWY apo | OatWY apo | |
|---|---|---|---|
| Data collection | CuKα | ALS 8.2.2 | ALS 8.2.2 |
| Wavelength | 1.5418 Å | 1.000 Å | 1.000 Å |
| Resolutiona | 40 to 2.7 Å (2.80 to 2.70) | 40 to 1.95 Å (2.02 to 1.95) | 40 to 2.20 Å (2.28 to 2.20) |
| Space group | P222 | F4132 | P222 |
| a | 78.86 Å | 199.63 Å | 79.09 Å |
| b | 95.16 Å | 199.63 Å | 94.81 Å |
| c | 101.13 Å | 199.63 Å | 100.85 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° | 90, 90, 90° |
| Rsym (%)a | 12.5 (50.3) | 8.6 (45.3) | 5.2 (26.9) |
| I/σ(I)a | 18.5 (3.6) | 68.0 (10.8) | 42.8 (6.5) |
| Completenessa | 100% (99.8%) | 100% (100%) | 100% (100%) |
| Unique reflectionsa | 40,415 (4058) | 25,297 (2489) | 39,235 (3848) |
| Redundancya | 7.7 (7.1) | 46.9 (47.6) | 8.1 (8.2) |
| Refinement | |||
| Average B factor (Å2) | |||
| Protein | 11.1 | 18.1 | |
| Ligand | |||
| Water | 26.4 | 26.9 | |
| Ramachandran statistics | |||
| Favored regions | 98.2% | 96.9% | |
| Additionally allowed regions | 1.8% | 3.1% | |
| Disallowed regions | 0.0% | 0.0% | |
| Rwork | 17.2% | 18.5% | |
| Rfree | 20.9% | 23.4% | |
| r.m.s.b bonds | 0.014 Å | 0.012 Å | |
| r.m.s. angles | 1.536° | 1.329° | |
a Values in parentheses represent the highest resolution shell.
b r.m.s. means root mean square.
TABLE 2.
Data collection and refinement statistics for OatWY (continued)
| OatWY + CoA | OatWY + acetyl-CoA | OatWY + S-CoAa | |
|---|---|---|---|
| Data collection | CuKα | ALS 4.2.2 | CLS 08ID |
| Wavelength | 1.5418 Å | 0.9790 Å | 0.9790 Å |
| Resolutionb | 40 to 2.20 Å (2.28–2.20) | 40 to 2.35 Å (2.48–2.35) | 40 to 1.90 Å (1.97–1.90) |
| Space group | P222 | P222 | P222 |
| a | 79.07 Å | 78.30 Å | 78.67 Å |
| b | 94.52 Å | 94.52 Å | 94.39 Å |
| c | 100.89 Å | 100.59 Å | 100.87 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° | 90, 90, 90° |
| Rsymb | 5.7% (46.4%) | 9.6% (44.1%) | 5.1% (28.6%) |
| I/σ(I)b | 34.5 (3.4) | 15.0 (3.2) | 52.8 (7.2) |
| Completenessb | 99.7% (97.4%) | 100% (100%) | 99.0% (95.2%) |
| Unique reflectionsb | 39,161 (3761) | 32,150 (4618) | 59,306 (5651) |
| Redundancyb | 6.0 (5.6) | 4.8 (4.9) | 10.0 (8.9) |
| Refinement | |||
| Average B factor (Å2) | |||
| Protein | 21.9 | 12.5 | 15.1 |
| Ligand | 50.8 | 50.4 | 47.9 |
| Water | 31.2 | 24.9 | 27.5 |
| Ramachandran statistics | |||
| Favored regions | 98.3% | 97.3% | 97.6% |
| Additionally allowed regions | 1.7% | 2.7% | 2.4% |
| Disallowed regions | 0.0% | 0.0% | 0.0% |
| Rwork | 19.5% | 18.3% | 19.3% |
| Rfree | 24.2% | 23.1% | 23.1% |
| r.m.s.c bonds | 0.008 Å | 0.009 Å | 0.011 Å |
| r.m.s. angles | 1.104° | 1.468° | 1.342° |
aS-CoA denotes S-(2-oxopropyl)-CoA.
b Values in parentheses represent the highest resolution shell.
c r.m.s. means root mean square.
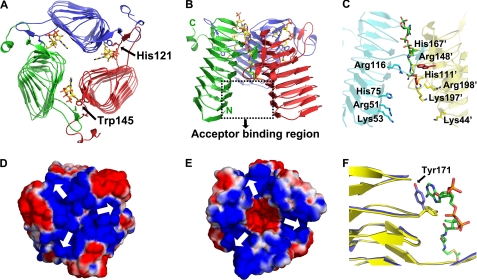
Each monomer of the OatWY homotrimer is composed of three domains. The C-terminal extension (residues 192–215) consists of a loop region followed by a short 310 helix. The N-terminal domain (residues 6–191) is composed of a left-handed β-helical (LβH) motif containing seven turns of parallel β-helix. LβH domains are characterized by repeating left-handed connections of parallel β-strands. Each helical turn consists of three β-strands connected by short loops and displays a signature motif composed of the repeating hexapeptide sequence ((LIV)(GAED)X2(STAV)X). LβH folds are stabilized by both intramolecular β-sheet hydrogen bonds and an inner hydrophobic core resulting from the packing of leucine and isoleucine residues at position 1 (typically termed “i”) of each hexapeptide motif and have been observed in various acyltransferases (41–46). Our DNA sequencing results revealed that the OatWY construct has a random mutation at position 67 (N67I) relative to that reported in the EMBL data base (accession number Y13969). This residue is located in position i of the interior core region of the LβH domain, which is characterized by an extremely hydrophobic environment. Interestingly, the observed Ile-67 mutation aligns and packs well within the Leu/Ile hydrophobic core typical of LβH domains. In the OatWY homotrimer, a long extended loop region (residues 118–136) projects from the central motif of each LβH domain to interact with the C-terminal extension (residues 192′–215′) of an adjacent monomer (Fig. 2, A and B). Through this intermolecular interface, each monomeric subunit is intimately associated with the other subunits of the homotrimer via a network of main chain and water-mediated hydrogen bonds and electrostatic interactions (a salt bridge between Arg-148 and Glu-92′ of the adjacent monomer for example). An electrostatic surface calculation of the OatWY trimer shows a predominantly electronegative character in the region of the protruding loop118–136 and the C-terminal extension, whereas the interior channel formed between the three LβH domains of the homotrimer is predominantly electropositive (Fig. 2, D and E).
FIGURE 2.
Quaternary structure and electrostatic surface map of OatWY. A, structure of OatWY-acetyl-CoA complex viewed parallel to the 3-fold axis, and B, structure viewed perpendicular to the 3-fold axis of the OatWY trimer (the three subunits are colored in blue, red, and green, with the N- and C-terminal ends indicated in the latter). The potential acceptor binding region is outlined by the black dotted box. Carbon atoms in the acetyl-CoA are represented as yellow, and non-carbon atoms are colored according to atom type (nitrogen, blue; oxygen, red; phosphorus, orange; and sulfur, gold). Two essential catalytic residues (His-121 and Trp-145′) are shown as stick models and are indicated by arrows. C, potential acceptor binding region. Surface-exposed residues potentially involved in polysaccharide acceptor binding are displayed in yellow for residues from one monomer and in cyan for residues from adjoining monomer. Bound CoA is shown as a stick model, and two conserved catalytic residues (His-121 and Trp-145) are represented as red sticks. Electrostatic surface representations (D, top view, and E, bottom view) along with the homotrimeric structure of OatWY show the overall electropositive charge (blue) at the monomer interface of the OatWY trimer. Arrows in the electrostatic surface maps indicate the location of substrate-binding sites between adjoining subunits. Electronegative surface charge is colored in red. F, movement of Tyr-171 upon donor binding. Superposition of the apo (yellow) and donor-bound (blue) OatWY structures. Tyr-171 is shown in blue; carbon atoms in the acetyl-CoA molecule are represented as green, and non-carbon atoms are colored according to atom type (nitrogen, blue; oxygen, red; phosphorus, orange; and sulfur, gold).
Donor-binding Site
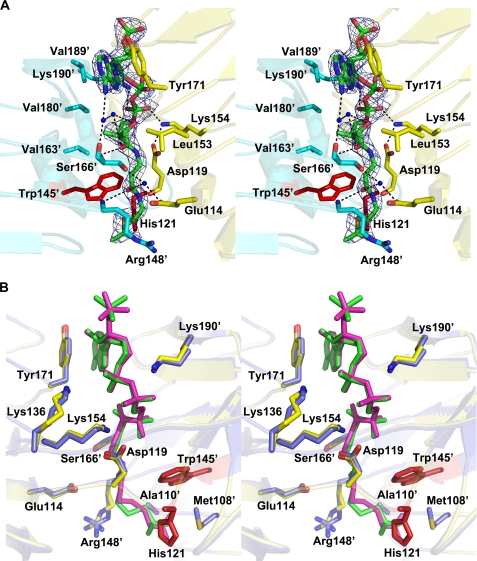
The co-substrate structures of OatWY in complex with the substrate acetyl-CoA and its analogs (CoA, S-(2-oxopropyl)-CoA) identify the donor-binding site as localized to the interface of adjacent monomers in the homotrimer. Specifically, the CoA moiety occupies a prominent cleft created collectively by the LβH domains localized to the C-terminal region of each monomer (Fig. 2, A and B). The donor forms polar and hydrophobic contacts with residues in both flanking monomers. Binding of donor appears to induce little overall conformational change in the enzyme as the apo and co-substrate structures can be overlapped with a root mean square deviation of 0.24–0.34 Å for all 630 C-α atoms of the homotrimer. However, a few critical and we believe functionally important differences do arise upon donor binding, the most significant being the movement of the aromatic side chain of Tyr-171. This tyrosine is oriented such that the side chain hydroxyl points toward the LβH domain, thereby blocking the cleft created by interfacing subunits in the apo structure. In contrast, the aromatic side chain of Tyr-171 flips in the co-substrate structure, to contact directly the adenine ring of the coenzyme A via hydrophobic interactions (Fig. 2F). In addition, the amine side chain of Lys-190′ is ordered in the complex structure via a direct electrostatic interaction with the ribose 3′-phosphate group of the CoA molecule. In general, the binding of CoA is stabilized by a prominent set of electrostatic interactions. In addition to Lys-190′, the ribose 3′-phosphate group is also bound via a direct interaction with Lys-154. The ribose 2′-phosphate group forms water-mediated hydrogen bonds to Lys-136 and directly interacts with the side chain of Lys-154. The carbonyl oxygen of the phosphopantothenyl arm forms hydrogen bonds to the main chain amide group of Ser-166′. The amide nitrogen located at the middle of the pantothenyl arm forms a direct hydrogen bond to Asp-119, and the amide nitrogen of the CoA hydrogen bonds with the carbonyl oxygen of Asp-119. The phosphopantothenyl arm of CoA is also stabilized by hydrophobic interactions with the side chains of apolar residues, including Val-163′, Val-180′, and Val-189′ from one monomer and Ile-123 and Leu-153 from the adjoining monomer (Fig. 3A). Importantly, the donor acetyl-CoA-bound structure (as compared with that with CoA) facilitates formation of an additional hydrogen bond between the acyl oxygen of acetyl-CoA and His-121, which we believe is one of the catalytic residues (see below). Interestingly, our structure with the donor substrate analog S-(2-oxopropyl)-CoA, an inhibitor of OatWY activity, binds in an identical fashion to that of acetyl-CoA except for the loss of this hydrogen bond to His-121, an interaction disfavored by the propyl functionality of the nonhydrolyzable S-(2-oxopropyl)-CoA.
FIGURE 3.
A, stereoview of the donor-binding site with electron density of acetyl-CoA in a refined 2Fo − Fc map contoured at 1.5σ. Bound acetyl-CoA molecule is shown using green for carbon and phosphorus atoms, with other atoms colored according to atom type (nitrogen, blue; oxygen, red; and sulfur, gold). Interacting residues from one monomer are shown in yellow, and interacting residues from the adjoining monomer are shown in cyan. Water molecules are depicted as blue spheres, and His-121 and Trp-145′ are displayed as red sticks. Polar contacts are shown as dotted lines. B, stereoview of a structural alignment between the complexes of OatWY with acetyl-CoA and a nonhydrolyzable CoA analog. Key residues that interact with the donor substrate are represented in yellow for the acetyl-CoA complex and in blue for the S-(2-oxopropyl)-CoA complex. Acetyl-CoA and S-(2-oxopropyl)-CoA molecules are depicted in green and magenta, respectively. His-121 and Trp-145′ are represented in red.
Acceptor Specificity of OatWY
A series of activity assays was carried out to examine the acceptor specificity of OatWY. All activity profiles were monitored by the spectrophotometric procedure described previously (24) and in the presence of various acceptor candidates, including monomeric sialic acid (Neu5Ac), activated sialic acid (CMP-Neu5Ac), glucose, [Glc-(α1→4)-Sia] disaccharide, colominic acid (a homopolymer of sialic acid with α2,8-linkages), and the OatWY-specific polysaccharide ([→6)-Glc-(α1→4)-Sia-(α2→]n (Table 3). OatWY specifically catalyzed the O-acetyltransferase reaction in the presence of the N. meningitidis serogroup Y polysaccharide. Other substrates, including colominic acid, showed insignificant activities except for the disaccharide molecule, which acted as an acceptor with ∼19% activity of the wild type enzyme. S-(2-Oxopropyl)-CoA was also tested as a potential donor, along with the OatWY polysaccharide. The activity level of the reaction mixture containing the S-(2-oxopropyl)-CoA was dramatically reduced.
TABLE 3.
Acceptor specificity of OatWY
| Acceptora | Activity |
|---|---|
| % | |
| Polysaccharide [Glc-(α1→4)-Sia]n | 100.0 ± 0.3 |
| Disaccharide [Glc-(α1→4)-Sia] | 19.2 ± 0.2 |
| Colominic acid [α(2→8)Sia]n | 2.1 ± 0.2 |
| Neu5Ac | 0.3 ± 0.2 |
| CMP-Neu5Ac | 0.3 ± 0.1 |
| Glucose | 0.0 ± 0.2 |
| S-(2-oxopropyl)-CoAb | 2.4 ± 0.1 |
| (−)-Control (water) | 0.7 ± 0.1 |
a Concentrations of acceptors were adjusted to 5 mm sialic acid equivalent to that described under “Experimental Procedures.”
b Instead of acetyl-CoA, the S-(2-oxopropyl)-CoA was added to the assay mixture in the presence of the natural acceptor ([Glc-(α1→4)-Sia]n).
Identification of Potential Catalytic Residues
To investigate the importance and roles of the highly conserved residues localized within the active site of OatWY (His-121 and Trp-145′), two mutant forms of the enzyme, H121A and W145A, were generated and analyzed for catalytic activity. In addition, a Y171A mutant was also generated to investigate potential effects on donor binding. The results showed that all three mutations dramatically reduced activity; the histidine mutant has only 2.0% of the activity of wild type protein. The tryptophan mutant showed 1.8% of the activity compared with wild type enzyme. Finally, the tyrosine mutant showed 2.1% of the activity of wild type OatWY (Table 4).
TABLE 4.
Comparison of activities of wild type and mutant OatWY
| Enzyme | Activity |
|---|---|
| % | |
| Wild type | 100.0 ± 1.0 |
| H121A | 2.0 ± 0.4 |
| W145A | 1.8 ± 0.7 |
| Y171A | 2.1 ± 0.7 |
Kinetic Analysis
A more complete kinetic analysis of OatWY was performed to further characterize the nature of the enzymatic reaction. All kinetic experiments were carried out at 25 °C, and data were fitted to the Michaelis-Menten equation. Kinetic parameters for wild type OatWY enzyme are summarized in Table 5. The Km value for donor (acetyl-CoA) was 0.21 mm, and the Km value for acceptor (Y polysaccharide) was 2.55 mg/ml, which is equivalent to 5.4 mm of the total [Glc-(α1→4)-Sia] disaccharide. Static light-scattering results showed that the estimated mass range of the polysaccharide is around 100–230 kDa, which is equivalent to 200–400 disaccharide units (data not shown). Based on this mass information, a Km value of 11–26 μm can be calculated for the polysaccharide. Specificity constants (kcat/Km) were 6.3 s−1 mm−1 for acetyl-CoA and 80–190 s−1 mm−1 for the polysaccharide.
TABLE 5.
Kinetic parameters of OatWY
| Acetyl-CoA |
Acceptora |
||||
|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km |
| μm | s−1 | mm−1s−1 | μm | s−1 | mm−1s−1 |
| 210 ± 24 | 1.3 ± 0.1 | 6.3 | 11 ± 1–26 ± 3 | 2.1 ± 0.1 | 80–190 |
aAcceptor was the N. meningitidis serogroup Y polysialic acid, ([Glc-(α1→4)-Sia]n).
DISCUSSION
Capsular polysialic acids that decorate the cell surfaces of various bacteria act as important virulence factors in these pathogenic species that inflict serious human disease, including sepsis and meningitis (47, 48). O-Acetylation, the only common modification of the bacterial sialic acid moiety, alters the physiological properties of the resulting modified bacterial polysaccharides (15, 49–51). In this study, we provide the first structural characterization of a polysialic acid O-acetyltransferase, which is that of OatWY from N. meningitidis, providing a foundation for understanding previous biochemical analyses as well as the new kinetic specificity and mutagenesis data presented here.
The OatWY enzyme structure was solved as an intimately associated homotrimer held together by numerous noncovalent interactions, which localize primarily to the C-terminal extension (residues 192–215) and the inserted loop region (residues 118–136) from the β-helix coils of the LβH domain. As typical, the LβH domains of the trimer are stabilized by an extensive intermolecular hydrogen bonding network between β-strands and further stabilized by hydrophobic interactions formed by the regular arrangement of Leu and Ile packed within the interior core of the LβH domain.
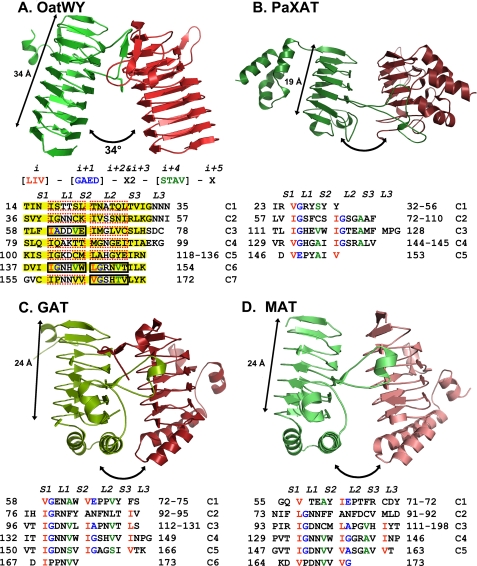
The LβH domains in OatWY show characteristic features of the repeating β-helix fold motif, each of which is composed of three β-strands (S1, S2, and S3) connected by short loop regions of 1–3 amino acids (L1, L2, and L3) (52). The LβH fold is found in various bacterial acetyltransferases, including xenobiotic acetyltransferase from Pseudomonas aeruginosa (PaXAT) (41), N-acetyltransferase from C. jejuni (PglD) (53), virginiamycin acetyltransferase D from Enterococcus faecium (VatD) (46), and lac operon galactoside acetyltransferase (GAT) (42), UDP-N-acetylglucosamine acyltransferase (44), and maltose acetyltransferase (MAT) (43) from E. coli. These proteins are usually composed of an α-helical or α/β domain in addition to the LβH domain, the former playing roles in oligomerization and “capping” of the LβH domain. Interestingly, OatWY appears to be a minimalized form of these enzymes, with the LβH domain being the only major structural domain and the small C-terminal extension composed only of a loop terminated by a short 310 helix.
A tandemly repeated hexapeptide sequence typically defines the LβH fold in bacterial acetyltransferases (41–43). The aliphatic residues (Leu, Ile, or Val) at position i of the hexapeptide motif ((LIV)(GAED)X2(STAV)X) project into the interior of the β-helical domain. Interestingly, OatWY contains an unusual set of hexapeptide-repeating motifs, with atypical residues at position i + 1 or i + 4 except for those hexapeptides initiating within sequence motifs 61 (IADDVE), 140 (IGNHVW), 146 (LGRNVT), and 164 (VGSHTV) (Fig. 4A). The first five coils of OatWY are also longer and more sequence variable than the more regularly repeating hexapeptides found in other bacterial acetyltransferases (Fig. 4). The irregular hexapeptide motifs of OatWY result in a larger size for the first five β-strand coils and lead in turn to a less compact overall LβH structure. In addition, the size of the LβH domain in OatWY (7 β-helical coils) is significantly extended compared with other bacterial acetyltransferase structures, which typically utilize ∼5 β-helical coils in the formation of the LβH domain.
FIGURE 4.
Structural comparison of OatWY with other bacterial acetyltransferases containing LβH motifs (A, polysialic acid O-acetyltransferase OatWY; B, xenobiotic acetyltransferase PaXAT (PDB access code 2XAT); C, GAT (PDB access code 1KRU); D, MAT (PDB access code 1OCX)). Only two monomeric subunits of the trimer are represented for clarity (in green and red). Angles between the 3-fold axis and LβH domains are illustrated with two-headed arrows along with the measured overall span of the LβH domains. Below the OatWY structure in A is the signature sequence of the hexapeptide-repeating motif (LIV)(GAED)X2(STAV)X displayed with positional indicators (i, i + 1, i + 2…) as well as the hexapeptide sequence motifs that define the OatWY LβH domain. Sheets and loops composed of the LβH domain are represented as S1–3 and L1–3, respectively. Sheets (S1–S3) are boxed with a yellow color, and the hexapeptide sequences that obey the rule ((LIV)(GAED)X2(STAV)X) are boxed with solid black rectangles, and those that do not with dashed red rectangles. The defining hexapeptide repeating motifs of each of the other acetyltransferases are also represented in a sequence table below.
The potential functional implications of these alterations in sequence and number of repeating patterns of the LβH motif can be illustrated by comparison of the geometric features and overall span of the trimeric LβH domain in OatWY and other bacterial acetyltransferases. In OatWY, the axis of the LβH domain makes an angle of 34° with the 3-fold axis of the trimer, strikingly larger than any angle created by other bacterial acetyltransferases containing the β-helical fold and in general very unusual for the family of hexapeptide repeat-containing proteins. Other bacterial sugar acetyltransferases reported to date typically show a nearly parallel arrangement of β-helical domain axes relative to the trimeric axis. For example, in the UDP-N-acetylglucosamine acyltransferase LpxA there is a 1–2° angle between axes (54), whereas the angle in the lac operon GAT or MAT is 10–12° (42, 43). In addition to the more splayed nature of the OatWY homotrimer, the greater number of LβH repeats in OatWY results in a longer span of the LβH domain (∼34 Å compared with the 18–20 Å range in other bacterial acetyltransferases) (Fig. 4). It seems likely that these differences in span and dramatic inclination of the angle in the OatWY trimer relate to its substrate specificity, because it binds to extended polymers of polysialic acids, which are much larger than the monosaccharide acceptor sugar substrates of other characterized sugar acetyltransferase families. Supporting this hypothesis, the polysialic acid O-acetyltransferase NeuO from E. coli K1, a close homolog of OatWY, was recently shown to display optimal activity against a polysialic acid longer than 14 sialic acid residues (22).
Our co-substrate structures show that the acetyl coenzyme A and its analogs define the OatWY active site as created at the interface of adjacent monomeric subunits, supporting the requirement of an active homotrimeric form for the enzyme. The donor substrate binds within a deep cleft that is characterized by the presence of two highly conserved residues, His-121 and Trp-145′. The CoA is held by several noncovalent interactions to the active site, including a striking set of hydrophobic contacts between the adenine ring and the aromatic side chain of Tyr-171. Comparison with the apo structure shows that Tyr-171 likely plays a key and unique structural role in promoting enzyme activity, because donor binding induces a significant movement of the Tyr-171 side chain from a position where it blocks entry of the catalytic cleft (apo-form) to one that allows the observed ring stacking with the adenine ring of the coenzyme A moiety (donor-bound form; Fig. 2F). The potential importance of the observed tyrosine-mediated donor binding for OatWY function is further confirmed by our mutagenesis studies, which showed that a Y171A mutant form of the enzyme had dramatically reduced activity. The active site of OatWY is also uniquely characterized by a number of positively charged residues including Arg-116, Arg-148′, and His-167′, which can potentially bind to and position the negatively charged terminal sugar of the incoming polysaccharide acceptor molecule for acetylation. Surface-exposed residues in the potential acceptor binding region (Fig. 2C) also provide a positively charged surface patch, including that arising from the side chains of Arg-51, Lys-53, His-75, and Lys-98 from one interacting subunit, and Lys-44′, Lys-85′, His-111′, Arg-197′, and Lys-198′ from the adjacent subunit. This capacious acceptor site, created by the large angle subtended by the monomer of OatWY with the trimeric axis (as described above), presents an appropriately sized and charged surface to interact with its negatively charged poly-glucosylsialic acid acceptor substrate.
The structure of OatWY in complex with substrate acetyl-CoA shares many of the features identified in the CoA-bound structure, interactions that ideally position the acetyl group in close proximity to the conserved His-121 and Trp-145′, in a manner compatible with a role in mediating the transfer reaction. Sequence alignments of OatWY with other bacterial acetyltransferases, including MAT, GAT, VatD, PaXAT, and NeuO, clearly show that these residues are well conserved (supplemental Fig. 2). Based on previous studies of bacterial acetyltransferases (42) and our current structural, mutagenesis, and kinetic information, a mechanism for acetyl transfer reaction to the poly-glucosylsialic acid substrate can now be proposed (Fig. 5). Trp-145′ is well positioned to interact in a typical hydrophobic stacking interaction with the six-membered ring of the sialic acid, thereby orienting the polysaccharide properly for acetyl transfer. Upon donor binding, Tyr-171 alters its original conformation to mediate hydrophobic interactions with the adenine group of the acetyl-CoA. The catalytic residue, His-121 from the adjoining subunit, abstracts the proton from the hydroxyl group (O-7 or O-9) of the polysialic acid with assistance from the main chain carbonyl oxygen of Arg-197′ in the adjacent subunit. This promotes the nucleophilic attack of the hydroxyl group on the carbonyl carbon of the acetyl-CoA, hence transfer of the acetyl group to the sialic acid. Our complex structure with the nonhydrolyzable acetyl-CoA analog, S-(2-oxopropyl)-CoA, further supports this mechanistic proposal (Fig. 3B). The primary structural and functional difference between the S-(2-oxopropyl)-CoA complex and that of the substrate acetyl-CoA is the presence of a methylene group between the sulfhydryl group of the CoA and the carbonyl carbon of the acetyl moiety, a chemical feature that renders the substrate inert, thereby inhibiting the enzymatic reaction competitively. Our structure shows this methylene group of the nonhydrolyzable S-(2-oxopropyl)-CoA is stabilized through additional hydrophobic contacts with Met-108 and Ala-110 in the enzyme active site.
FIGURE 5.
Mechanistic scheme of OatWY polysialic acid acetyltransferase. In the schematic above, the two interacting subunits, which define a complete active site, are represented as gray boxes and serve to illustrate the conformational role of Tyr-171 in allowing access to substrate acetyl-CoA. In the dotted box below, a proposed mechanistic scheme is shown highlighting the role of His-121 from one subunit and Trp-145′ from the second subunit that make up the active site.
Our generated activity profiles with various sugar compounds, including Neu5Ac, CMP-Neu5Ac, glucose, [Glc-(α1→4)-Sia] disaccharide, colominic acid, and Y polysaccharide ([→6)-Glc-(α1→4)-Sia-(α2→]n), indicate that OatWY specifically reacts with Y meningococcal polysaccharides. These results clearly show that OatWY is highly specific for the unique form of the heteropolymeric polysialic acid described above and does not bind to the homopolymeric polysaccharide of E. coli K1 or N. meningitidis serogroup B (colominic acid, [α(2→8)Sia]n). Other substrate candidates also did not show detectable activities. The exquisite acceptor specificity of OatWY was further investigated by kinetic analysis and yielded a Km value for the polysaccharide of 2.55 mg/ml (∼11–26 μm, when calculated on the basis of the average molecular mass of 100–230 kDa estimated from the light-scattering analysis). Because the sequence of OatWY from W-135 meningococci was reported to be 100% identical to the one from Y meningococci (20), we can predict that the W polysaccharide ([→6)-Gal-(α1→4)-Sia-(α2→]n) would also be an acceptor for OatWY.
Molecular modeling of a truncated piece of the polysaccharide acceptor model from Y meningococci, specifically [→6)-Glc-(α1→4)-Sia-(α2→]4, with the crystal structure of OatWY showed that the acceptor substrate can be readily accommodated, in terms of size and potential electrostatic interactions within the proposed acceptor binding region. The surface-exposed residues described above, His-75 and Lys-98 from one subunit and His-111′, Arg-148′, and Arg-197′ from the adjoining subunit, provide many direct polar and electrostatic contacts to the negatively charged C-2 carboxylate and hydroxyl groups of the modeled acceptor polysialic acid. In addition to the major interactions with the positively charged residues, negatively charged and polar residues such as Ser-73 and Glu-92 are also poised appropriately for acceptor binding in our model. Other positively charged surface residues “downstream” from our modeled acceptor (localized in the N-terminal region of the LβH domain, including Arg-51 and Lys-44′) may potentially provide additional electrostatic interactions in the longer, physiological polysialic acceptors. Future direct support illustrating acceptor binding will hopefully be achieved once the production of homogeneous mixtures of suitably sized sugar polymers becomes feasible.
Supplementary Material
Acknowledgments
We thank Dr. Harry Jennings at National Research Council Canada for the sample of N. meningitidis serogroup Y polysaccharide; Drs. Raz Zarivach, Matthew Caines, and Leo Yen-Cheng Lin for fruitful discussions; and Dr. Thomas Spreter for performing the static light-scattering analysis. We also thank Drs. Igor D'angelo, Francesco Rao, and Liam Worrall for synchrotron data collection, and the staff at the Advanced Light Source beamline 4.2.2/8.2.2 (Berkeley, CA), the Canadian Light Source beamline 08ID-1 (Saskatoon, SK), and the X-ray Crystallography Hub at the Centre for Blood Research (University of British Columbia). We thank the Michael Smith Foundation for Health Research for infrastructure support (to N. C. J. S. and S. G. W.) and Canada Foundation for Innovation and British Columbia Knowledge Development Fund for infrastructure support.
This work was supported in part by the Canadian Institutes of Health Research (to N. C. J. S., S. G. W., and W. W. W.), Howard Hughes Medical Institute International Scholar program (to N. C. J. S.) for operating funds, and the Michael Smith Foundation for Health Research for scholarship (to N. C. J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- MES
- 4-morpholineethanesulfonic acid
- HPLC
- high pressure liquid chromatography
- LβH
- a left-handed β-helical
- GAT
- galactoside acetyltransferase
- MAT
- maltose acetyltransferase.
REFERENCES
- 1.Ryan K. J., Ray C. G., Sherris J. C. (eds) (2004) Sherris Medical Microbiology: An Introduction to Infectious Diseases, 4th Ed., pp. 327–333, McGraw-Hill Inc., New York [Google Scholar]
- 2.Bilukha O. O., Rosenstein N. (2005) MMWR Recomm. Rep. 54, 1–21 [PubMed] [Google Scholar]
- 3.Maiden M. C., Ibarz-Pavón A. B., Urwin R., Gray S. J., Andrews N. J., Clarke S. C., Walker A. M., Evans M. R., Kroll J. S., Neal K. R., Ala'aldeen D. A., Crook D. W., Cann K., Harrison S., Cunningham R., Baxter D., Kaczmarski E., Maclennan J., Cameron J. C., Stuart J. M. (2008) J. Infect. Dis. 197, 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens D. S. (2007) FEMS Microbiol. Rev. 31, 3–14 [DOI] [PubMed] [Google Scholar]
- 5.Virji M. (2009) Nat. Rev. Microbiol. 7, 274–286 [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee A. K., Jennings H. J., Kenny C. P., Martin A., Smith I. C. (1975) J. Biol. Chem. 250, 1926–1932 [PubMed] [Google Scholar]
- 7.Bhattacharjee A. K., Jennings H. J., Kenny C. P., Martin A., Smith I. C. (1976) Can. J. Biochem. 54, 1–8 [DOI] [PubMed] [Google Scholar]
- 8.Troy F. A., 2nd (1992) Glycobiology 2, 5–23 [DOI] [PubMed] [Google Scholar]
- 9.Kleene R., Schachner M. (2004) Nat. Rev. Neurosci. 5, 195–208 [DOI] [PubMed] [Google Scholar]
- 10.Moxon E. R., Kroll J. S. (1990) Curr. Top. Microbiol. Immunol. 150, 65–85 [DOI] [PubMed] [Google Scholar]
- 11.Lemercinier X., Jones C. (1996) Carbohydr. Res. 296, 83–96 [DOI] [PubMed] [Google Scholar]
- 12.Klein A., Roussel P. (1998) Biochimie 80, 49–57 [DOI] [PubMed] [Google Scholar]
- 13.Fusco P. C., Farley E. K., Huang C. H., Moore S., Michon F. (2007) Clin. Vaccine Immunol. 14, 577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M. L., Slauch J. M. (1999) FEMS Immunol. Med. Microbiol. 26, 83–92 [DOI] [PubMed] [Google Scholar]
- 15.Frasa H., Procee J., Torensma R., Verbruggen A., Algra A., Rozenberg-Arska M., Kraaijeveld K., Verhoef J. (1993) J. Clin. Microbiol. 31, 3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhasin N., Albus A., Michon F., Livolsi P. J., Park J. S., Lee J. C. (1998) Mol. Microbiol. 27, 9–21 [DOI] [PubMed] [Google Scholar]
- 17.Lewis A. L., Hensler M. E., Varki A., Nizet V. (2006) J. Biol. Chem. 281, 11186–11192 [DOI] [PubMed] [Google Scholar]
- 18.Lewis A. L., Nizet V., Varki A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11123–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houliston R. S., Endtz H. P., Yuki N., Li J., Jarrell H. C., Koga M., van Belkum A., Karwaski M. F., Wakarchuk W. W., Gilbert M. (2006) J. Biol. Chem. 281, 11480–11486 [DOI] [PubMed] [Google Scholar]
- 20.Claus H., Borrow R., Achtman M., Morelli G., Kantelberg C., Longworth E., Frosch M., Vogel U. (2004) Mol. Microbiol. 51, 227–239 [DOI] [PubMed] [Google Scholar]
- 21.Deszo E. L., Steenbergen S. M., Freedberg D. I., Vimr E. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5564–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergfeld A. K., Claus H., Vogel U., Mühlenhoff M. (2007) J. Biol. Chem. 282, 22217–22227 [DOI] [PubMed] [Google Scholar]
- 23.Bergfeld A. K., Claus H., Lorenzen N. K., Spielmann F., Vogel U., Mühlenhoff M. (2009) J. Biol. Chem. 284, 6–16 [DOI] [PubMed] [Google Scholar]
- 24.Alpers D. H., Appel S. H., Tomkins G. M. (1965) J. Biol. Chem. 240, 10–13 [PubMed] [Google Scholar]
- 25.Kim A. R., Rylett R. J., Shilton B. H. (2006) Biochemistry 45, 14621–14631 [DOI] [PubMed] [Google Scholar]
- 26.Øtwinowski Z., Minor W. (1997) Method. Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27.Leslie A. (1992) Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 28.North Atlantic Treaty Organization. Scientific Affairs Division (1998) Direct Methods for Solving Macromolecular Structures (Fortier S. ed) pp. 131–142, Kluwer Academic Publishers, Norwell, MA [Google Scholar]
- 29.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick G. M. (2002) Z. Kristallogr. 217, 644–650 [Google Scholar]
- 31.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 35.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 36.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott O., Olson A. J. (2009) J. Compt. Chem., in press [Google Scholar]
- 38.Sanner M. F. (1999) J. Mol. Graph. Model. 17, 57–61 [PubMed] [Google Scholar]
- 39.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 40.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beaman T. W., Sugantino M., Roderick S. L. (1998) Biochemistry 37, 6689–6696 [DOI] [PubMed] [Google Scholar]
- 42.Wang X. G., Olsen L. R., Roderick S. L. (2002) Structure 10, 581–588 [DOI] [PubMed] [Google Scholar]
- 43.Lo Leggio L., Dal Degan F., Poulsen P., Andersen S. M., Larsen S. (2003) Biochemistry 42, 5225–5235 [DOI] [PubMed] [Google Scholar]
- 44.Raetz C. R., Roderick S. L. (1995) Science 270, 997–1000 [DOI] [PubMed] [Google Scholar]
- 45.Beaman T. W., Binder D. A., Blanchard J. S., Roderick S. L. (1997) Biochemistry 36, 489–494 [DOI] [PubMed] [Google Scholar]
- 46.Kehoe L. E., Snidwongse J., Courvalin P., Rafferty J. B., Murray I. A. (2003) J. Biol. Chem. 278, 29963–29970 [DOI] [PubMed] [Google Scholar]
- 47.Vogel U., Hammerschmidt S., Frosch M. (1996) Med. Microbiol. Immunol. 185, 81–87 [DOI] [PubMed] [Google Scholar]
- 48.Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orskov F., Orskov I., Sutton A., Schneerson R., Lin W., Egan W., Hoff G. E., Robbins J. B. (1979) J. Exp. Med. 149, 669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fattom A. I., Sarwar J., Basham L., Ennifar S., Naso R. (1998) Infect. Immun. 66, 4588–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szu S. C., Li X. R., Stone A. L., Robbins J. B. (1991) Infect. Immun. 59, 4555–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iengar P., Joshi N. V., Balaram P. (2006) Structure 14, 529–542 [DOI] [PubMed] [Google Scholar]
- 53.Rangarajan E. S., Ruane K. M., Sulea T., Watson D. C., Proteau A., Leclerc S., Cygler M., Matte A., Young N. M. (2008) Biochemistry 47, 1827–1836 [DOI] [PubMed] [Google Scholar]
- 54.Williams A. H., Immormino R. M., Gewirth D. T., Raetz C. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10877–10882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.